Abstract
The adaptation of Saccharomyces cerevisiae to situations in which cell wall integrity is seriously compromised mainly involves the cell wall integrity (CWI) pathway. However, in a recent work (Bermejo, C., Rodriguez, E., García, R., Rodríguez-Peña, J. M., Rodríguez de la Concepción, M. L., Rivas, C., Arias, P., Nombela, C., Posas, F., and Arroyo, J. (2008) Mol. Biol. Cell 19,1113 -1124) we have demonstrated the co-participation of the high osmotic response (HOG) pathway to ensure yeast survival to cell wall stress mediated by zymolyase, which hydrolyzes the β-1,3 glucan network. Here we have characterized the role of both pathways in the regulation of the overall yeast transcriptional responses to zymolyase treatment using whole genome expression profiling. A main group of yeast genes is dependent on both MAPKs, Slt2 and Hog1, for their induction. The transcriptional activation of these genes depends on the MAPKKK Bck1, the transcription factor Rlm1, and elements of the sho1 branch of the HOG pathway, but not on the sensors of the CWI pathway. A second group of genes is dependent on Slt2 but not Hog1 or Pbs2. However, the induction of these genes is dependent on upstream elements of the HOG pathway such as Sho1, Ste50, and Ste11, in accordance with a sequential activation of the HOG and CWI pathways. Zymolyase also promotes an osmotic-like transcriptional response with the activation of a group of genes dependent on elements of the Sho1 branch of HOG pathway but not on Slt2, with the induction of many of them dependent on Msn2/4. Additionally, in the absence of Hog1, zymolyase induces an alternative response related to mating and filamentation as a consequence of the cross-talk between these pathways and the HOG pathway. Finally, in the absence of Slt2, zymolyase increases the induction of genes associated with osmotic adaptation with respect to the wild type, suggesting an inhibitory effect of the CWI pathway over the HOG pathway. These studies clearly reveal the complexity of the signal transduction machinery responsible for regulating yeast adaptation responses to cell wall stress.
Yeast cell integrity depends on an external envelope, the cell wall, whose mechanical strength allows cells to support turgor pressure and affords them protection against extreme environmental conditions. Because this structure is essential for survival, stress conditions that alter the cell wall lead to the activation of a cellular response that allows cells to adapt and survive (2, 3). This response is mainly characterized by activation of a transcriptional adaptation program that been extensively studied in the last few years by means of DNA microarray experiments. Transcriptional responses in mutants deleted in genes that are important for cell wall biogenesis as well as those activated in the presence of cell wall-perturbing agents have been characterized (4-7). Such responses include the transcriptional activation of specific genes for each particular stress condition but also the induction of a cluster of genes that are co-induced under all the conditions mentioned above. This common group of genes represents the transcriptional fingerprint of cell wall stress adaptation responses and includes mainly genes related to cell wall remodeling, metabolism, and signaling (5, 8). The final consequences of these responses are an increase in the amount of several cell wall proteins (CWPs) synthesized by the cell, an increase in β-glucan and chitin contents, changes in the association among cell wall polymers, and the relocalization of important proteins from the cell wall construction machinery to the lateral cell wall, all of them required for proper cell wall remodeling under cell wall stressing circumstances.
The regulation of these adaptive responses to cell wall stress is mainly mediated by the cell wall integrity pathway (see Ref. 2 for a recent review). A pair of membrane proteins, Mid2 and Wsc1, act as the main sensors of this pathway (9, 10). Under conditions of activation, these sensors interact with the guanine nucleotide exchange factor Rom2, activating the small GTPase Rho1, which then interacts and activates Pkc1 (2). The main role of activated Pkc1 is to trigger a MAPK3 module. Phosphorylation of the MAPKKK Bck1 activates a pair of redundant MAPKKs (Mkk1 and Mkk2), which finally phosphorylate the MAPK Slt2. The phosphorylated form of this protein acts mainly on two transcription factors: the MCM1-Agamous-Deficiens-serum response factor box transcription factor Rlm1 (11) and SCB binding factor (12). The transcriptional activation of most genes induced in response to Congo Red and heat shock, both of them stresses activating this pathway, mainly depends on Rlm1 (5, 13).
Different cell wall-interfering compounds such Calcofluor White or Congo Red, both of which bind to cell wall polysaccharides, caspofungin, which inhibits β-1,3 glucan synthase activity, or zymolyase, which degrades the β-1,3 glucan network, lead to the activation of the MAPK Slt2 (1, 4, 14-16). This activation is responsible for the transcriptional program described above that finally allows cells to become adapted. However, differences in the regulation of cell wall stress responses have been found, depending on the actual type of cell wall stress. We have recently shown that in contrast to the yeast transcriptional response to Congo Red, which depends almost completely on the MAPK Slt2 and the transcription factor Rlm1 (5), adaptation to cell wall stress caused by zymolyase requires the participation of both the cell wall integrity (CWI) and the high osmotic response (HOG) pathways (1). Cellular responses to this stress require essential components of the CWI pathway, namely, the redundant MAPKKs Mkk1/Mkk2, the MAPKKK Bck1, and Pkc1, but they do not require upstream elements, including the sensors Mid2 and Wsc1 and the guanine nucleotide exchange factors of this pathway (1). Moreover, activation of the MAPK of the CWI pathway (Slt2) depends on the elements of the Sho1 branch of the HOG pathway (i.e. Sho1, Ste20, Ste50, Ste11, and Pbs2), and it is blocked in a double mutant strain in which genes encoding the mucin-like proteins Msb2 and Hkr1, recently described as potential osmosensors of the Sho1 branch of the HOG pathway (17), are deleted. Therefore, zymolyase-mediated cell wall damage should be sensed through the Sho1 branch of the HOG pathway, leading to the sequential activation of the MAPK Slt2 of the CWI pathway, which in turns activates the transcription factor Rlm1 (1). Transcriptional activation of CRH1, one of the targets of the cell wall stress response that encodes a transglycosidase activity responsible for Chitin-β-1,6 glucan cross-linking (18-20), by zymolyase requires this connection between the HOG and CWI pathways (1).
Here, to globally characterize how yeast regulates transcriptional responses to damage to the β-1,3 glucan network caused by zymolyase and to investigate the role of the CWI and HOG pathways in this regulation in more detail, genome-wide transcriptional responses to zymolyase were characterized in Saccharomyces cerevisiae wild type and mutant strains deleted in different elements of both pathways by using DNA microarrays. Data analysis revealed a complex level of regulation of these responses and a completely collaborative participation of both pathways to assess cell integrity under these cell wall stress circumstances.
EXPERIMENTAL PROCEDURES
Yeast Strains—All of the experiments were performed with the S. cerevisiae BY4741 strain (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and mutant derivatives provided by Euroscarf (Frankfurt, Germany). Single mutant strains present the corresponding gene completely deleted and replaced by the geneticin resistance-codifying KanMX4 module. The yeast strains used in this work were: sho1Δ, ste50Δ, ste11Δ, ssk1Δ, pbs2Δ, hog1Δ, wsc1Δ, mid2Δ, bck1Δ, slt2Δ, rlm1Δ, fus3Δ, and kss1Δ. The double mutants hog1Δslt2Δ (strain CR001) and msn2Δmsn4Δ (RG001) have been described previously (1).
Culture Conditions—Depending on the experimental approaches used, yeast cells were grown on YPD (2% glucose, 2% peptone, 1% yeast extract) or SD medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose) supplemented with the required amino acids. For routine cultures, yeast cells were grown overnight at 24 °C to an optical density of 0.8-1 (A600). The culture was refreshed to optical density of 0.2 at 600 nm and grown at 24 °C for 3 h. Next, the culture was divided into two parts. One part was allowed to continue growing under the same conditions (the non-treated culture) while the other one was supplemented with zymolyase from Arthrobacter luteus (MP Biomedicals, Inc.) to a final concentration of 0.8 unit/ml. The cells were collected at the indicated times and processed, depending on the experimental approach, as described below.
