Abstract
Abnormal accumulation and activation of receptor tyrosine kinase Ron (recepteur d'origine nantais) has been demonstrated in a variety of primary human cancers. We show that RNA interference-mediated knockdown of Ron kinase in a highly tumorigenic colon cancer cell line led to reduced proliferation as compared with the control cells. Decreased Ron expression sensitized HCT116 cells to growth factor deprivation stress-induced apoptosis as reflected by increased DNA fragmentation and caspase 3 activation. In addition, cell motility was decreased in Ron knockdown cells as measured by wound healing assays and transwell assays. HCT116 cells are heterozygous for gain of function mutant PIK3CA H1047R. Analysis of signaling proteins that are affected by Ron knockdown revealed that phosphatidylinositol 3-kinase (PI3K) activity of the mutant PI3K as well as AKT phosphorylation was substantially reduced in the Ron knockdown cells compared with the control cells. Moreover, we demonstrated in vivo that knockdown of Ron expression significantly reduced lung metastasis as compared with the control cells in the orthotopic models. In summary, our results demonstrate that Ron plays an essential role in maintaining malignant phenotypes of colon cancer cells through regulating mutant PI3K activity. Therefore, targeting Ron kinase could be a potential strategy for colon cancer treatment, especially in patients bearing gain of function mutant PI3K activity.
The receptor tyrosine kinase Ron (recepteur d'origine nantais) belongs to the Met proto-oncogene family (1, 2). Mature Ron is a 180-kDa heterodimer composed of a 40-kDa extracellular α-chain and a 150-kDa transmembrane β-chain with tyrosine kinase activity (2). Macrophage-stimulating protein is the only ligand that has been identified for Ron (3, 4). Upon ligand binding, Ron dimerizes, becomes autophosphorylated, and transduces a variety of signals that regulate different downstream pathways including Ras/mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K),3 c-Jun N-terminal kinase (JNK), β-catenin, and nuclear factor-κB (5-12). Ron can be activated through ligand-dependent or -independent mechanisms (3, 13, 14), which lead to responses important for tumorigenesis and metastasis, including cell scattering, proliferation, motility, and survival (15, 16).
Ron is normally expressed at relatively low levels in cells of epithelial origin (4). Recent studies have shown that Ron is overexpressed in 47% of breast tumor tissues as compared with benign epithelium and that elevated Ron expression was strongly associated with invasive activity by tumors (17). In addition, Ron is moderately expressed in normal colorectal mucosa, but is significantly increased in the majority of primary human colorectal adenocarcinoma samples (18). Ron overexpression has also been demonstrated in head and neck tumors (19). Furthermore, splice variants of Ron have been identified in human colon cancer. These variants were found to confer constitutive Ron activity, transformation, and tumorigenicity to NIH3T3 cells (18). Collectively, these studies suggest that the aberrant and constitutive activation of Ron plays an important role in the progression of human carcinomas to invasive/metastatic phenotypes.
As indicated above, activated Ron transduces a variety of signals that regulate different pathway cascades including the PI 3-kinase pathway (11). Recent studies have shown that PIK3CA, which encodes the catalytic subunit of PI 3-kinase p110α, is mutated in ∼⅓ of colorectal cancers (20). It is also mutated at a high frequency in other cancers including breast, ovarian, and hepatocellular carcinomas (21-23), making it one of the most common mutations described to date in human cancers. We have identified 4 colon cancer cell lines bearing gain of function PIK3CA mutations including HCT116 cells. We have shown that the mutant PI3K is constitutively active during growth factor deprivation stress (GFDS) (24) and that mutant PI3K promotes metastasis of colon cancer in an orthotopic model in vivo (25). However, it is not well understood whether the mutant PI3K is subject to regulation and, if it is, what are the mechanisms. We hypothesized that Ron kinase regulates the activity of mutant PI 3-kinase and that this regulation has an impact on the metastatic potential of HCT116 cells.
Tumorigenicity of colorectal cancer results from the pathological transformation of normal colonic epithelium to an adenoma and ultimately an invasive cancer. Convergence of many epigenetic and genetic events during tumorigenesis eventually results in the deregulation of pivotal kinases that play critical roles in cell proliferation, survival, invasion, and metastasis (26). Treatment of advanced disease with molecularly targeted drugs has not had a large clinical impact in part because colorectal cancer is a disease associated with a high degree of heterogeneity leading to complicated phenotypes (27-29). Therefore, there is a need to identify new targets and develop novel target-specific therapies. One of the hallmarks of colorectal cancer is the ability to invade adjacent structures and to metastasize into remote organs (30, 31). Colorectal cancer is the second leading cause of cancer mortality in the United States (32). The majority of these deaths are from metastatic disease. Thus, targeting invasion and metastasis is an important strategy for colorectal cancer treatment. However, the mechanisms that control invasion and metastasis of colon cancer are largely unknown. Ron kinase has been shown to play a role in epithelial to mesenchymal transition (33), a process important for tumor progression, invasion, and metastasis. Therefore, we hypothesized that Ron kinase plays an important role in controlling colon cancer malignant phenotypes such as invasion and metastasis, and could be a potential target for colorectal cancer treatment.
It has been shown that mammary-specific Ron overexpression induces highly metastatic mammary tumors in mice (34) and that Ron signaling augments mammary tumor formation and metastasis in a murine model of breast cancer (35). Metastasis is a complex, multistep process that requires a tumor cell to fulfill two rate-limiting steps: invasion and distant colony formation (36-38). The role of Ron in both of these steps of colon cancer metastasis has not been determined. Many in vivo models of metastasis such as tail vein injection only allow for the study of the ability of a tumor cell to form distant colonies. They circumvent the process of invasion, which is a prerequisite for the second rate-limiting step of metastasis, distant colony formation. In the absence of transgenic animal models of metastasis, orthotopic models of colon cancer allow for the study of invasion in the bowel wall in addition to distant colony formation in other organs.
We reduced Ron expression in a highly tumorigenic and metastatic colon cancer cell line, HCT116, using stable transfection of Ron-specific shRNAs. The work presented here demonstrates that inhibition of Ron expression resulted in the reversal of several properties associated with the malignant phenotype in HCT116 cells including survival, migration, and in vivo metastasis. Although it has been shown by Xu et al. (39) that silencing of Ron alters oncogenic phenotypes of human colon carcinoma cells, this is the first report demonstrating that Ron knockdown inhibited the activation of gain of function mutant PI3K (thus indicating that this mutant enzyme is not independent of upstream activation) and reduced metastasis in an in vivo orthotopic model of colon cancer. Because mutant PI3K promotes metastasis of colon cancer (25), our results suggest that targeting Ron kinase could be a potential strategy for treatment of colon cancer patients with metastatic carcinoma.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents—The human colon carcinoma HCT116 cell line was established in tissue culture from a primary tumor as described previously (40). HCT116 cells were cultured in SM medium (McCoy's 5A serum-free medium (Sigma) with pyruvate, vitamins, amino acids, and antibiotics) supplemented with 10 ng/ml epidermal growth factor, 20 μg/ml insulin, and 4 μg/ml transferrin as described previously (41). Cells were maintained at 37 °C in a humidified atmosphere of 5% CO2.
Antibody for Ron was purchased from Santa Cruz Biotechnology Inc. Antibody for actin was purchased from Sigma. Poly-(ADP-ribose) polymerase, AKT, pAKT (Ser473), and caspase 3 antibodies were obtained from Cell Signaling Technology.
