Abstract
Srv2/cyclase-associated protein is expressed in virtually all plant, animal, and fungal organisms and has a conserved role in promoting actin depolymerizing factor/cofilin-mediated actin turnover. This is achieved by the abilities of Srv2 to recycle cofilin from ADP-actin monomers and to promote nucleotide exchange (ATP for ADP) on actin monomers. Despite this important and universal role in facilitating actin turnover, the mechanism underlying Srv2 function has remained elusive. Previous studies have demonstrated a critical functional role for the G-actin-binding C-terminal half of Srv2. Here we describe an equally important role in vivo for the N-terminal half of Srv2 in driving actin turnover. We pinpoint this activity to a conserved patch of surface residues on the N-terminal dimeric helical folded domain of Srv2, and we show that this functional site interacts with cofilin-actin complexes. Furthermore, we show that this site is essential for Srv2 acceleration of cofilin-mediated actin turnover in vitro. A cognate Srv2-binding site is identified on a conserved surface of cofilin, suggesting that this function likely extends to other organisms. In addition, our analyses reveal that higher order oligomerization of Srv2 depends on its N-terminal predicted coiled coil domain and that oligomerization optimizes Srv2 function in vitro and in vivo. Based on these data, we present a revised model for the mechanism by which Srv2 promotes actin turnover, in which coordinated activities of its N- and C-terminal halves catalyze sequential steps in recycling cofilin and actin monomers.
Remodeling of cell shape during cell motility, cell division, and cell morphogenesis requires not only the rapid assembly of new actin filaments but also the coordinated disassembly of older filaments. Dynamic turnover provides cells with the plasticity necessary to remodel actin networks rapidly in response to cues, and replenishes the pool of assembly competent ATP-bound actin monomers available for new growth. Although major advancements have been made in determining the mechanisms that promote actin assembly (1, 2), comparatively little is known about the mechanisms governing actin disassembly and turnover. The rate-limiting step in filament disassembly is dissociation of subunits from filament ends (3). actin depolymerizing factor/cofilin (referred to herein as cofilin) accelerates this step by severing and depolymerizing older (ADP-rich) actin filaments (4-7). Cofilin remains bound to dissociated ADP-actin monomers and strongly inhibits nucleotide exchange (ATP for ADP) on monomeric actin (8). This leads to an accumulation of cofilin-bound ADP-actin monomers and depletes available cofilin and ATP-actin monomers. For this reason, cells require a mechanism for rapidly displacing cofilin from ADP-G-actin to maintain rapid actin turnover.
Recent studies suggest that this function is performed by Srv2/cyclase-associated protein (CAP),2 which here we refer to as Srv2 (9, 10). Srv2 is expressed ubiquitously in all animal, plant, and fungal organisms and cell types examined, and it plays critical roles in cell polarity, cell motility, cytokinesis, and endocytosis (11). Early biochemical analyses suggested that Srv2 functioned as an actin-monomer sequestering protein (12-15). However, more recent studies on animal, fungal, and plant Srv2 proteins have revised this view, showing that Srv2 instead promotes the rapid recycling of cofilin from ADP-actin monomers, and it catalyzes the conversion of ADP-actin monomers back to an ATP-bound state (9, 10, 16). Thus, Srv2 functions universally (across distant species) to accelerate cofilin-mediated actin turnover.
Despite being a ubiquitous core component of the actin turnover machinery, the underlying mechanism by which Srv2 recycles actin monomers from cofilin to accelerate actin turnover has remained poorly understood. Deciphering this mechanism presents a serious challenge, because Srv2 is a multidomain protein with numerous binding partners and forms a high molecular weight complex of unknown architecture (10, 12, 17). From its N to C terminus, Srv2 is comprised of a coiled coil domain, a dimeric helical folded domain (which we refer to here as the HFD) (18-21), a first polyproline motif (P1) that interacts with profilin (22), a WH2 domain (the function of which remains uncertain), a second polyproline motif (P2) that binds to the SH3 domain of Abp1 (23), and finally a dimeric folded domain comprised entirely of β-sheets that binds to G-actin and is critical for Srv2 function in vivo (21, 24). Native Saccharomyces cerevisiae Srv2 forms a stable ∼600-kDa complex consisting of a 1:1 molar ratio of Srv2 to actin (10), and gel filtration analysis indicates that full-length Srv2 oligomerizes (9). However, the functional importance of Srv2 oligomerization has never been addressed in vitro or in vivo.