Plasmids—To obtain the collection of plasmids bearing transcriptional fusions to the LacZ gene used in this work, we replaced the 1.1-kb fragment (EcoRI/BamHI) containing the promoter of CRH1 from the pCRH1-LACZ plasmid (1) by the promoter region of selected genes obtained from genomic DNA by PCR amplification (the oligonucleotide sequences are available upon request). Thus, the following plasmids were generated: pAFR1-LacZ (-842/-7), pSED1-LacZ (-999/-1), pCWP1-LacZ (-978/-5), pMLP1-LacZ (-1186/-1), pHOR2-LacZ (-954/-19), and pHSP12-LacZ (-589/+9). The numbers in parentheses indicate the upstream fragment with respect to the open reading frame start codon used for each gene.
β-Galactosidase Assays—Yeast cell extracts were prepared by harvesting cells by centrifugation from 5 ml of an exponentially growing culture. Then the cells were resuspended in 250 μl of breaking buffer (100 mm Tris-HCl, pH.8, 1 mm dithiothreitol, 20% glycerol), and glass beads (Glasperlen ca. 1 mm, Sartorius AG, Germany) were added to break cells in a Fast-Prep system (FP120/BIO101 ThermoSavant). Finally, extracts were clarified by centrifugation, and protein concentrations were measured using the Bradford method. β-Galactosidase assays were performed using the crude extracts obtained as described previously (21), scaling the protocol to a 96-well microtiter plate format. 10 μl of cell extract was mixed with 90 μl of Z buffer plus β-mercaptoethanol (0.03%) and 20 μl of o-nitrophenyl-β-d-galactopyranoside (4 mg/ml in Z buffer). The absorbance of the enzymatic reaction was measured at 415 nm on a microplate reader (model 680; Bio-Rad) after at least 10 min of incubation at 30 °C and the addition of 50 μl of 1 m Na2CO3 to stop the reaction. β-Galactosidase activity was expressed as nmoles of o-nitrophenyl-β-d-galactopyranoside converted/min/mg of protein. The experiments were performed at least in triplicate from independent yeast transformants.
RNA Isolation, cDNA Synthesis, and Chip Hybridization—Total RNA was isolated from exponentially growing cells (5 × 108) with the “mechanical disruption protocol” using the RNeasy MIDI kit (Qiagen), following the instructions of the manufacturer. RNA concentrations were determined by measuring absorbance at 260 nm. RNA purity and integrity were assessed using RNA Nano Labchips in an Agilent 2100B Bioanalyzer (Agilent Technologies, Palo Alto, CA) following the manufacturer's instructions. Next, cDNA was synthesized from 25-30 μg of total RNA by reverse transcription using the CyScribe™ post-labeling kit (Amersham Biosciences), incorporating Cy3-dUTP or Cy5-dUTP into the cDNA corresponding to each sample to be compared. Both labeled cDNA populations were combined, dried in a vacuum trap, and used as a hybridization probe after resuspension in 15 μl of hybridization solution (50% formamide, 6× SSC, 0.5% SDS, 5× Denhardt's solution, 20 μg of poly(A) (P-9403; Sigma), and 100 μg/ml salmon sperm (Invitrogen). The printed microarrays, including the complete set of 6306 open reading frames coded by the S. cerevisiae genome, used in this study were provided by the Microarray Centre of the University Health Network, Toronto, Canada. The slides were prehybridized in prehybridization solution (6× SSC, 0.5% SDS, 1% bovine serum albumin (A-7906; Sigma)) for 1 h and were then hybridized overnight with labeled probe at 42 °C in a Lucidea SlidePro Automated Hybridization Station (Amersham Biosciences). Before scanning, the chips were washed and dried in the Hybridization Station. For each condition tested, comparison of treated and untreated samples, the total RNA from two different cultures was analyzed, and in addition, for each sample two different hybridizations were performed, including fluorochrome swapping to minimize transcriptional changes because of technical variability (7). This therefore corresponded to four DNA microarrays analyzed for each condition.
Microarray Image Analysis, Data Processing, and Statistical Methods—All of these processes were basically carried out following the protocols described previously by García et al. (5). The genes were considered to be up- or down-regulated when their expression ratio under the conditions tested was >2 or <0.5, respectively. To determine whether the gene induction observed in the wild type strain after treatment with zymolyase was blocked, in the mutant strains used along this work we used the relationship between the responses of each mutant versus those of the wild type strain. Thus, a value of mutant ratio/wild type ratio of 0.65 was considered the threshold for defining a significant reduction in gene induction. In any case, genes whose ratio (zymolyase ±) was <1.6 in any of the mutant backgrounds tested were deemed as not being up-regulated. Clustering analysis was performed using MEV (Multiexperiment viewer) version 4.2 software from TIGR (22).
The microarray data described here follow the minimum information about a microarray experiment recommendations and have been deposited at the NCBI gene expression and hybridization array data repository with accession numbers GSM248993, GSM248994, GSM248995, GSM249020, GSM249086, GSM249088, GSM249091, GSM249098, GSM249099, GSM249101, GSM249103, GSM249104, GSM249105, GSM249106, GSM249125, GSM249126, GSM249127, GSM249128, GSE9930, GSE9931, GSE9932, GSE9933, GSE9934, and GSE12684.
Promoter Analysis—The YEASTRACT program was used to find associations between transcription factors and genes induced under the conditions studied (23). Documented associations between a transcription factor and target gene are supported by published data concerning: (i) a change in the expression of the target gene caused by a deletion (or mutation) in the gene encoding transcription factor and (ii) binding of the transcription factor to the promoter region of the target gene, as supported by band shift, footprinting, or chromatin immunoprecipitation assays. Potential associations between a transcription factor and a target gene are based on the occurrence of the transcription factor-binding site in the promoter region of the target gene.
Quantitative RT-PCR Assays—Total RNA was isolated from cells (5 × 107) with the mechanical disruption protocol using the RNeasy MINI kit (Qiagen), following the manufacturer's instructions. RNA concentration, purity, and integrity were determined as described above. Real time RT-qPCR assays were performed as previously described (5), using an ABI 7700 instrument (Applied Biosystems). For quantification, the abundance of each gene was determined using the amount of the standard transcript of ACT1 for input cDNA normalization, and the final data on relative gene expression between the two conditions tested were calculated following the 2-ΔΔCt method, as described by Livak and Schmittgen (24). The primer sequences are available upon request.
RESULTS
Transcriptional Profiles of WT, slt2Δ, and hog1Δ Cells in Response to Zymolyase Treatment—The finding that the transcriptional activation of CRH1 by zymolyase was dependent on both pathways, the CWI and HOG pathways (1), prompted us to study whether this dual regulation was unique to CRH1 or, as expected, a general mechanism for regulating gene expression responses to this specific cell wall stress. To accomplish this, using DNA microarrays we characterized the global pattern of yeast gene expression after zymolyase treatment in WT cells as well as in strains deleted in SLT2, encoding the MAPK of the CWI pathway and HOG1, which encodes the MAPK of the HOG pathway.
In a previous work (5) we described the transcriptome of wild type yeasts in the presence of several cell wall stresses, including 5 units/ml of zymolyase. However, the mutant strains slt2Δ and hog1Δ proved to be hypersensitive to this concentration of zymolyase, so we repeated these experiments using a concentration of zymolyase compatible with the growth of these mutants (0.8 unit/ml). First, the global pattern of gene expression of a wild type strain BY4741 growing in the absence or presence of 0.8 unit/ml for 3 h was determined. At this concentration, 77 genes were induced, and 10 were repressed by zymolyase treatment. As shown in Fig. 1, the main functional groups of genes include cell wall biogenesis, metabolism, signal transduction, stress, transport, and morphogenesis. Moreover, the response at 0.8 unit/ml was very similar to that observed at 5 units/ml (5) (see supplemental Table S1 for details). We then addressed the involvement of the HOG and CWI pathways in the regulation of this response. Strains slt2Δ and hog1Δ were grown in the presence of 0.8 unit/ml of zymolyase for 3 h, and their transcriptional profiles were analyzed using DNA microarrays. The ratio of expression for each gene in the wild type and mutant strains was calculated, and levels of Hog1 and Slt2 dependence were established as described under “Experimental Procedures.”