Ron Expression Profiling—Total RNA was prepared from human cancer cell lines using column purification with Qiagen RNeasy or Promega SV systems. cDNA was prepared by random-primed reverse transcription (Applied Biosystems cDNA Archive kit). Q-PCR was performed using SYBR Green chemistry and gene expression calculated using the comparative Ct method. The primers for Q-PCR are ACATCACCATCTGTGGCCAGCATC (forward) and TCCATCCCCTCGGGCACTCAGATT (reverse). Ct value derived from Q-PCR was normalized to glyceraldehyde-3-phosphate dehydrogenase to give a ΔCt value.
Transfection of Ron siRNA—Transient transfection of HCT116 cells was performed with 50 or 75 nm Ron siRNA (Dharmacon) using GeneSilencer (Genlantis). Total RNA was prepared 24 h post-transfection and gene knockdown was determined by reverse transcriptase-PCR. The sequences of reverse transcriptase-PCR primers are the same as those of Q-PCR. Caspase 3 induction was measured 48 h post-transfection using Caspase Glo from Promega.
To enable in vivo studies, three retroviral vector-based shRNAs against different regions of Ron were developed. Their sequences were GAAAGAGTCCATCCAGCTA, CACACCAACACTGACCCTT, and GTAGATGGTGAATGTCATA, respectively. A control vector with a scrambled sequence was also generated. 293GP packing cells (Clontech) were co-transfected with vesicular stomatitis virus-G and empty vector (designated pSR), scrambled shRNA vector (designated SC) or Ron shRNA vectors. The viruses were harvested 48 h later and used to infect HCT116 cells. Puromycin (1.0 μg/ml) was used to select infected cells. Individual pools were then isolated by plating the pooled cells at limiting dilution.
Colony Formation Assays and Soft Agarose Assays—The Ron knockdown clones and the control cells were plated in 24-well plates at a density of 300 cells/well to determine the effects of Ron knockdown on clonogenic potential at low seeding density. Two weeks later, cell colonies were visualized by staining with3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide, followed by solubilization of the dye in dimethyl sulfoxide and quantifying at 570 nm.
Soft agarose assays were used to determine the transformation properties of the cells as described previously (42, 43). Briefly, HCT116 control and Ron knockdown cells were suspended at 3000 cells/ml in 1 ml of 0.4% SeaPlaque-agarose in McCoy's 5A serum-free medium and plated on top of 1 ml of 0.8% agarose in the same medium in 6-well plates. Plates were incubated for 2-3 weeks at 37 °C with 5% CO2 in a humidified incubator. Cell colonies were visualized by staining with 0.5 mol of p-iodonitrotetrazolium violet (Sigma).
Apoptosis Assays—Apoptosis assays were performed to determine the response of the control cells and Ron knockdown clones to GFDS. Cells were seeded in 96-well plates and allowed to grow to 80% confluence. The cells were then changed to medium without serum or growth factors (SM) for 5 days. Apoptosis assays were then performed using a DNA fragmentation enzyme-linked immunosorbent assay kit as described in the manufacturer's protocol (Invitrogen).
Western Blot Analysis—Cells were lysed in Nonidet P-40 lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.5% Nonidet P-40, 50 mm sodium fluoride, 1 mm NaVO3, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, 25 μg/ml aprotinin, 25 μg/ml trypsin inhibitor, and 25 μg/ml leupeptin) at 4 °C. The supernatants were cleared by centrifugation. Protein (30 to 100 μg) was fractionated on an acrylamide denaturing gel and transferred onto a nitrocellulose membrane (Amersham Biosciences) by electroblotting. The membrane was blocked with 5% nonfat dry milk in TBST (50 mm Tris, pH 7.5, 150 mm NaCl, 0.05% Tween 20) for 1 h at room temperature or overnight at 4 °C and washed in TBST. The membrane was then incubated with primary antibodies for 1 h at room temperature or overnight at 4 °C. After washing with TBST for 15 min, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences) for an hour at room temperature. After further washing in TBST for 15 min, the proteins were detected by the enhanced chemiluminescence system (Amersham Biosciences).
PI 3-Kinase Assay—The cells were washed with phosphate-buffered saline and lysed in lysis buffer (137 mm NaCl, 20 mm Tris, pH 7.4, 1 mm MgCl2, 1 mm CaCl2, 1% Nonidet P-40, 100 μm NaVO4, and 1 mm phenylmethylsulfonyl fluoride). Protein concentrations were determined by the BCA protein assay reagent (Pierce). Lysates (400 μg of protein) were incubated with p85 antibody (Upstate) at 4 °C overnight, followed by a further incubation with protein A-agarose for 2 h. Immune complexes were washed twice with each wash buffer (PI3K wash 1, phosphate-buffered saline, 1% Nonidet P-40/NaVO4; PI3K wash 2, 100 mm Tris pH 7.4, 5 mm LiCl/NaVO4; PI3K wash 3, 10 mm Tris, pH 7.4, 150 mm NaCl, 5 mm EDTA/NaVO4). After the last wash was removed, PI 3-kinase assays were performed as described previously (44). Briefly, samples were resuspended in 50 μl of PI3K buffer (20 mm Tris, pH 7.5, 100 mm NaCl, and 0.5 mm EGTA), and 10 μg of phosphatidylinositol was added. After 10 min at room temperature, 10 μCi of [32P]ATP and MgCl2 to a final concentration of 20 μm was added. After 10 min at room temperature, lipids were extracted, first with 150 μl of CHCl3: methanol:HCl (10:20:0.2) and 100 μl of pure CHCl3. The second extraction used 80 μl of methanol, 1 n HCl (1:1). Samples were spotted on 1% potassium oxalate-treated TLC plates (Analtech, Newark, DE) and developed in CHCl3:methanol: NH4OH:H2O (129:114:15:21).
Wound Healing Assays and Transwell Motility Assays—Confluent cell monolayers were manually wounded by scraping the cells with a pipette tip to perform wound healing assays. Wound size was verified under a microscope to ensure that all wounds were the same width. The cell culture medium was then replaced with fresh medium, and wound closure was monitored by microscopy 24 h later.
Transwell motility assays were performed utilizing 8-μm pore, 6.5-mm polycarbonate transwell filters (Corning Costar Corp.). After trypsinizing the cells, single cell suspensions were seeded onto the upper surface of the filters in supplemental McCoy's 5A medium in the absence of growth factors and allowed to migrate toward McCoy's 5A medium with 10% fetal bovine serum. After 18 h incubation, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide was added to the medium. The cells on the upper surface of the filter were removed with a cotton swab, and the cells that had migrated to the underside of the filter were visualized under the microscope followed by solubilization of the dye in dimethyl sulfoxide and quantifying at 570 nm.
Orthotopic Implantation—The control cells and Ron knockdown clones were transfected with green fluorescence protein (GFP). Exponentially growing GFP-labeled control cells (pSR or SC) or RON knockdown clones (5 × 106 cells) were inoculated subcutaneously onto the dorsal surfaces of separate BALB/c nude male mice. Once xenografts were established, they were excised and minced into 1-mm3 pieces. Two of these pieces were then orthotopically implanted into other BALB/c nude mice. For operative procedures, animals were anesthetized with isoflurane inhalation. A 1-cm laparotomy was performed and both the cecum and ascending colon were exteriorized. Using ×7 magnification and microsurgical techniques, the serosa was disrupted by scraping in two locations. The 1-mm3 pieces of xenograft were subserosally implanted using an 8-0 nylon suture at the two points of serosal disruption. The bowel was then returned to the peritoneal cavity and the abdomen was closed with 5-0 vicryl suture.