The C terminus of Srv2 binds to ADP-G-actin with high affinity (Kd ∼20 nm) and to ATP-G-actin with 100-fold lower affinity (Kd ∼2 μm) (24). It is also sufficient to accelerate nucleotide exchange on actin monomers (9). It has been proposed that these unique properties allow Srv2 to function as a middleman in recycling actin monomers, promoting a rapid handoff of monomers from cofilin to profilin. These established activities of the C terminus of Srv2 have dominated recent models for the Srv2 mechanism. However, one earlier study pointed to a functional role for the N terminus of human Srv2 (CAP1), showing that it binds to cofilin-actin complexes and helps promote actin turnover in vitro (9). Since this study, there has been no further investigation of the underlying mechanism of the N-terminal half of Srv2/CAP and, importantly, no test of its in vivo role.
Here, we have purified full-length recombinant Srv2 protein, reconstituted the 600-kDa Srv2-actin complex, and dissected its biochemical activities and genetic functions. This work reveals a critical in vivo function for the N-terminal half of Srv2 in driving cofilin-mediated actin turnover. We have pinpointed the activity to a conserved surface patch on Srv2 and shown that this surface binds cofilin-actin complexes. Furthermore, we have identified a conserved, cognate Srv2-binding site on cofilin. From these and other data, we propose that the Srv2 mechanism of promoting actin turnover involves coordinated functions of its N- and C-terminal halves, which drive sequential steps of actin monomer and cofilin recycling.
EXPERIMENTAL PROCEDURES
Yeast Strains, Cell Growth, and Plasmids—Standard methods were used for all DNA manipulations and for growth and transformation of yeast strains. To construct a TRP1-marked SRV2 integration plasmid (pSRV2+), the SRV2 coding region plus 500 bp upstream and downstream of genomic DNA was PCR-amplified and subcloned into HindIII and SacII sites of pBSSK. NotI and SpeI sites were introduced 80 bases downstream of the stop codon by site-directed mutagenesis. The TRP1 marker was PCR-amplified and subcloned into the NotI and SpeI sites. The resulting pSRV2+ plasmid was used as template for PCR-based site-directed mutagenesis to generate srv2 alleles. For purification of wild type and mutant His6-tagged full-length Srv2 proteins, open reading frames of the corresponding integration plasmids were PCR-amplified and subcloned into NcoI and NotI sites of pHAT2. A similar strategy was used to construct plasmids expressing His6-tagged Srv2-ΔCC, C-Srv2, and N-Srv2 polypeptides. All plasmids were sequenced. TRP1 marked srv2 alleles were integrated into the haploid yeast strain BGY330 (srv2Δ::HIS3). Insertion cassettes were obtained by digesting integration plasmids with SacII and then were transformed into BGY330. Transformants were selected by growth on Trp- media and loss of growth on His- media. Successful integrations were verified by PCR amplification of the SRV2 coding region from genomic DNA and diagnostic restriction analysis. The coding regions of COF1, cof1-5, and cof1-9 were subcloned into pGEX2T for expression as GST fusion proteins (pAM50, pPL32, and pPL18).