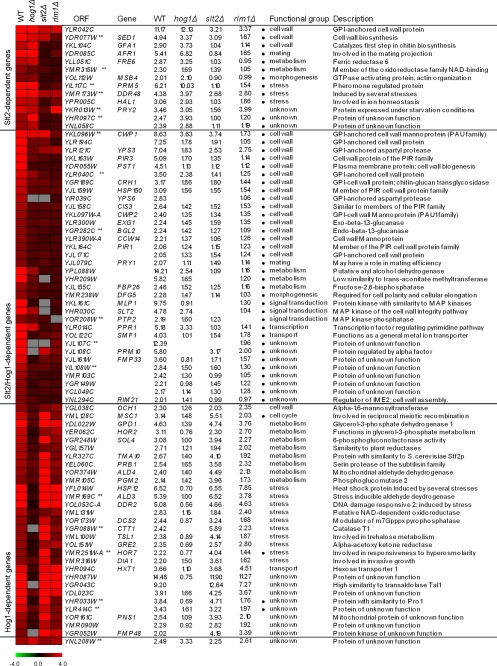
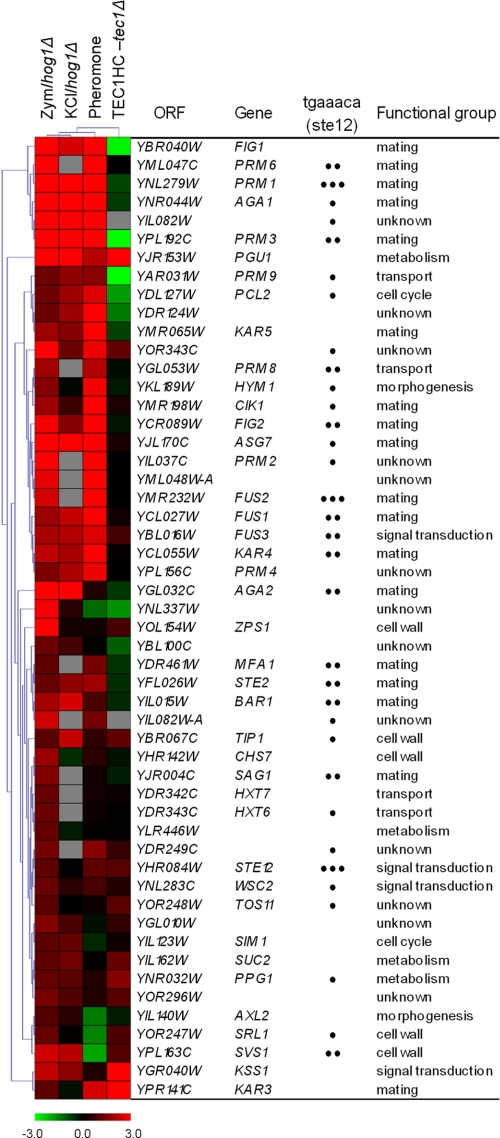
FIGURE 1.
Classification of S. cerevisiae genes induced by zymolyase treatment depending on their regulation by Slt2 and Hog1 MAPKs. Open reading frames whose transcripts were induced at least 2-fold in the WT BY4741 strain after zymolyase treatment (0.8 unit/ml for 3 h) and their corresponding ratios in the hog1Δ, slt2Δ, and rlm1Δ under the same conditions are shown. Microarray data corresponding to average values of induction after the significance test was performed (see “Experimental Procedures”) are shown for all genes, except for PRY2, YHR097C, BGL2, and ALD3, where RT-qPCR data are indicated. Genes whose expression was validated by RT-qPCR are labeled with **. Functional categories and description were assigned based on the information provided by the YPD Proteome Bioknowledge® Library. Genes were grouped together on the basis of their dependence for activation by zymolyase on the Slt2 and Hog1 MAPKs, considering a mutant/wild type ratio of <0.65 as a threshold. Those genes dependent on Rlm1 for activation by zymolyase are labeled with a black dot. Clustering was obtained using MEV (Multiexperiment viewer) version 4.2 software from TIGR (22). The degree of color saturation represents the expression log2 ratio value, as indicated by the scale bar. Gray denotes missing values.
On the basis of the dependence on these MAPKs for their induction, the genes induced by zymolyase in a WT strain were classified in three different groups (Fig. 1). A main group, including 34 genes, was found to be dependent on both the Slt2 and Hog1 MAPKs. A second group of 13 genes were dependent on Slt2 but not on Hog1. A third group of 29 genes proved to be dependent on the MAPK Hog1 but not on Slt2. Finally, an additional gene (YNL208W) was neither dependent on Hog1 nor Slt2.
The gene expression data of 18 of the 77 genes, belonging to the different groups, were checked by RT-qPCR to validate the microarray results. A very good correlation was found between both data sets (supplemental Table S2), except for slight variations in the case of BGL2, PRY2, YHR097C, and ALD3. The details of the gene expression data, based on microarrays and RT-qPCR experiments, together with a clustering analysis to visualize the different groups based on the dependence on the Slt2 and Hog1 MAPKs, are shown in Fig. 1.
To predict potential transcription factors involved in the regulation of the response, genes included in the different groups were analyzed for documented regulatory associations using YEASTRACT (see “Experimental Procedures”). The main regulatory associations are shown in Table 1.
TABLE 1.
Regulatory associations for genes included in the genome-wide transcriptional response to zymolyase
DRA, percentage of genes showing documented regulatory associations with transcription factors as deduced from analysis with YEASTRACT (see “Experimental Procedures” for details). TF-binding sites, percentage of genes in each group showing in their promoters at least one DNA-binding domain for the indicated transcription factor. Those transcription factors with percentages higher than 50% in either documented regulatory associations or transcription factor-binding site analysis are shown.
| Transcription factor | DRA | TF-binding sites |
|---|---|---|
| % | % | |
| Slt2-dependent genes | ||
| Ste12 | 69.2 | 30.8 |
| Tec1 | 53.8 | 38.5 |
| Swi4 | 53.8 | 23.1 |
|
Rlm1 |
46.2 |
69.2 |
| Slt2/Hog1-dependent genes | ||
| Rlm1 | 67.6 | 67.6 |
|
Sok2 |
50.0 |
50.0 |
| Hog1-dependent genes | ||
| Sok2 | 82.8 | 34.5 |
| Met4 | 75.9 | 13.8 |
| Msn4 | 69.0 | 86.2 |
| Msn2 | 65.5 | 86.2 |
| Yap1 | 65.5 | 31.0 |
| Aft1 | 62.1 | 10.3 |
| Cst6 | 58.6 | 24.1 |
|
Hsf1 |
51.7 |
27.6 |
| Alternative genes induced in hog1Δ | ||
| Ste12 | 78.4 | 63.5 |
| Tec1 | 52.9 | 34.6 |
|
Sok2 |
52.9 |
46.2 |
| Alternative genes induced in slt2Δ | ||
| Sok2 | 73.3 | 43.3 |
| Yap1 | 63.3 | 33.3 |
| Msn4p | 56.7 | 86.7 |
| Msn2p | 56.7 | 86.7 |
| Rpn4 | 56.7 | 0.0 |
| Aft1 | 53.3 | 1.0 |
| Met4 | 50.0 | 3.3 |
Genes Regulated by Slt2 and Hog1—The genomic approach described above allowed us to conclude that dual regulation by the Slt2 and Hog1 MAPKs in response to zymolyase was not exclusive to CRH1 but that it functioned for many other yeast genes (see Fig. 1 for details). As expected, many of the genes within this group (67.6%) were potentially regulated by Rlm1 (Table 1). Using DNA microarrays, the transcriptional profile of an rlm1Δ strain growing in the presence of 0.8 unit/ml zymolyase was determined and compared with the profile of the WT growing under the same conditions. The results from this analysis are shown in Fig. 1. Considering the same threshold as that used in the analysis of hog1Δ and slt2Δ strains to define the dependence of the induction of each gene on these MAPKs, the whole set of genes included in the groups of Slt2/Hog1- and Slt2-dependent genes was also dependent on Rlm1 for induction. In contrast, as expected only the induction of four genes within the group of Hog1-dependent genes but not Slt2-dependent genes, MSC1, YLR414C, HOR7, and YHR033W, was dependent on Rlm1 (Fig. 1).