Subsequently, animals were anesthetized with a 1:1 mixture of ketamine (10 mg/ml) and xylazine (1 mg/ml) by intraperitoneal injection (0.01 ml/mg) and weekly GFP fluorescence imaging was performed for up to 5 weeks. Specifically, GFP fluorescence imaging was performed using a light box illuminated by fiber optic lighting at 470 nm (Illumatool BLS, Lightools Research, Encinitas, CA) using a Retiga EXi color CCD camera (QImaging, Burnaby BC, Canada). High-resolution images consisting of 1,360 × 1036 pixels were captured directly using a MS Windows-based PC. Images were visually optimized for contrast and brightness using commercial software (Adobe Photoshop, CS2 Adobe, San Jose, CA). Excitation of GFP in the light box facilitated identification of primary and metastatic disease by direct near-real time visualization of fluorescence in live animals.
Thirty-five days post-implantation, animals were euthanized. Organs were explanted, imaged, and immediately placed in buffered 10% formalin. Tissues were then processed and embedded in paraffin. Slides were cut for hematoxylin and eosin staining.
Immunohistochemistry—Primary tumors established from the control and Ron knockdown cells were harvested and placed in 10% neutral buffered formalin fixative for 12 to 24 h and then embedded in paraffin. Deparaffinized tissue specimens were subjected to immunohistochemical staining for the detection of AKT phosphorylation using an indirect detection method (45). The catalyzed signal amplification system was used for the phosphospecific antibodies (Dako Corporation, Carpinteria, CA). The antibody staining was accompanied by a negative control in which slides were incubated with a matching blocking peptide to the primary antibody. Specimens were processed on the same day to eliminate any variability in conditions. Slides were digitally photographed using the same settings.
TUNEL, Hematoxylin and Eosin, and Ki67 Staining—Slides were cut from paraffin-embedded blocks and stained with Apotag (Oncor, Gaithersburg, MD) terminal nucleotidyl transferase-mediated nick end labeling (TUNEL) assay kit.
The apoptotic rate was determined quantitatively by counting the number of positively stained apoptotic bodies per 75-μm2 field at ×20 magnification. Six and nine animal slides for the control and RON knockdown clones, respectively, were analyzed. Three histologically similar fields viewed at ×20 were randomly selected from each slide for analysis. This procedure was performed by two blinded observers. The ratio of the average number of apoptotic cells to the total number of cells counted was used to represent apoptotic rates.
Slides cut from paraffin-embedded blocks were also used for hematoxylin and eosin stains and for immunohistochemical characterizations. Serial sections were cut to complement the hematoxylin and eosin sections and were stained with an IgG1 rabbit polyclonal antibody for Ki67 (Dako Corporation). Ki67 is a nonhistone nuclear antigen present in late G1, G2, and S phases of the cell cycle but not G0. The optimal dilution of 1:100 was used. Three- to 4-μm sections were cut, deparaffinized in xylene, and rehydrated in descending grades of ethanol. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in water. Immunostaining was done using a variation of the avidin-biotin-peroxidase method. Slides were counterstained with methyl green.
The proliferation rate was determined quantitatively by utilization of NIH Image J (pubic domain software). Image settings were as follows: threshold range 10-192; pixel size 20-5000. Six and nine slides from each control and Ron knockdown animal, respectively, were analyzed. Three histologically similar fields viewed at ×20 were randomly selected for analysis. The mean proliferation was determined for each group.
Statistical Analysis—Graphpad software (San Diego, CA) was utilized for statistical analysis. Statistical significance was determined using two-tailed Student's t test with a p value less than 0.05.
RESULTS
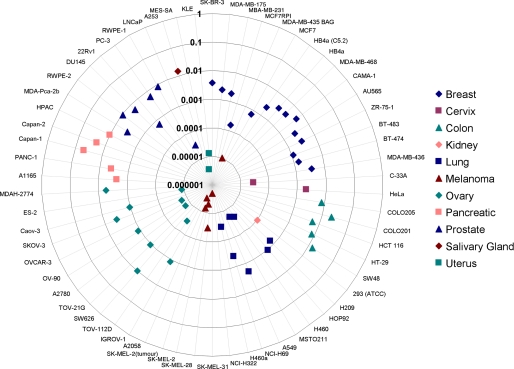
Ron Knockdown Reduced Clonogenic and Anchorage-independent Growth in HCT116 Colon Cancer Cells—To characterize the role of Ron in oncogenesis, we first generated the expression profiles of Ron in different tumor cell lines of the NCI 60 panel. As shown in Fig. 1, among the cell lines tested, Ron is highly expressed in colon, pancreatic, breast, and prostate cancer cells, suggesting that Ron overexpression is broadly associated with oncogenic phenotypes of various origins.
FIGURE 1.
Ron is highly expressed in colon, breast, pancreatic, and prostate cancers. RNA from cells in the NCI 60 panel was isolated and Ron mRNA expression was measured using Q-PCR. Values were normalized to glyceraldehyde-3-phosphate dehydrogenase control.
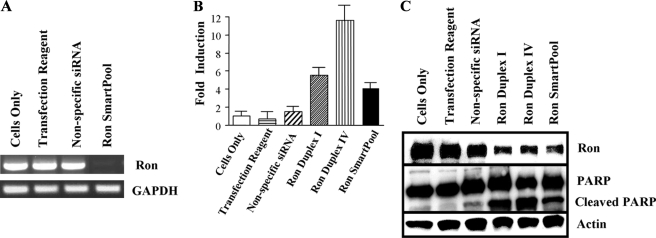
We focused our study on the role of Ron in colon cancer. Chemically synthesized siRNAs against Ron were transiently transfected into the HCT116 colon cancer cell line. HCT116 cells have a high level of Ron expression (Fig. 1) and are highly tumorigenic in athymic mice with tumor formation less than 15 days after inoculation (42). Ron siRNA transfection almost completely abrogated Ron mRNA expression and knocked down 60-70% of Ron protein expression (Fig. 2, A and C). As a result, Ron siRNA-transfected cells displayed increased apoptosis as measured by caspase activation and poly(ADP-ribose) polymerase cleavage (Fig. 2, B and C).
FIGURE 2.
Knockdown of Ron expression by small interfering RNA-induced apoptosis. HCT116 cells were transfected with either a nonspecific siRNA or Ron-specific siRNA (Smartpool, Dharmacon). A, message levels of Ron were determined at 24 h. B, Ron Smartpool and two individual siRNA duplexes were transfected into HCT116 cells and caspase 3 induction was measured 48 h later. Values are the means of three replicates. Error bars indicate S.E. C, protein expression of Ron and poly(ADP-ribose) polymerase (PARP) cleavage was determined in HCT116 cells 48 h post siRNA transfection. Actin was used to normalize sample loading. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
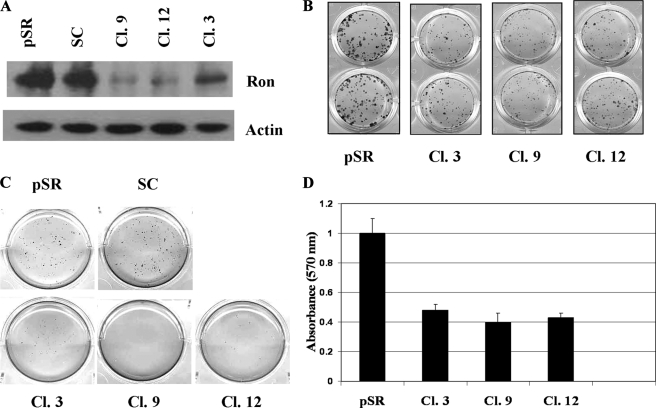
Vector-based Ron shRNAs were used to stably knockdown Ron expression in HCT116 cells to enable in vivo studies and to generate strains with stable properties. As shown in Fig. 3A, Ron protein expression was markedly reduced in some of the Ron shRNA-transfected clones as compared with the empty vector or scrambled shRNA transfected control cells (designated pSR or SC, respectively). Three different shRNAs that target three different sites of Ron cDNA were used to generate stable Ron knockdown clones to eliminate off-target effects of shRNAs. MET protein expression was not affected by Ron shRNAs indicating the specificity of Ron shRNAs (data not shown). Colony formation assays were used to compare the ability of the control cells and Ron knockdown cells to form colonies when plated at sparse densities. Knockdown of Ron expression resulted in reduced colony formation. Ron knockdown clones had fewer and smaller colonies than the control cells (Fig. 3B, top panel). Quantitation of the colonies indicated that the ability of Ron knockdown clones to form colonies was less than half of that of the control cells (Fig. 3B, bottom panel).