Protein Purification—Full-length N-terminally His6-tagged Srv2 proteins were expressed in Escherichia coli BL21-RP (DE3) cells. Cultures were grown to log phase at 37 °C and then induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside for 16 h at 25 °C. Each 1 liter of cells was pelleted and resuspended in 15 ml of Buffer A (50 mm phosphate buffer pH 8.0, 300 mm NaCl, 1 mm DTT) supplemented with 30 mm imidazole and a standard mixture of protease inhibitors (25). Cells were lysed by sonication, and the lysate was clarified by centrifugation at 16,000 rpm in an SA600 rotor for 15 min and applied to 300 μl of nickel-nitrilotriacetic acid-agarose resin (Qiagen, Valencia, CA). After 1.5 h of incubation at 4 °C, the resin was transferred to an empty 1-ml poly-prep chromatography column (Bio-Rad) and washed with 30 ml of Buffer A supplemented with 60 mm imidazole. Proteins were eluted with Buffer A supplemented with 250 mm imidazole in three fractions of 300 μl each. The second fraction contained ∼80% of the Srv2 protein and was further purified using a Superose 6 gel filtration column (GE Healthcare) equilibrated with Buffer B (20 mm Tris, pH 8.0, 50 mm NaCl, and 1 mm DTT). Peak fractions were concentrated 4-fold using a Centricon-10 device (Millipore, Billerica, MA), aliquoted, snap-frozen in liquid N2, and stored at -80 °C.
Rabbit skeletal muscle actin was purified as described (26). ADP-actin was prepared as described (3, 24). Yeast actin was purified as described (10). GST-Cof1, Cof1-5, and Cof1-9 were expressed in E. coli BL21(DE3) cells and purified as described for GST-Srv2 fragments (24). For actin-binding assays, Cof1, Cof1-5, and Cof1-9 were cleaved from GST and purified as described (22). Wild type and mutant N-Srv2 His6 fusion proteins used in supernatant depletion assays were purified as described (27).
Nucleotide Exchange Assays—Nucleotide exchange rate on actin monomers was determined by following incorporation of etheno-ATP. Briefly, 2 μm rabbit muscle actin in G-buffer (10 mm Tris, pH 7.5, 0.2 mm CaCl2, 0.2 mm DTT, no ATP) was mixed with Tris/NaCl buffer (20 mm Tris, pH 8.0, 50 mm NaCl) alone or proteins in Tris/NaCl buffer and added to 50 μm etheno-ATP (Molecular Probes, Eugene, OR). The reaction was monitored at 350 nm excitation and 410 nm emission at 25 °C in a fluorescence spectrophotometer for at least 600 s (Photon Technology International, Lawrenceville, NJ). SigmaPlot was used to fit the data to a hyperbolic function of the following form: f = y0 + a·x/(b + x). Exchange rates were determined from the initial, linear slopes of the curves. To study Srv2 effects on cofilin inhibition of nucleotide exchange, G-actin (2 μm final) and Cof1 (10 μm, final) were mixed and preincubated for 5 min and then added to the other reaction components.
G-actin Binding Assays—Interactions of full-length Srv2 protein with rabbit muscle ADP-G-actin and ATP-G-actin were measured by change in fluorescence of NBD-labeled actin as described (22). Because of the greatly reduced affinity of Srv2 for ATP-NBD-actin compared with ADP-NBD-actin, the ATP-NBD-actin binding curves did not plateau, as noted in previous studies (24). Therefore, we estimated the Kd value for the Srv2-ATP-actin complex assuming that the maximal increase of NBD-fluorescence is the same for ADP-NBD-actin and ATP-NBD-actin.
Phosphate Release Assays—Kinetics of Pi release during steady state turnover of F-actin was measured by Pi release using EnzChek kit (Molecular Probes). Variable concentrations of Srv2 and/or Cof1 were mixed with polymerization buffer (2 mm MgCl2, 0.5 mm ATP, 50 mm KCl), 0.2 mm 2-amino-mercap-to-7-methylpurine ribonucleoside, and 0.1 unit of purine nucleoside phosphorylase. Actin polymerization was initiated by addition of rabbit muscle actin monomers (7.7 μm final), with the exception of Fig. 5C where instead yeast actin was used. Absorbance at 360 nm was monitored at 25 °C. After actin assembly reached steady state, a constant rate of Pi production was observed, and data were collected for 15 min and analyzed to determine slope (relative rate of Pi release).
FIGURE 5.