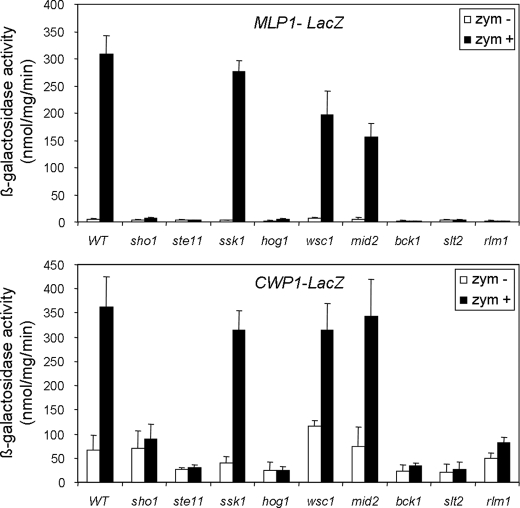
We have previously shown that the induction of CRH1, one of the genes included in the group of Slt2/Hog1-dependent genes, by zymolyase requires essential elements of the CWI pathway, including Rho1, Pkc1, Bck1, Mkk1/Mkk2, Slt2, and the transcription factor Rlm1, as well as elements of the Sho1 branch of the HOG pathway such as Hog1, Pbs2, Ste11, Ste20, Ste50, and Sho1, but not the transcription factors regulated by Hog1 such as Hot1, Msn2/4, Sko1, or Smp1 (1). Because Rlm1 was required for transcriptional activation of the whole set of these genes within this group, the most likely hypothesis was that all these genes were being regulated by the same mechanism as the one described for CRH1. To test this, reporter constructions including the promoter of the MLP1 and CWP1 genes fused to lacZ were obtained, and transcriptional activation after zymolyase treatment was studied in strains deleted in different elements of both the HOG and CWI pathways. As shown in Fig. 2, the induction of CWP1 and MLP1 was dependent on different elements of the Sho1 branch of the HOG pathway, such Ste11 and Sho1, but not on the Sln1 branch (Ssk1). Moreover, the induction of these genes also depended on the MAPKKK Bck1 but not on the sensors Wsc1 and Mid2 of the CWI pathway (Fig. 2). All of these data are in agreement with a model of sequential activation of the HOG and CWI pathways necessary for adaptation to zymolyase-mediated cell wall damage, in which the sensing of damage should be exerted through the Sho1 branch of the HOG pathway (see “Discussion” for further details).
FIGURE 2.
MLP1 and CWP1 induction by zymolyase in mutants of the CWI and HOG pathways. Expression of MLP1-LacZ and CWP1-LacZ was measured in wild type (BY4741) and sho1Δ, ste11Δ, ssk1Δ, hog1Δ, wsc1Δ, mid2Δ, bck1Δ, slt2Δ, and rlm1Δ mutants growing in the absence (white bars) or presence of 0.8 unit/ml zymolyase for 3 h (black bars). Three independent experiments were carried out to calculate the means and standard deviations.
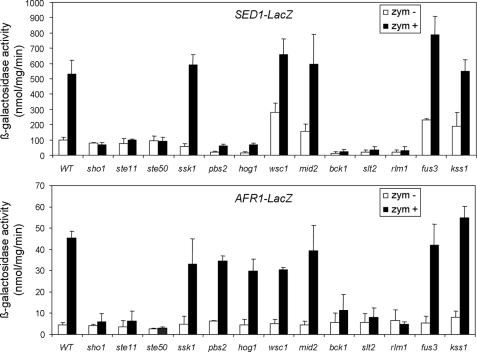
Genes Regulated by Slt2 but Not Hog1—The induction by zymolyase of a group of 13 genes proved to be dependent on Slt2 but not on Hog1. As expected, all of these genes except PRY2 were also dependent on Rlm1 (Fig. 1). By using fusions of the promoters of SED1 and AFR1 to lacZ and mutant strains deleted in different elements of the CWI pathway, we observed that the induction of both genes by zymolyase was dependent not only on Slt2 and Rlm1 but also on the MAPKKK Bck1 of the CWI pathway. However, none of these genes was dependent on the two main sensors of this pathway, Mid2 and Wsc1, for their induction by zymolyase treatment (Fig. 3).
FIGURE 3.
SED1 and AFR1 induction by zymolyase in mutants of the CWI, HOG, mating, and filamentation pathways. Expression of SED1-LacZ and AFR1-LacZ was determined in wild type BY4741 and sho1Δ, ste11Δ, ste50Δ, ssk1Δ, pbs2Δ, hog1Δ, wsc1Δ, mid2Δ, bck1Δ, slt2Δ, rlm1Δ, fus3Δ, and kss1Δ mutant cells growing in the absence (white bars) or presence of 0.8 unit/ml zymolyase (black bars). The results are represented as the means with SDs derived from three independent experiments.
A high percentage of genes within this group contained documented regulatory associations with transcription factors of the mating and filamentation pathways: Ste12 and Tec1, respectively (Table 1). However, as shown in Fig. 3 the induction of SED1 and AFR1 by zymolyase was not dependent on the MAPKs Fus3 or Kss1, suggesting that these MAPK pathways are not involved in their regulation.
Our microarray data revealed that the genes within this group were not dependent on the MAPK Hog1. However, because the transcriptional activation of SED1 and AFR1 was not dependent on the sensors Wsc1 and Mid2, we decided to check the possible involvement of upstream elements of the HOG pathway. As expected, deletion of HOG1 had no effect on the induction of SED1 and AFR1 by zymolyase (Fig. 3). Additionally, the deletion of PBS2 or elements of the Sln1 branch of the HOG pathway, such as SSK1, was dispensable for the transcriptional activation of these reporters. However, the deletion of upstream elements of the Sho1 branch such as SHO1, STE11, or STE50 completely abrogated the induction of these genes by zymolyase (Fig. 3). It is worth noting that in the case of SED1, basal expression levels were very reduced in hog1Δ and pbs2Δ cells, but activation was still observed in these mutants in the presence of zymolyase. To check the behavior of additional genes within this group, an analysis of PRM5 and DDR48 (both dependent on Slt2 but not on Hog1) gene expression was carried out by RT-qPCR in WT, hog1Δ, sho1Δ, pbs2Δ, and ste11Δ mutants treated or not with zymolyase. HOG1 and PBS2 but not STE11 and SHO1 were dispensable for their induction by zymolyase (data not shown), confirming that in addition to elements of the CWI pathway, the activation of genes within this group requires upstream elements of the HOG pathway (Sho1 and Ste11) but not the MAPKK Pbs2 or the MAPK Hog1.
Genes Regulated by Hog1 but Not Slt2—As stated above, 29 genes were dependent on the Hog1 MAPK but not on Slt2 for their induction. To investigate whether these genes overlapped with those induced under hyperosmotic conditions, we compared our response to the already characterized genome-wide transcriptional responses to osmotic stress, i.e. 0.5 m KCl (25) and 0.7 m NaCl (26). As shown in Table 2, of the 29 genes 27 were also transcriptionally activated by either KCl and/or NaCl, respectively. Only two genes, OCH1 and YHR033W, were induced by zymolyase but not induced under hyperosmotic stress. Additionally, of the 27 overlapping genes, 25 had been characterized as Hog1-dependent for induction by either NaCl or KCl (Table 2).
TABLE 2.