FIGURE 3.
Clonogenic and anchorage-independent growth was reduced in HCT116 cells stably transfected with Ron shRNA. A, Ron knockdown clones and the control cells were lysed and subjected to Western blot analyses with an antibody to Ron. Actin was used as a loading control. B and D, cells were plated in 24-well plates at 300 cells/well. Cell colonies were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide and visualized after 2 weeks of incubation (B). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide stain was dissolved in dimethyl sulfoxide. Relative cell numbers were then determined by the resultant absorbance at 595 nm. Values are the means of four replicates (D). Error bars indicate S.E. C, anchorage-independent colony formation in soft agarose by HCT116 control and Ron knockdown cells was determined as described under “Experimental Procedures.”
We further compared the anchorage-independent growth of the control cells and Ron knockdown cells by soft agarose assays. Fig. 3C showed that Ron knockdown clones had greatly impaired colony formation in soft agarose as compared with the control cells, indicating that knockdown of Ron expression impaired the anchorage independent growth of HCT116 cells. These results suggested that high levels of Ron expression in HCT116 cells might contribute to the malignant phenotype of these cells.
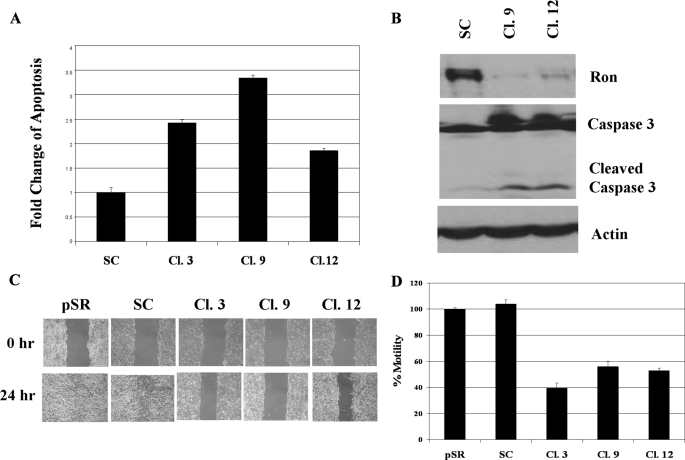
Knockdown of Ron Expression-sensitized HCT116 Cells to Growth Factor Deprivation Stress-induced Apoptosis—To determine whether Ron plays an important role in cell survival, we compared the responses of the control cells and Ron knockdown cells to GFDS. DNA fragmentation assays showed that stressed Ron knockdown cells displayed increased apoptosis as compared with the control cells during GFDS (Fig. 4A). These results were further confirmed by increased caspase 3 activation in the Ron knockdown clones (Fig. 4B), indicating that reduced Ron expression rendered HCT116 cells more sensitive to GFDS-induced apoptosis. This suggests that Ron-mediated resistance to apoptosis induced by exposure to environmental stresses such as growth factor and nutrient restriction that are thought to be prevalent prior to vascularization of growing tumors might play an important role in the malignancy of HCT116 cells.
FIGURE 4.
Ron knockdown-sensitized HCT116 cells to GFDS-induced apoptosis and impaired their motility. Ron knockdown clones and control cells were grown to 80-90% confluence and the cells were then deprived of growth factors for 5 days. A, DNA fragmentation assays were performed to determine apoptosis as described under “Experimental Procedures.” Error bars indicate S.E. B, cells were harvested and Western blot analyses were performed with Ron or caspase 3 antibody. Actin was used as a loading control. C, Ron knockdown clones and the control cells were grown to confluence. Motility was examined in wound healing assays as described under “Experimental Procedures.” D, Ron knockdown clones and the control cells were starved in growth factor-deprived medium overnight. They were then plated in the transwells and transwell assays were performed as described under “Experimental Procedures.”
Knockdown of Ron Impaired Motility of HCT116 Cells—Differentiated colonic epithelial cells possess a cobblestone-like structural-functional polarization and form tight junctions that mediate intracellular adhesion and cell-cell contact (46). The loss of this polarization is believed to be a critical step in epithelial tumorigenesis (47). Ron has been found to induce cell dissociation, migration, and invasion in a variety of cancer cells including those from breast, colon, and skin (10, 16, 17). Therefore, we compared the motility of Ron knockdown cells with that of the control cells. The rate of wound closure was used to measure cell motility in wound healing assays. Wound closure was almost complete in the control cells by 24 h, but was significantly delayed in Ron knockdown clones (Fig. 4C). Results from transwell assays showed that Ron knockdown clones exhibited only half of the motility of the control cells (Fig. 4D), confirming that Ron knockdown cells had markedly reduced motility compared with the control cells. These results along with increased apoptosis in Ron knockdown cells described above suggested that reduced Ron expression may lead to reduced tumorigenicity and metastasis in vivo.
Ron Kinase-regulated Activation of the Mutant PI3K—Activated Ron regulates a variety of different signaling pathways including the PI 3-kinase pathway (11). Recent studies have shown that PIK3CA is mutated in ∼⅓ of 234 colorectal cancers, but only in 2/76 adenomas (20), suggesting that PIK3CA mutations might contribute to tumor progression of colon cancer. HCT116 cells have been identified as bearing the gain of function PIK3CA mutation H1047R (24). We have shown that the mutant PI3K is constitutively active during GFDS, which rendered HCT116 cells resistant to GFDS-induced apoptosis (24). Furthermore, we demonstrated that mutant PI3K promotes metastasis of colon cancer in an orthotopic model in vivo (25). However, it is not clear whether the mutant PI3K is subject to upstream regulation or and, if it is, what are the mechanisms used. We hypothesized that Ron kinase controls activation of mutant PI 3-kinase in HCT116 cells.
To test this hypothesis, we first compared the PI 3-kinase activity between the Ron knockdown and control cells under GFDS. Sustained PI3K activity under stress conditions has been shown to play an important role in enabling PI3K mutant cells to survive environmental stresses (24). Because environmental restriction on growth appears to be common in solid tumors due to inadequate vascularization (48, 49), the ability of PI3K mutant cells to withstand these stresses is considered a key factor in tumor development and progression. Therefore, mutational activation of PIK3CA is likely to be one of the mechanisms of tumor progression in colon carcinomas and initial clinical data indicate that mutant PI3K is associated with a poorer prognosis in patients (50).
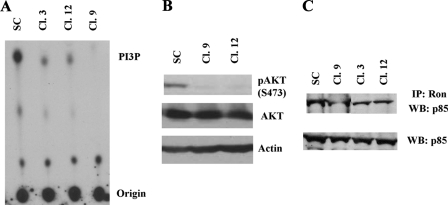
We found that PI 3-kinase activity was markedly reduced in Ron knockdown clones as compared with the control cells during GFDS (Fig. 5A). In addition, phosphorylation of AKT, one of the downstream mediators of PI3K, was also reduced in Ron knockdown cells as compared with the control cells (Fig. 5B), further confirming that the mutant PI3K signaling pathway was inhibited by Ron knockdown. These results indicated that the mutant PI3K in HCT116 cells was regulated by upstream Ron kinase.
FIGURE 5.