Comparison of Hog1 and Msn2/4-dependent zymolyase reponses to hyperosmotic stress response
Those genes whose induction by zymolyase is dependent on Hog1 but not Slt 2 are included in the table, and their ratios of induction are compared with the ones found in a msn2/4Δ mutant under the same conditions (this work) or with previously characterized osmotic stress responses (0.5 m KCl (25) and 0.7 m NaCl (26)). The far-right column shows the number of STRE-binding sites present in the promoter of the corresponding genes.
|
Open reading frame |
Gene |
Zym |
0.5 m KCl |
0.7 m NaCl |
msn2/4Δ |
STRE sites |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | hog1Δ | WT | hog1Δ | WT | hog1Δ | Zym | 0.7 m NaCl | |||
| YGL038C | OCH1 | 2.30 | 1.26 | 3.60 | ||||||
| YML128C | MSC1 | 3.14 | 1.48 | a | b | a | b | b | b | 2 |
| YDL022W | GPD1 | 4.63 | 1.39 | a | b | a | b | 8.35 | 4 | |
| YER062C | HOR2 | 3.11 | 0.76 | a | b | a | b | b | 1 | |
| YGR248W | SOL4 | 3.08 | 1.00 | a | b | a | b | b | b | 1 |
| YGL157W | 2.71 | 1.21 | a | b | a | b | 3.48 | 2 | ||
| YLR327C | TMA10 | 2.67 | 1.40 | a | b | a | b | b | ||
| YEL060C | PRB1 | 2.54 | 1.65 | a | 3.55 | 1 | ||||
| YOR374W | ALD4 | 2.40 | 1.40 | a | b | a | b | 3.97 | 2 | |
| YMR105C | PGM2 | 2.14 | 1.42 | a | b | a | b | b | 5 | |
| YFL014W | HSP12 | 6.52 | 0.70 | a | b | a | b | b | 7 | |
| YMR169C | ALD3 | 5.39 | 1.00 | a | b | a | b | b | b | |
| YOL053C-A | DDR2 | 5.08 | 0.56 | a | b | a | b | b | b | 3 |
| YML131W | 2.83 | 1.15 | a | b | a | b | b | 1 | ||
| YOR173W | DCS2 | 2.44 | 0.87 | a | b | a | b | 2 | ||
| YGR088W | CTT1 | 2.42 | a | b | a | b | b | b | 1 | |
| YML100W | TSL1 | 2.38 | 0.89 | a | b | a | b | b | b | |
| YOL151W | GRE2 | 2.35 | 0.69 | a | b | a | b | b | ||
| YMR251W-A | HOR7 | 2.22 | 0.77 | a | b | a | 3.36 | 4 | ||
| YMR316W | DIA1 | 2.20 | 1.50 | a | b | 1.92 | ||||
| YHR094C | HXT1 | 3.66 | 1.10 | a | 5.90 | 1 | ||||
| YHR087W | 14.48 | 0.75 | a | b | b | 2 | ||||
| YGR043C | 9.20 | a | b | a | b | b | ||||
| YDL023C | 3.91 | 1.66 | a | b | 9.85 | |||||
| YHR033W | 3.84 | 0.69 | 8.39 | 1 | ||||||
| YLR414C | 3.43 | 1.61 | a | b | a | 8.81 | ||||
| YOR161C | 2.54 | 1.09 | a | b | a | 3.98 | ||||
| YMR090W | 2.29 | 0.92 | a | b | a | b | b | b | 1 | |
| YGR052W | 2.02 | a | b | a | b | b | 3 | |||
Genes induced at least 2-fold in response 0.5 m KCl and 0.7 m NaCl, respectively.
Genes whose induction is abrogated in the corresponding mutant (hog1Δ or msn2/4Δ) under the indicated conditions.
Prediction of the transcription factors involved in the regulation of genes within this group with YEASTRACT (Table 1) had revealed that most of these genes, 69 and 65.5%, respectively, showed a documented regulatory association with Msn4 and Msn2. Moreover, other statistically significant regulatory associations included transcription factors related to stress responses, such as Yap1 (27), Hsf1 (28) and Sok2 (29), all of them being functionally associated with Msn2/4 in the transcriptional regulation of stress adaptation responses. To characterize the role of Msn2/4 in the regulation of the transcriptional response to zymolyase functionally, we compared the global transcriptional profile of a double mutant, msn2Δ msn4Δ, growing in the presence of 0.8 unit/ml of zymolyase to that of a WT strain under the same conditions. We found that, within the groups including Slt2- and Slt2/Hog1-dependent genes, only three genes (FRE6, PPR1, and YMR103C) required Msn2/4 for induction. However, of the 29 genes specifically dependent on Hog1, the transcriptional activation of 17 of them was dependent on Msn2/4 (see Table 2 for details), highlighting the importance of Msn2/4 in the transcriptional regulation of this set of genes in response to zymolyase. Interestingly, seven of these genes had previously been described as being dependent on Msn2/4 for induction by hyperosmotic stress. Most of the genes induced by zymolyase in a Msn2/4-dependent manner (69%) had one or more Msn2/4 putative DNA-binding sites (STRE boxes) in their promoters (see Table 2 for details). It is worth mentioning the presence of a group of genes that were found to be induced by zymolyase in a msn2Δ msn4Δ mutant but not in the WT strain (supplemental Table S3).
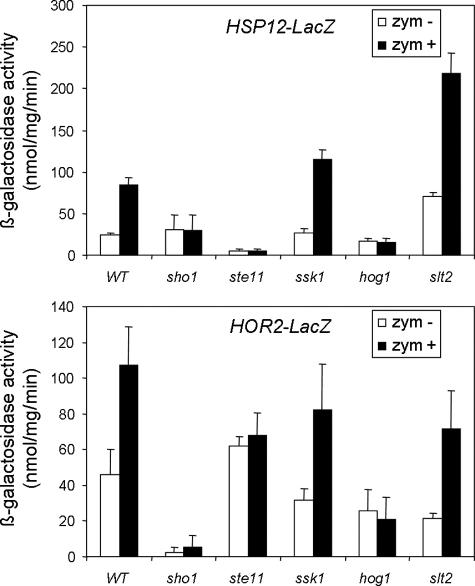
Experiments with lacZ reporters of HOR2 and HSP12, both of them dependent on Hog1, but not on Slt2, for induction by zymolyase, revealed that this induction was not dependent on the CWI pathway or on the SLN1 branch of the HOG pathway but was completely dependent on the Sho1 branch of the HOG pathway, because the induction of these genes was lost in hog1Δ, ste11Δ, and sho1Δ mutant strains (Fig. 4).
FIGURE 4.
Induction of HSP12 and HOR2 depends on the Sho1 branch of the HOG pathway but not on the CWI pathway. The expression of HSP12-LacZ and HOR2-LacZ was studied in wild type BY4741, and sho1Δ, ste11Δ, ssk1Δ, hog1Δ, and slt2Δ cells growing in the absence (white bars) or presence of 0.8 unit/ml zymolyase for 3 h (black bars). The means and S.D. values are derived from three independent experiments.
In the Absence of HOG1, Zymolyase Activates the Mating and Filamentation Pathways—In the absence of HOG1, zymolyase treatment led to the induction of 52 genes and the repression of six genes, respectively, whose expression did not change in the wild type strain (supplemental Table S4). A good part of the genes induced alternatively in a hog1Δ mutant by zymolyase were functionally related to mating. Moreover, most of these genes possess pheromone response elements (tgaaaca) (30) in their promoters (Fig. 5). The transcription factor Ste12 binds to the pheromone response elements to regulate genes required for mating and, together with Tec1, to regulate the genes required for invasive and pseudohyphal growth (31, 32). It has previously been shown that in the absence of Hog1 or Pbs2, hyperosmotic stress leads to inappropriate activation of both the pheromone response pathway (25, 33, 34) and the filamentation/invasion pathway (35). We therefore compared the alternative response to zymolyase in a hog1Δ strain to the transcriptional profiles of hog1Δ cells treated with 0.5 m KCl (25), wild type cells growing in the presence of α-factor (32), or cells overexpressing Tec1 (31) (Fig. 5). Hierarchical clustering of these profiles revealed the existence of a big cluster of genes, including those with the higher expression values, shared not only by the zymolyase and osmotic stress response in a hog1Δ strain but also by the response to α-factor in a wild type strain (Fig. 5). The majority of the genes within this cluster are not induced or even repressed in cells overexpressing Tec1. However, there is also a cluster of co-regulated genes that includes genes with lower induction ratios (see the lower part of the clustering in Fig. 5) between the zymolyase-induced response in a hog1Δ strain and the transcriptional profile of cells overexpressing Tec1 (Fig. 5). Consistent with this co-regulation, Kss1, the MAPK of the filamentation/invasion response pathways was clearly hyperphosphorylated by zymolyase in hog1Δ and pbs2Δ mutants (data not shown). All of these data suggest that in the absence of Hog1, zymolyase-mediated cell wall stress, like osmotic stress, activates both the mating and filamentation pathways.
FIGURE 5.
Hierarchical clustering of the genes alternatively induced by zymolyase in a hog1Δ strain and other related conditions. Each column represents a different condition. Each row represents the ratio of expression for each gene, as indicated in the color scale. The induced transcriptional response to zymolyase in a hog1Δ strain but not present in a wild type strain (Zym/hog1Δ) was compared with that documented in: (a) hog1Δ cells treated for 180 min with 0.5 m KCl (KCl/hog1Δ) (25); (b) wild type cells treated with 50 nm α-factor for 30 min (Pheromone) (32); and (c) cells overexpressing Tec1 versus a tec1Δ mutant (TEC1HC-tec1Δ) (31). The number of pheromone response elements (tgaaaca), in the promoters of the genes is indicated by black dots.
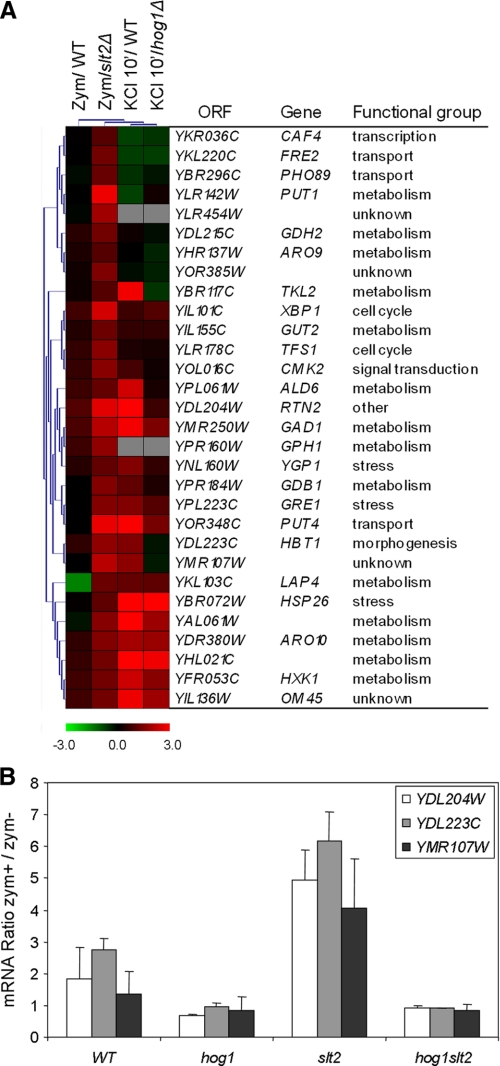
Alternative Response to Zymolyase in a slt2Δ Strain—Interestingly, in the absence of SLT2 zymolyase also led to an alternative response not elicited by a wild type strain (supplemental Table S5). Analysis of the regulatory associations of the 30 genes included in this response revealed an enrichment in genes putatively regulated by transcription factors related to stress such as Sok2, Yap1, Msn2, and Msn4 (Table 1). Hierarchical analysis of these genes together with the genome-wide transcriptional profile of yeast WT cells in response to hyperosmotic stress conditions (25) revealed that the majority of them were also induced by 0.5 m KCl in a WT strain (Fig. 6A). Moreover, many of them had been characterized as Hog1-dependent under hyperosmotic stress (Fig. 6A). These data clearly suggest an inhibitory effect of the CWI pathway on the HOG pathway. Thus, in the absence of Slt2, the HOG pathway is more activated by zymolyase. To check whether Hog1 was responsible for this transcriptional activation, the levels of expression of some of these genes were quantified by RT-qPCR in WT, hog1Δ, slt2Δ, and hog1Δslt2Δ strains. As shown in Fig. 6B, the genes YDL204W, YDL223C, and YMR107W were clearly induced by zymolyase in a slt2Δ strain with respect to the WT, and this transcriptional activation was completely abrogated in a hog1Δslt2 strain, indicating that the activation of these genes in the slt2Δ strain is dependent on Hog1.
FIGURE 6.
Zymolyase induces a higher osmotic-like response in the absence of SLT2. A, hierarchical clustering of the genes induced by zymolyase in a slt2Δ but not in a wild type strain (Zym/slt2Δ) with other transcriptional profiles: (a) wild type cells treated with 0.8 unit/ml zymolyase for 3 h (Zym/WT) (this work); (b) wild type cells treated with 0.5 m KCl for 10 min. (KCl 10′/WT); and (c) hog1Δ cells treated for 10 min with 0.5 m KCl (KCl 10′/hog1Δ) (25). Each column represents a different condition, and each row represents the ratio of expression for each gene, as indicated in the color scale. B, analysis by quantitative RT-qPCR of the level expression of selected genes alternatively induced by zymolyase in a slt2Δ mutant. The expression of YDL204W, YDL223C, and YMR107W was determined in wild type and hog1Δ, slt2Δ, and hog1Δslt2Δ mutant strains, respectively. The results are expressed as the ratios of zymolyase-treated versus untreated cells.
DISCUSSION
The adaptation of yeast to cell wall stress is mainly regulated through the CWI pathway. Activation of this pathway leads to the hyperphosphorylation of the MAPK Slt2, which in turn activates the transcription factor Rlm1 that finally leads to the transcriptional response that modulates adaptation to conditions interfering with cell wall stability. Several authors have recently suggested that activation of the CWI pathway by different stresses (heat shock, hypo-osmotic shock, actin perturbation, or cell wall stress) does not operate only in a linear “top down” manner but also as a consequence of lateral inputs that impact this MAPK pathway at different levels (1, 36). Regarding the activation of the CWI pathway by cell wall stress, it seems that different cell wall stresses involve different mechanisms. Thus, cell wall perturbations elicited by Congo Red, a compound that binds to chitin, are mainly sensed through Mid2 and activate a transcriptional response that almost completely depends on the MAPK Slt2 and the transcription factor Rlm1 (5). However, activation of the CWI pathway by alterations to the β-1,3 glucan network caused by zymolyase treatment (a main β-1,3 glucanase activity but also with some protease activity) (37) requires not only elements of the CWI but also a competent HOG pathway. Zymolyase activates both MAPKs: Hog1 and Slt2. Slt2 hyperphosphorylation under these conditions depends on the elements of the Sho1 branch of the HOG pathway, and it requires essential components of the CWI pathway such Mkk1/Mkk2, Bck1, and Pkc1, but not already known upstream elements, including the sensors and guanine nucleotide exchange factors of this pathway (1). Based on these results, we proposed a model in which damage to the cell wall must in this case be sensed through the sensors of the Sho1 branch of the HOG pathway (Msb2 and/or Hkr1) (17) and that the sequential activation of the HOG and CWI pathways must be necessary for the adaptation response to this specific cell wall damage. Hyperactivation of Slt2, as a consequence of previous activation of the elements of the Sho1 branch of the HOG pathway, must finally be responsible, through Rlm1, for the transcriptional adaptation response. This and other examples in which one MAPK pathway activates another MAPK pathway (38-40) illustrates that although pathway specificity is absolutely necessary, the MAPK pathways do not always compete and are often coordinated in a positive manner (41).