Knockdown of Ron kinase inhibited the PI 3-kinase pathway. Ron knockdown clones and control cells were grown to 80-90% confluence and the cells were then deprived of growth factors for 5 days. A, cell lysates were harvested and subjected to PI 3-kinase assays as described under “Experimental Procedures.” B, cell lysates harvested in A were subjected to Western blot analysis with pAKT (S473) or AKT antibody. Actin was used as a loading control. C, cell lysates from A were subjected to Western blot (WB) analyses with a p85 antibody or immunoprecipitated (IP) with a Ron antibody followed by Western blot analyses with a p85 antibody.
The p85 subunit of PI3K stabilizes p110α but also inhibits its lipid kinase activity. It has been shown that binding of Cdc42 GTPase to p85 activates PI3K activity (51). Ron kinase has been implied to activate PI3K activity through binding to the p85 regulatory subunit (12). To determine whether this is the case in the HCT116 cell, cell lysates from Scrambled or Ron shRNA-transfected cells were immunoprecipitated with anti-Ron antibody followed by Western blot analyses with anti-p85 antibody. As shown in Fig. 5C, there was less p85 binding to Ron kinase in Ron knockdown clones than in scrambled shRNA-transfected control cells under GFDS, whereas total p85 expression levels were very similar among those cells. These results suggest that Ron association with the p85 subunit could be one of the mechanisms of Ron-mediated PI3K activation.
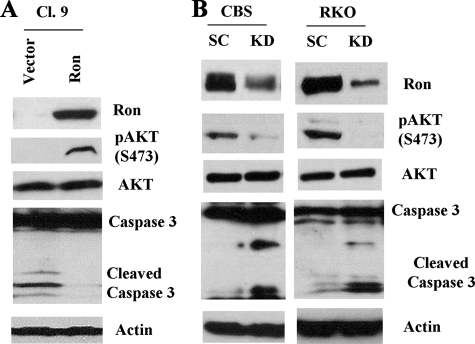
To rescue the phenotype caused by Ron knockdown, we overexpressed Ron in one of the Ron knockdown clones (Cl.9). Fig. 6A showed that Ron expression was restored in Cl.9 as compared with empty vector-expressing control cells. As a result, these Ron-expressing cells displayed higher AKT phosphorylation at Ser473 and were more resistant to GFDS-induced apoptosis as reflected by reduced caspase 3 cleavage as compared with the vector control cells (Fig. 6A). These results indicate that Ron activates PI3K/AKT and is responsible for stress resistance in HCT116 cells.
FIGURE 6.
A, Ron was overexpressed in one of the Ron knockdown clones (Cl.9). The vector or Ron expressing cells were deprived of growth factors for 5 days. Western blot analyses were performed with antibodies to Ron, pAKT (S473), or caspase 3. Actin was used as a loading control. B, CBS and RKO cells were transfected with either a nonspecific scrambled shRNA (SC) or a Ron-specific shRNA (KD). Transfected cells were grown to 80-90% confluence and then deprived of growth factors for 5 days. Cell lysates were harvested and Western blot analyses were performed with antibodies to Ron, pAKT (S473), AKT, or caspase 3. Actin was used as a loading control.
To ensure that Ron-mediated PI3K activation and cell survival are not specific to HCT116 cells, Ron expression was knocked down in another mutant PIK3CA H1047R bearing colon cancer cell line (RKO) and one wild type PIK3CA bearing cell line (CBS). Knockdown of Ron in each of these cell lines resulted in reduced AKT phosphorylation as well as increased caspase 3 cleavage under GFDS (Fig. 6B), indicating that Ron regulates activation of both mutant and wild type PI3K in colon cancer cells in response to stress.
Reduced Metastasis of HCT116 Cells by Ron Knockdown—We have shown that mutant PI3K promotes metastasis in HCT116 colon cancer cells (25). If mutant PI3K is under control of Ron activation, knockdown of Ron kinase would be expected to reduce metastasis in vivo. We used an orthotopic model approach to determine the effect of Ron kinase knockdown on the metastasis of HCT116 cells. To perform orthotopic implantation of Ron-inhibited HCT116 cells, subcutaneous xenografts were first generated. Xenograft formation was significantly reduced in animals bearing Ron knockdown cells as compared with those with the control cells by day 28 after subcutaneous injection (data not shown). The xenografts formed by the control cells and Ron knockdown clones were used for subsequent orthotopic implantation.
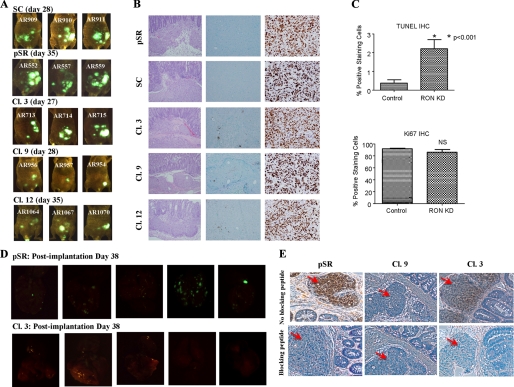
As shown in Table 1, animals implanted with xenografts formed by either the control cells (pSR or SC) or Ron knockdown cells (Cl.3, -9, and -12) demonstrated 100% primary growth at the site of implantation with clear invasion of the bowel on histologic evaluation (Fig. 7B, left panel). Fluorescence imaging, however, showed a greater tumor burden and tumor spread in the control animals as compared with the animals implanted with the HCT116 Ron knockdown clones (Fig. 7A). The difference in the size of the primary tumors between the control animals and those with Ron knockdown clones was likely due to increased apoptosis in the Ron knockdown cells because TUNEL staining assays of primary tumors showed that there were significantly more apoptotic cells in the tumors of Ron knockdown cells than in those of the control cells (Fig. 7B, middle panel, and Fig. 7C, top panel, p < 0.001). In addition, Ki67 staining of the primary tumors indicated that there was little difference in cell proliferation between the control and Ron knockdown cells (Fig. 7, B, right panel, and C, bottom panel, p = 0.2345), which is consistent with effects on cell survival rather than proliferation as a key factor in determining metastasis. Comparison of the explanted lungs from pSR vector control animals with those orthotopically implanted with one of the representative Ron knockdown clones in Fig. 7D indicates a clear increase in tumor burden in the lungs of pSR vector-control animals. Table 1 summarizes the differences in distant colony formation as a consequence of Ron knockdown as documented by histological analysis. The lungs and liver of each animal were serially sectioned at 1-mm intervals and the presence or absence of metastatic disease was determined by microscopic histological analysis. The control animals demonstrated distant metastases in 81% of the animals. The animals with Ron knockdown clones, however, demonstrated distant colonies in only 33% of the animals.
TABLE 1.
Knockdown of Ron reduces liver or lung metastasis
| Primary | Liver | Lung | Liver or Lung | |
|---|---|---|---|---|
| Clone 12 | 10/10 | 0/10 | 3/10 | 3/10 (30%) |
| Clone 9 | 10/10 | 2/10 | 2/10 | 4/10 (40%) |
| Clone 3 | 9/9 | 3/9 | 2/9 | 3/9 (33%) |
| 29/29 (100%) | 5/29 (17%) | 7/29 (24%) | 10/29 (33%) | |
| Control | 36/36 (100%) | 15/36 (42%) | 24/36 (67%) | 29/36 (81%) |
FIGURE 7.
Exponentially growing cells (5 × 106) were inoculated subcutaneously in athymic nude mice. The xenografts formed by the control cells and Ron knockdown clones were used for subsequent orthotopic implantation. A, fluorescence imaging of the animals on the indicated days. B, primary tumors established from the control and RON knockdown cells were processed for hematoxylin and eosin (left panel), TUNEL (middle panel), and Ki67 (right panel) staining. Both TUNEL and Ki67 images were captured at ×20 magnification. C, primary tumors established from the control and RON knockdown cells were analyzed to assess the proliferation and apoptotic rate. NIH Image J software was utilized to enumerate positive staining cells and total number of cells. The percentage of positive staining cells was calculated. Error bars indicate S.E. D, 38 days post-implantation, animals were euthanized. Organs were explanted and imaged. E, immunohistochemistry was performed for pAKT (S473) on primary tumors of control or Ron knockdown cells. The blocking peptide was used to show the specificity. The red arrows indicate tumor cells.
To assess PI3K/AKT activation in primary tumors from animals exposed to control or Ron knockdown cells, AKT phosphorylation was determined by immunohistochemistry. As shown in Fig. 7E, AKT phosphorylation was reduced in primary tumors of Ron knockdown cells as compared with those of control cells.
Taken together, these in vivo results demonstrated an important role for Ron in tumor invasion and distant colony formation indicating the potential for treatment by Ron antagonism in patients with metastases. Identifying the role of Ron in the complex process of metastasis may ultimately allow for the development of additional novel therapeutic approaches.
DISCUSSION
The scatter factors including hepatocyte growth factor and macrophage stimulating protein are different from other cytokines. They are involved in the control of cell dissociation, migration, and branching morphogenesis in addition to protecting cells from apoptosis (42, 52-56). Scatter factors appear to have an important role in mediating the metastatic process. Met and Ron tyrosine kinases are the receptors for hepatocyte growth factor and macrophage stimulating protein, respectively. The role of hepatocyte growth factor and Met in metastasis has been extensively studied (see Ref. 57). However, the role of Ron in determining metastasis has not been fully elucidated. We have shown here that reduced Ron expression in human colon carcinoma cells led to decreased anchorage-independent growth, increased sensitivity to GFDS-induced apoptosis, impaired motility, and most importantly markedly reduced metastasis in an orthotopic model.
There are often “off-target” effects when using siRNA to knockdown the specific protein expression. We employed three different shRNA constructs that targeted Ron at different regions. Although the knockdown effect was not exactly the same with different constructs, the overall outcome of Ron knockdown was very similar in all three Ron knockdown clones as demonstrated by modulation of in vitro and in vivo properties. This indicated that the results we obtained in this study were not due to off-target effects.
HCT116 cells bear one allele of gain of function mutant PIK3CA H1047R. We have shown previously that the mutant PI3K is constitutively active during GFDS (24) and that mutant PI3K promotes metastasis of colon cancer in the orthotopic model in vivo (25). There is a recent study demonstrating that there was no difference between PI 3-kinase activity and downstream signaling by wild type and mutant PIK3CA carrying colon cancer cells under normal cell culture conditions (58). We also showed that PI 3-kinase activity and signaling are very similar between the wild type and mutant PI3K cells under normal conditions (GFDS 0 h) (24). However, there was a significant difference in PI3K signaling between those two cell types under GFDS (GFDS 48 or 96 h) (24). These results indicated that PI 3-kinase activity was sustained in the mutant cells under GFDS, whereas it was rapidly decayed in the wild type cells. Sustained PI3K activity under stress conditions would enable mutant cells to survive environmental stresses, which might play an important role in tumor development and progression. Therefore, exploration of mechanisms controlling activation of mutant PI3K would help understand tumor progression in colon carcinomas. We showed in this study that knockdown of Ron expression inhibited constitutive activation of the mutant PI3K in HCT116 cells during GFDS, which led to reduced metastasis in vivo. These results indicate that the mutant PI3K is subject to regulation by upstream Ron kinase and that mutant PI3K bearing cells are sensitive to Ron kinase inhibition. This suggests that co-existence of Ron kinase and mutant PI3K might facilitate the development of metastasis in colon cancer and that inhibition of Ron kinase might be an efficient way to treat cancer patients with activated Ron kinase or mutant PI3K.
Both HCT116 and RKO cells harbor mutant PI3K H1047R (in the kinase domain), which has been proposed to have a direct effect on the conformation of the activation loop of the p110α catalytic subunit, enhancing its interaction with phosphatidylinositide substrates (59). Therefore H1047R mutant PI3K might have higher affinity for substrates and ATP than the wild type enzyme, especially when the cells are deprived of growth factors and nutrients (i.e. under GFDS). The p85 subunit stabilizes p110α but also inhibits the lipid kinase activity of p110α. Because H1047R mutation will not affect binding of p110α to p85 (59), Ron might regulate activation of mutant PI3K H1047R through interacting with the p85 subunit as it does with the wild type PI3K. Our results that p85 associated with Ron kinase was reduced in Ron knockdown HCT116 cells as compared with the control cells support this hypothesis.
Metastasis is a complex, multistep process that requires a tumor cell to be able to fulfill the two rate-limiting steps of invasion and distant colony formation. In vivo approaches such as tail vein injection allow for the study of the ability of a tumor cell to form distant colonies. However, they do not allow for the study of invasion, which is ultimately necessary prior to distant colony formation. In the absence of transgenic animal models of metastasis, this orthotopic model of colon cancer allows for the study of invasion in the bowel wall and distant colony formation in the liver and lungs. Metastatic spread to the liver and lungs recapitulates the pattern of colorectal cancer metastases in humans. Therefore, our results demonstrate the potential key role of Ron kinase in colon cancer progression.
Recently, in vivo and in vitro observations indicated that the metastatic potential of tumors is associated with an increased resistance to apoptosis (60-62). We showed that the Ron knockdown cells displayed decreased survival capacity under stress conditions, which might contribute to decreased metastasis in the orthotopic model. However, we also showed that Ron knockdown impaired the motility of HCT116 cells (Fig. 4, C and D). Therefore, decreased invasion of the Ron knockdown cells might also contribute to reduced metastatic potential in vivo. More studies are needed to determine whether decreased survival or invasion alone would change the metastatic potential of those cells in vivo.
As shown in Fig. 7A, Ron knockdown cells formed smaller primary tumors than the control cells. TUNEL and Ki67 staining showed that the primary tumors from Ron knockdown cells had more apoptotic cells than those of the control cells, whereas the numbers of cycling cells were very similar (Fig. 7, B and C). These results are consistent with in vitro data that cell cycle profiles analyzed by flow cytometry showed that there was not much difference between the control and Ron knockdown cells (data not shown). Therefore, the difference in the sizes of primary tumors of control animals and those implanted with Ron knockdown xenografts is likely due to the different survival capacity of the control and Ron knockdown cells rather than a difference in proliferation.
One could argue that the reduced metastasis in animals implanted with Ron knockdown xenografts is the consequence of smaller primary tumors. We cannot rule out that possibility. However, there is no correlation between the size of primary tumors and metastatic potential in colon and pancreatic cancers (63, 64). Furthermore, the American Joint Committee on Cancer staging for colorectal cancer, which is often used to predict prognosis, is not based on the size of the primary tumor. Staging is determined by the depth of invasion in the bowel wall, absence or presence of lymph node metastases, and the absence or presence of distant metastases (65). Therefore, it is very likely that the metastatic potential is determined by the characteristics of cancer cells (i.e. survival capacity, invasiveness, etc.) instead of the size of primary tumors.
Colorectal carcinoma is one of the most common types of adult malignancy and the second leading cause of cancer related mortality in the United States. The treatment of advanced colorectal cancer has been very ineffective. Traditional chemotherapy and radiotherapy are limited by their lack of specificity. Cancer-specific target-based therapy, which involves recognition of suitable targets that are important for cell proliferation, survival, invasion, and especially metastasis may provide additional specificity to these approaches. Ron has been implicated as a potential target for colorectal cancer. Our studies have verified the role of Ron in metastasis of colon cancer and provided in vitro and in vivo models to test Ron inhibitors for colorectal cancer treatment. Additionally, combination of the inhibition of Ron with other molecularly targeted therapies that have impact on cell proliferation and survival may provide added benefit due to the impact of Ron inhibition of epithelial to mesenchymal transition, invasion, and metastasis.