Here, the characterization of genome-wide transcriptional profiles to cell wall stress mediated by zymolyase in mutants of the HOG and CWI pathways served us to further characterize the impact of the connection between both pathways in the regulation of the yeast transcriptional responses under these conditions. The transcriptional profiling of the HOG-CWI connection described here perfectly matches the model described above concerning the sequential activation of Hog1 and Slt2 for adaptation to zymolyase. We have previously described that the overphosphorylation of Slt2 by zymolyase is completely abrogated in sho1Δ, ste50Δ, ste20Δ, and ste11Δ mutants. In addition, although most of the Slt2 activation by zymolyase is lost in hog1Δ and pbs2Δ mutants, these mutants still show a slight phosphorylation of this MAPK (1). Accordingly, a main group of genes up-regulated in the presence of zymolyase depends on both MAPKs: Hog1 and Slt2. In addition to CRH1, genes within this group such MLP1 and CWP1 depend on Bck1, Ste11, and Sho1, but not on the Sln1 branch of the HOG pathway or on the Mid2 and Wsc1 sensors of the CWI pathway. In agreement, the transcriptional induction of the genes within this group is dependent on the transcription factor Rlm1 but not on the transcription factor Msn2/4, regulated by Hog1. Moreover, other transcription factors regulated by this MAPK, such as Hot1, Sko1, and Smp1, are not involved in the induction of CRH1, one of the genes included in this group (1).
Additionally, we found a group of 13 genes whose induction by zymolyase was dependent on Slt2 but not on Hog1. The induction of these genes should be related to the residual Slt2 activation found in hog1Δ and pbs2Δ mutants referenced above. Interestingly, the induction of genes such as SED1, AFR1, DDR48, and PRM5, belonging to this group, did not require Pbs2 or Hog1 but was completely dependent on the presence of upstream elements of the Sho1 branch of the HOG pathway like Sho1, Ste50, and Ste11. Furthermore, transcriptional activation of these genes by zymolyase did not require Mid2, Wsc1, or Ssk1.
In sum, all of these results reinforce the notion that cells sense cell wall damage under these conditions through the Sho1 branch of the HOG pathway and hence that signaling is connected to the CWI pathway. Elements like Hkr1/Msb2, Sho1, Opy2, Ste50, Ste11, Hog1, and Pbs2 are required for this signaling mechanism, although the transcriptional activation of a group of genes being dependent on Slt2 but not on Hog1 or Pbs2 clearly indicates the existence of a direct branch connection between upstream elements of the HOG pathway and the CWI pathway to regulate responses to cell wall stress provoked by zymolyase.
Hog1 is activated by zymolyase, although to a much lower extent than osmotic stress. Apparently, in contrast to what happens under osmotic stress (42), Hog1 is not translocated from the cytoplasm to the nucleus by zymolyase treatment (1). However, here we detected a group of genes whose induction by zymolyase was dependent on Hog1 but not on Slt2. As deduced from our genome-wide profile of the msn2Δmsn4Δ mutant, the transcription factor Msn2/4 is responsible for the induction of ∼60% of these genes. The osmotic-like response mediated by zymolyase includes 29 genes, 25 of them also being induced after 45 min of osmotic shock (0.7 m NaCl) (26). This represents ∼14% of the osmotic response (181 genes). On comparing both responses in detail, we found that a high proportion of the genes included in this osmotic-like response elicited by zymolyase (68%) were included within the group of genes with higher induction ratios under osmotic stress (ratios ≥ 10). Therefore, the cell wall damage caused by zymolyase must involve two different transcriptional responses: (a) an osmotic transcriptional adaptation response, which would include genes that respond to low levels of Hog1 hyperphosphorylation, and (b) a main response, related to adaptation to cell wall integrity defects regulated by Slt2 and Rlm1. The later response requires previous activation of a complex that includes elements of the Sho1 branch of the HOG pathway.
Additional concerns about interaction between MAPK signaling pathways emerged from the nature of transcriptional responses induced by zymolyase in hog1Δ and slt2Δ mutants. In the case of hog1Δ cells, zymolyase induced the expression of a cluster of genes that were not induced in a wild type strain. Most of the genes within this cluster are also induced by hyperosmotic stress in a hog1Δ strain (25, 43). Moreover, this cluster included genes that are up-regulated by α-factor (32) and related to different aspects of the mating process (pheromone recognition, signaling, cell adherence, cell fusion, nuclear fusion, etc.), as well as some genes related to the filamentation pathway (Fig. 5). This is in accordance with previously described activation of Fus3 and Kss1 by hyperosmotic stress in the absence of HOG1 or PBS2 (25, 33, 35). Our data show that the same mechanism of regulation is also operating in response to zymolyase treatments. This suggests that the pheromone and filamentation pathways are inappropriately activated by the cell wall damage mediated by zymolyase in cells lacking a functional HOG pathway, probably, as suggested for hyperosmotic stress, because of the existence of a HOG-mediated inhibitory effect of mating and filamentation activity that could insulate the mating and pheromone pathways from activation by osmotic stress (33).
In addition to the interaction between the mating and filamentation pathways and the HOG pathway, we found an additional connection between the HOG and CWI pathways. In the absence of the MAPK Slt2, the transcriptional response induced by zymolyase and related to the osmotic stress adaptation response was clearly increased. In other words the zymolyase induced many more genes coincident with the osmotic stress response in a slt2Δ than in a wild type strain. As expected, this additional response would depend on Hog1, because the induction by zymolyase of several genes included in this group was abrogated in a double hog1Δslt2Δ mutant. The mechanism involved is a consequence of the hyperactivation of the HOG pathway, as deduced from the fact that Hog1 is more hyperphosphorylated by zymolyase in a slt2Δ than in a wild type strain (1). Accordingly, our data suggest that just as the HOG pathway interacts with the mating and filamentation pathways, the CWI pathway could have some inhibitory effect on the HOG pathway under conditions of cell wall stress because of the presence of zymolyase. This inhibitory effect could provide a mechanism to ensure that, under these conditions, signaling through the Sho1 branch of the HOG pathway will be basically devoted to the activation of the CWI pathway and the corresponding cell wall adaptation response and not to the activation of a transcriptional osmotic response.
The precise mechanism by which the MAPK Slt2 actively inhibits the activation of the Hog1 MAPK is not known and will need further investigation. One possibility is that the down-regulation might be exerted through a negative regulator of the HOG pathway controlled by the CWI pathway. Interestingly, in agreement with this idea, the expression of PTP2, coding for a protein phosphatase involved in dephosphorylating Hog1 (44), is induced by zymolyase in a Slt2-dependent manner. In the absence of Slt2, PTP2 is not induced by zymolyase, correlating with a higher degree of Hog1 phosphorylation under these circumstances. Although we cannot rule out an alternative mechanism by which the effect of Slt2 on the HOG pathway might be mediated through the upstream elements of this pathway, we favor the first hypothesis because zymolyase, which almost failed to induce the expression of the YDL204W and YDL233C genes in a wild type strain, led to a high transcriptional activation of these genes not only in a slt2Δ but also in a ptp2Δ mutant strain.4 Interestingly, it has previously been shown that heat shock activates both the Slt2 and Hog1 MAPKs, Ptp2 and Ptp3 being involved in the prevention of the hyperactivation of Hog1 under these circumstances as well as in blocking inappropriate cross-talk between the HOG and CWI pathways (45).
Supplementary Material
Acknowledgments
We thank all members of the Universidad Complutense de Madrid-Parque Cientifico de Madrid Genomic Unit for RT-qPCR experiments. We also thank María Molina, Francesc Posas, and members of the Research Unit 4 at the Department of Microbiology for fruitful discussions and Sonia Díez-Muñiz for technical assistance.
This work was supported by projects BIO2007-67821 from the Ministerio de Educación y Ciencia (Spanish government), EU STREP FUNGWALL LSBHCT-2004-511952 and S-SAL-0246/2006 from the Comunidad de Madrid.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1-S5.