This work was supported, in whole or in part, by National Institutes of Health Grants CA34432 and CA54807 (to M. G. B.) from the NCI. This work was also supported by a pilot project grant from Eppley Cancer Institute (to J. W.) and by grants from the Ralph Wilson Foundation and the Buswell Research Foundation (to A. R.).
Footnotes
The abbreviations used are: PI3K, phosphatidylinositol 3-kinase; GFDS, growth factor deprivation stress; TUNEL, terminal nucleotidyl transferase-mediated nick end labeling; shRNA, short hairpin RNA; siRNA, small interfering RNA; GFP, green fluorescent protein.
References
- 1.Rubin, J. S., Bottaro, D. P., and Aaronson, S. A. (1993) Biochim. Biophys. Acta 1155 357-371 [DOI] [PubMed] [Google Scholar]
- 2.Ronsin, C., Muscatelli, F., Mattei, M. G., and Breathnach, R. (1993) Oncogene 8 1195-1202 [PubMed] [Google Scholar]
- 3.Wang, M. H., Ronsin, C., Gesnel, M. C., Coupey, L., Skeel, A., Leonard, E. J., and Breathnach, R. (1994) Science 266 117-119 [DOI] [PubMed] [Google Scholar]
- 4.Gaudino, G., Follenzi, A., Naldini, L., Collesi, C., Santoro, M., Gallo, K. A., Godowski, P. J., and Comoglio, P. M. (1994) EMBO J. 13 3524-3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santoro, M. M., Penengo, L., Orecchia, S., Cilli, M., and Gaudino, G. (2000) Oncogene 19 5208-5211 [DOI] [PubMed] [Google Scholar]
- 6.Li, B. Q., Wang, M. H., Kung, H. F., Ronsin, C., Breathnach, R., Leonard, E. J., and Kamata, T. (1995) Biochem. Biophys. Res. Commun. 216 110-118 [DOI] [PubMed] [Google Scholar]
- 7.Zhou, Y. Q., Chen, Y. Q., Fisher, J. H., and Wang, M. H. (2002) J. Biol. Chem. 277 38104-38110 [DOI] [PubMed] [Google Scholar]
- 8.Liu, Q. P., Fruit, K., Ward, J., and Correll, P. H. (1999) J. Immunol. 163 6606-6613 [PubMed] [Google Scholar]
- 9.Santoro, M. M., Gaudino, G., and Marchisio, P. C. (2003) Dev. Cell 5 257-271 [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y. Q., Zhou, Y. Q., Angeloni, D., Kurtz, A. L., Qiang, X. Z., and Wang, M. H. (2000) Exp. Cell Res. 261 229-238 [DOI] [PubMed] [Google Scholar]
- 11.Wang, M. H., Montero-Julian, F. A., Dauny, I., and Leonard, E. J. (1996) Oncogene 13 2167-2175 [PubMed] [Google Scholar]
- 12.Xiao, Z. Q., Chen, Y. Q., and Wang, M. H. (2000) Biochem. Biophys. Res. Commun. 267 669-675 [DOI] [PubMed] [Google Scholar]
- 13.Danilkovitch-Miagkova, A., and Leonard, E. J. (2001) Histol. Histopathol. 16 623-631 [DOI] [PubMed] [Google Scholar]
- 14.Follenzi, A., Bakovic, S., Gual, P., Stella, M. C., Longati, P., and Comoglio, P. M. (2000) Oncogene 19 3041-3049 [DOI] [PubMed] [Google Scholar]
- 15.Waltz, S. E., McDowell, S. A., Muraoka, R. S., Air, E. L., Flick, L. M., Chen, Y. Q., Wang, M. H., and Degen, S. J. (1997) J. Biol. Chem. 272 30526-30537 [DOI] [PubMed] [Google Scholar]
- 16.Wang, M. H., Dlugosz, A. A., Sun, Y., Suda, T., Skeel, A., and Leonard, E. J. (1996) Exp. Cell Res. 226 39-46 [DOI] [PubMed] [Google Scholar]
- 17.Maggiora, P., Marchio, S., Stella, M. C., Giai, M., Belfiore, A., De Bortoli, M., Di Renzo, M. F., Costantino, A., Sismondi, P., and Comoglio, P. M. (1998) Oncogene 16 2927-2933 [DOI] [PubMed] [Google Scholar]
- 18.Zhou, Y. Q., He, C., Chen, Y. Q., Wang, D., and Wang, M. H. (2003) Oncogene 22 186-197 [DOI] [PubMed] [Google Scholar]
- 19.Lin, H. S., Berry, G. J., Fee, W. E., Jr., Terris, D. J., and Sun, Z. (2004) Arch. Otolaryngol. Head Neck Surg. 130 311-316 [DOI] [PubMed] [Google Scholar]
- 20.Samuels, Y., Wang, Z., Bardelli, A., Silliman, N., Ptak, J., Szabo, S., Yan, H., Gazdar, A., Powell, S. M., Riggins, G. J., Willson, J. K., Markowitz, S., Kinzler, K. W., Vogelstein, B., and Velculescu, V. E. (2004) Science 304 554. [DOI] [PubMed] [Google Scholar]
- 21.Campbell, I. G., Russell, S. E., Choong, D. Y., Montgomery, K. G., Ciavarella, M. L., Hooi, C. S., Cristiano, B. E., Pearson, R. B., and Phillips, W. A. (2004) Cancer Res. 64 7678-7681 [DOI] [PubMed] [Google Scholar]
- 22.Lee, J. W., Soung, Y. H., Kim, S. Y., Lee, H. W., Park, W. S., Nam, S. W., Kim, S. H., Lee, J. Y., Yoo, N. J., and Lee, S. H. (2005) Oncogene 24 1477-1480 [DOI] [PubMed] [Google Scholar]
- 23.Bachman, K. E., Argani, P., Samuels, Y., Silliman, N., Ptak, J., Szabo, S., Konishi, H., Karakas, B., Blair, B. G., Lin, C., Peters, B. A., Velculescu, V. E., and Park, B. H. (2004) Cancer Biol. Ther. 3 772-775 [DOI] [PubMed] [Google Scholar]
- 24.Wang, J., Kuropatwinski, K., Hauser, J., Rossi, M. R., Zhou, Y., Conway, A., Kan, J. L., Gibson, N. W., Willson, J. K., Cowell, J. K., and Brattain, M. G. (2007) Mol. Cancer Ther. 6 1143-1150 [DOI] [PubMed] [Google Scholar]
- 25.Guo, X. N., Rajput, A., Rose, R., Hauser, J., Beko, A., Kuropatwinski, K., Levea, C., Hoffman, R. M., Brattain, M. G., and Wang, J. (2007) Cancer Res. 67 5851-5858 [DOI] [PubMed] [Google Scholar]
- 26.Hanahan, D., and Weinberg, R. A. (2000) Cell 100 57-70 [DOI] [PubMed] [Google Scholar]
- 27.Cunningham, D., Humblet, Y., Siena, S., Khayat, D., Bleiberg, H., Santoro, A., Bets, D., Mueser, M., Harstrick, A., Verslype, C., Chau, I., and Van Cutsem, E. (2004) N. Engl. J. Med. 351 337-345 [DOI] [PubMed] [Google Scholar]
- 28.Perez-Soler, R., Chachoua, A., Hammond, L. A., Rowinsky, E. K., Huberman, M., Karp, D., Rigas, J., Clark, G. M., Santabarbara, P., and Bonomi, P. (2004) J. Clin. Oncol. 22 3238-3247 [DOI] [PubMed] [Google Scholar]
- 29.Saltz, L. B., Meropol, N. J., Loehrer, P. J., Sr., Needle, M. N., Kopit, J., and Mayer, R. J. (2004) J. Clin. Oncol. 22 1201-1208 [DOI] [PubMed] [Google Scholar]
- 30.Portera, C. A., Jr., Berman, R. S., and Ellis, L. M. (1998) Surg. Oncol. 7 183-195 [DOI] [PubMed] [Google Scholar]
- 31.Yokota, J. (2000) Carcinogenesis 21 497-503 [DOI] [PubMed] [Google Scholar]
- 32.Jemal, A., Siegel, R., Ward, E., Murray, T., Xu, J., and Thun, M. J. (2007) CA-Cancer J. Clin. 57 43-66 [DOI] [PubMed] [Google Scholar]
- 33.Wang, D., Shen, Q., Chen, Y. Q., and Wang, M. H. (2004) Oncogene 23 1668-1680 [DOI] [PubMed] [Google Scholar]
- 34.Zinser, G. M., Leonis, M. A., Toney, K., Pathrose, P., Thobe, M., Kader, S. A., Peace, B. E., Beauman, S. R., Collins, M. H., and Waltz, S. E. (2006) Cancer Res. 66 11967-11974 [DOI] [PubMed] [Google Scholar]
- 35.Peace, B. E., Toney-Earley, K., Collins, M. H., and Waltz, S. E. (2005) Cancer Res. 65 1285-1293 [DOI] [PubMed] [Google Scholar]
- 36.Chambers, A. F., Naumov, G. N., Vantyghem, S. A., and Tuck, A. B. (2000) Breast Cancer Res. 2 400-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chambers, A. F., Naumov, G. N., Varghese, H. J., Nadkarni, K. V., MacDonald, I. C., and Groom, A. C. (2001) Surg. Oncol. Clin. N. Am. 10 243-255, vii [PubMed] [Google Scholar]
- 38.Chambers, A. F., Groom, A. C., and MacDonald, I. C. (2002) Nat. Rev. Cancer 2 563-572 [DOI] [PubMed] [Google Scholar]
- 39.Xu, X. M., Wang, D., Shen, Q., Chen, Y. Q., and Wang, M. H. (2004) Oncogene 23 8464-8474 [DOI] [PubMed] [Google Scholar]
- 40.Brattain, M. G., Levine, A. E., Chakrabarty, S., Yeoman, L. C., Willson, J. K., and Long, B. (1984) Cancer Metastasis Rev. 3 177-191 [DOI] [PubMed] [Google Scholar]
- 41.Boyd, D. D., Levine, A. E., Brattain, D. E., McKnight, M. K., and Brattain, M. G. (1988) Cancer Res. 48 2469-2474 [PubMed] [Google Scholar]
- 42.Wang, J., Sun, L., Myeroff, L., Wang, X., Gentry, L. E., Yang, J., Liang, J., Zborowska, E., Markowitz, S., Willson, J. K., and Brattain, M. G. (1995) J. Biol. Chem. 270 22044-22049 [DOI] [PubMed] [Google Scholar]
- 43.Wang, J., Han, W., Zborowska, E., Liang, J., Wang, X., Willson, J. K., Sun, L., and Brattain, M. G. (1996) J. Biol. Chem. 271 17366-17371 [DOI] [PubMed] [Google Scholar]
- 44.Jackson, J. G., St. Clair, P., Sliwkowski, M. X., and Brattain, M. G. (2004) Cancer Res. 64 2601-2609 [DOI] [PubMed] [Google Scholar]
- 45.Nakane, P. K. (1975) Ann. N. Y. Acad. Sci. 254 203-211 [DOI] [PubMed] [Google Scholar]
- 46.Cereijido, M., Shoshani, L., and Contreras, R. G. (2000) Am. J. Physiol. 279 G477-G482 [DOI] [PubMed] [Google Scholar]
- 47.Martin, T. A., and Jiang, W. G. (2001) Histol. Histopathol. 16 1183-1195 [DOI] [PubMed] [Google Scholar]
- 48.Goldie, J. H., and Coldman, A. J. (1979) Cancer Treat. Rep. 63 1727-1733 [PubMed] [Google Scholar]
- 49.Hajra, K. M., and Liu, J. R. (2004) Apoptosis 9 691-704 [DOI] [PubMed] [Google Scholar]
- 50.Kato, S., Iida, S., Higuchi, T., Ishikawa, T., Takagi, Y., Yasuno, M., Enomoto, M., Uetake, H., and Sugihara, K. (2007) Int. J. Cancer 121 1771-1778 [DOI] [PubMed] [Google Scholar]
- 51.Zheng, Y., Bagrodia, S., and Cerione, R. A. (1994) J. Biol. Chem. 269 18727-18730 [PubMed] [Google Scholar]
- 52.Nakamura, T., Teramoto, H., and Ichihara, A. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 6489-6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoker, M., Gherardi, E., Perryman, M., and Gray, J. (1987) Nature 327 239-242 [DOI] [PubMed] [Google Scholar]
- 54.Gherardi, E., Gray, J., Stoker, M., Perryman, M., and Furlong, R. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 5844-5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zarnegar, R., and Michalopoulos, G. (1989) Cancer Res. 49 3314-3320 [PubMed] [Google Scholar]
- 56.Medico, E., Mongiovi, A. M., Huff, J., Jelinek, M. A., Follenzi, A., Gaudino, G., Parsons, J. T., and Comoglio, P. M. (1996) Mol. Biol. Cell 7 495-504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trusolino, L., and Comoglio, P. M. (2002) Nat. Rev. Cancer 2 289-300 [DOI] [PubMed] [Google Scholar]
- 58.Morrow, C. J., Gray, A., and Dive, C. (2005) FEBS Lett. 579 5123-5128 [DOI] [PubMed] [Google Scholar]
- 59.Huang, C. H., Mandelker, D., Schmidt-Kittler, O., Samuels, Y., Velculescu, V. E., Kinzler, K. W., Vogelstein, B., Gabelli, S. B., and Amzel, L. M. (2007) Science 318 1744-1748 [DOI] [PubMed] [Google Scholar]
- 60.Del Bufalo, D., Biroccio, A., Leonetti, C., and Zupi, G. (1997) FASEB J. 11 947-953 [DOI] [PubMed] [Google Scholar]
- 61.Glinsky, G. V., Glinsky, V. V., Ivanova, A. B., and Hueser, C. J. (1997) Cancer Lett. 115 185-193 [DOI] [PubMed] [Google Scholar]
- 62.Inbal, B., Cohen, O., Polak-Charcon, S., Kopolovic, J., Vadai, E., Eisenbach, L., and Kimchi, A. (1997) Nature 390 180-184 [DOI] [PubMed] [Google Scholar]
- 63.Lowney, J. K., Frisella, M. M., Lairmore, T. C., and Doherty, G. M. (1998) Surgery 124 1043-1048 [DOI] [PubMed] [Google Scholar]
- 64.Wolmark, N., Cruz, I., Redmond, C. K., Fisher, B., and Fisher, E. R. (1983) Cancer 51 1315-1322 [DOI] [PubMed] [Google Scholar]
- 65.Greene, F. L., Page, D. L., Fleming, I. D., Fritz, A. G., Charles, B. M., Haller, D. G., and Morrow, M. (2002) Colon and Rectum in AJCC Cancer Staging Handbook, 6th Ed. pp. 127-138, Springer-Verlag New York LLC, New York