Footnotes
The abbreviations used are: MAPK, mitogen-activated protein kinase; MAPKK, MAPK kinase; MAPKKK, MAPK kinase kinase; WT, wild type; RT, reverse transcription; qPCR, quantitative PCR; HOG, high osmotic response; CWI, cell wall integrity.
R. García, J. M. Rodríguez-Peña, C. Bermejo, C. Nombela, and J. Arroyo, unpublished results.
References
- 1.Bermejo, C., Rodríguez, E., García, R., Rodríguez-Peña, J. M., Rodríguez de la Concepción M. L., Rivas, C., Arias, P., Nombela, C., Posas, F., and Arroyo, J. (2008) Mol. Biol. Cell 191113 -1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levin, D. E. (2005) Microbiol. Mol. Biol. Rev. 69262 -291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Popolo, L., Gualtieri, T., and Ragni, E. (2001) Med. Mycol. 39 (Suppl. 1) 111-121 [PubMed] [Google Scholar]
- 4.Lagorce, A., Hauser, N. C., Labourdette, D., Rodríguez, C., Martin-Yken, H., Arroyo, J., Hoheisel, J. D., and Francois, J. (2003) J. Biol. Chem. 27820345 -20357 [DOI] [PubMed] [Google Scholar]
- 5.García, R., Bermejo, C., Grau, C., Pérez, R., Rodríguez-Peña, J. M., Francois, J., Nombela, C., and Arroyo, J. (2004) J. Biol. Chem. 27915183 -15195 [DOI] [PubMed] [Google Scholar]
- 6.Boorsma, A., De Nobel, H., ter Riet, B., Bargmann, B., Brul, S., Hellingwerf, K. J., and Klis, F. M. (2004) Yeast 21413 -427 [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez-Peña, J. M., Pérez-Díaz, R. M., Alvarez, S., Bermejo, C., García, R., Santiago, C., Nombela, C., and Arroyo, J. (2005) Microbiology 1512241 -2249 [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez-Peña, J. M., Diéz-Muñiz, S., Nombela, C., and Arroyo, J. (2008) J. Biotechnol. 133311 -317 [DOI] [PubMed] [Google Scholar]
- 9.Ketela, T., Green, R., and Bussey, H. (1999) J. Bacteriol. 1813330 -3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajavel, M., Philip, B., Buehrer, B. M., Errede, B., and Levin, D. E. (1999) Mol. Cell. Biol. 193969 -3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe, Y., Takaesu, G., Hagiwara, M., Irie, K., and Matsumoto, K. (1997) Mol. Cell. Biol. 172615 -2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baetz, K., Moffat, J., Haynes, J., Chang, M., and Andrews, B. (2001) Mol. Cell. Biol. 216515 -6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung, U. S., and Levin, D. E. (1999) Mol. Microbiol. 341049 -1057 [DOI] [PubMed] [Google Scholar]
- 14.De Nobel, H., Ruiz, C., Martín, H., Morris, W., Brul, S., Molina, M., and Klis, F. M. (2000) Microbiology 1462121 -2132 [DOI] [PubMed] [Google Scholar]
- 15.Reinoso-Martín, C., Schuller, C., Schuetzer-Muehlbauer, M., and Kuchler, K. (2003) Eukaryot. Cell 21200 -1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martín, H., Rodríguez-Pachón, J. M., Ruiz, C., Nombela, C., and Molina, M. (2000) J. Biol. Chem. 2751511 -1519 [DOI] [PubMed] [Google Scholar]
- 17.Tatebayashi, K., Tanaka, K., Yang, H. Y., Yamamoto, K., Matsushita, Y., Tomida, T., Imai, M., and Saito, H. (2007) EMBO J. 263521 -3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodríguez-Peña, J. M., Cid, V. J., Arroyo, J., and Nombela, C. (2000) Mol. Cell. Biol. 203245 -3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabib, E., Blanco, N., Grau, C., Rodríguez-Peña, J. M., and Arroyo, J. (2007) Mol. Microbiol. 63 921-935 [DOI] [PubMed] [Google Scholar]
- 20.Cabib, E., Farkas, V., Kosik, O., Blanco, N., Arroyo, J., and McPhie, P. (2008) J. Biol. Chem. 28329859 -29872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amberg, D. C., Burke, D. J., and Strathern, J. N. (2005) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, John Inglis, Cold Spring Harbor, NY
- 22.Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., Braisted, J., Klapa, M., Currier, T., Thiagarajan, M., Sturn, A., Snuffin, M., Rezantsev, A., Popov, D., Ryltsov, A., Kostukovich, E., Borisovsky, I., Liu, Z., Vinsavich, A., Trush, V., and Quackenbush, J. (2003) BioTechniques 34374 -378 [DOI] [PubMed] [Google Scholar]
- 23.Teixeira, M. C., Monteiro, P., Jain, P., Tenreiro, S., Fernandes, A. R., Mira, N. P., Alenquer, M., Freitas, A. T., Oliveira, A. L., and Sa-Correia, I. (2006) Nucleic Acids Res. 34D446 -D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak, K. J., and Schmittgen, T. D. (2001) Methods 25402 -408 [DOI] [PubMed] [Google Scholar]
- 25.O'Rourke, S. M., and Herskowitz, I. (2004) Mol. Biol. Cell 15532 -542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rep, M., Krantz, M., Thevelein, J. M., and Hohmann, S. (2000) J. Biol. Chem. 2758290 -8300 [DOI] [PubMed] [Google Scholar]
- 27.Kuge, S., and Jones, N. (1994) EMBO J. 13655 -664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imazu, H., and Sakurai, H. (2005) Eukaryot. Cell 41050 -1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shenhar, G., and Kassir, Y. (2001) Mol. Cell. Biol. 211603 -1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sengupta, P., and Cochran, B. H. (1990) Mol. Cell. Biol. 106809 -6812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madhani, H. D., Galitski, T., Lander, E. S., and Fink, G. R. (1999) Proc. Natl. Acad. Sci. U. S. A. 9612530 -12535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts, C. J., Nelson, B., Marton, M. J., Stoughton, R., Meyer, M. R., Bennett, H. A., He, Y. D., Dai, H., Walker, W. L., Hughes, T. R., Tyers, M., Boone, C., and Friend, S. H. (2000) Science 287873 -880 [DOI] [PubMed] [Google Scholar]
- 33.Hall, J. P., Cherkasova, V., Elion, E., Gustin, M. C., and Winter, E. (1996) Mol. Cell. Biol. 166715 -6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Rourke, S. M., and Herskowitz, I. (1998) Genes Dev. 122874 -2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davenport, K. D., Williams, K. E., Ullmann, B. D., and Gustin, M. C. (1999) Genetics 1531091 -1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrison, J. C., Zyla, T. R., Bardes, E. S., and Lew, D. J. (2004) J. Biol. Chem. 2792616 -2622 [DOI] [PubMed] [Google Scholar]
- 37.Zlotnik, H., Fernández, M. P., Bowers, B., and Cabib, E. (1984) J. Bacteriol. 1591018 -1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hahn, J. S., and Thiele, D. J. (2002) J. Biol. Chem. 27721278 -21284 [DOI] [PubMed] [Google Scholar]
- 39.García-Rodríguez, L. J., Valle, R., Durán, A., and Roncero, C. (2005) FEBS Lett. 5796186 -6190 [DOI] [PubMed] [Google Scholar]
- 40.Buehrer, B. M., and Errede, B. (1997) Mol. Cell. Biol. 176517 -6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, R. E., and Thorner, J. (2007) Biochim. Biophys. Acta 17731311 -1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrigno, P., Posas, F., Koepp, D., Saito, H., and Silver, P. A. (1998) EMBO J. 175606 -5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Rourke, S. M., and Herskowitz, I. (2002) Mol. Cell. Biol. 224739 -4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattison, C. P., and Ota, I. M. (2000) Genes Dev. 141229 -1235 [PMC free article] [PubMed] [Google Scholar]
- 45.Winkler, A., Arkind, C., Mattison, C. P., Burkholder, A., Knoche, K., and Ota, I. (2002) Eukaryot. Cell 1 163-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.