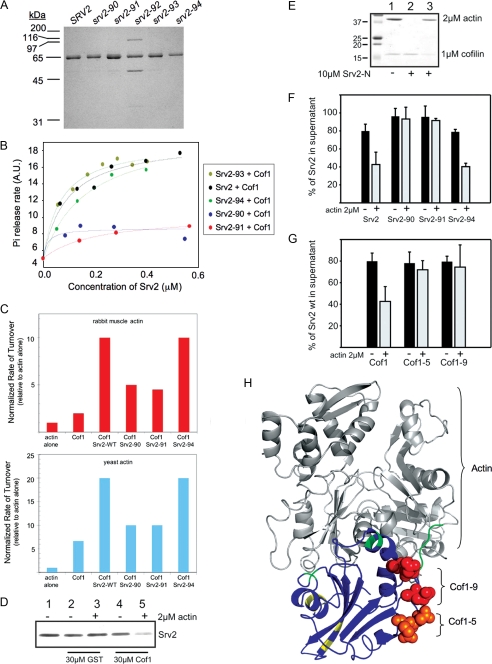
Srv2 binding to cofilin-actin complexes. A, Coomassie-stained gel of purified wild type and mutant full-length Srv2 proteins. B, concentration-dependent effects of wild type and mutant Srv2 proteins on rate of turnover of F-actin in the presence of Cof1 measured in Pi release assays. C, comparison of the effects of wild type and mutant Srv2 polypeptides and/or Cof1 on rate of actin turnover measured by Pi release for rabbit muscle actin (upper panel) versus yeast actin (lower panel). D, supernatant depletion pulldown assay measuring binding of full-length wild type Srv2 to GST-Cof1 on beads in the presence (+) and absence (-) of ADP-G-actin. Supernatants were analyzed on Coomassie-stained gels to compare levels of unbound Srv2. Lane 1, Srv2 loading control. Lanes 2 and 3, control reactions with GST alone on beads. Lanes 4 and 5, reactions with GST-Cof1 on beads. E, supernatant depletion pulldown assay using GST-N-Srv2 beads and soluble Cof1. Lane 1, actin and Cof1 loading controls. Lane 2, GST-N-Srv2 and Cof1 without actin. Lane 3, GST-N-Srv2 with actin and Cof1. F, supernatant depletion pulldown assays comparing binding of wild type and mutant Srv2 proteins to GST-Cof1 beads in the presence (+) and absence (-) of ADP-G-actin. G, supernatant depletion pulldown assays comparing binding of Srv2 to wild type and mutant GST-Cof1 beads in the presence (+) and absence (-) of ADP-G-actin. Data in E and F were averaged from four independent experiments; standard deviations shown as error bars. H, residues mutated in cof1-5 (orange) and cof1-9 (red) highlighted on the modeled structure of S. cerevisiae cofilin (Protein Data Bank code 1COF) bound to actin (44). The two major actin binding surfaces on Cof1 defined by mutagenesis studies (35) are shaded green and yellow. Note the Srv2-binding surface is separate from the actin-binding interface. A.U., arbitrary units.
Microscopy—To visualize actin organization, yeast cells were grown to log phase, fixed in 5% formaldehyde, and stained with Alexa488-phalloidin (Invitrogen). To assess co-localization of Srv2 and F-actin, fixed cells were processed as described (28) and probed with Alexa488-phalloidin and anti-Srv2 primary antibodies. Images were acquired on a Zeiss E600 microscope (Thornwood, NY) equipped with a Hammamatsu Orca ER CCD camera (Bridgewater, NJ) running Openlab software (Improvision Inc., Waltham, MA).
Immunoblotting—Yeast whole cell lysates were prepared (29), and Srv2 antibodies were affinity-purified (30) as described. Immunoblots were probed with 1:4000 anti-yeast C-Srv2 chicken polyclonal (24) or 1:20,000 anti-yeast tubulin rabbit polyclonal antibody (a gift from Frank Solomon, Massachusetts Institute of Technology).
Supernatant Depletion Pulldown Assays—Assays were performed as described (24). In Fig. 5, C, E, and F, GST-Cof1, GST-Cof1-5, GST-Cof1-9, and GST alone (30 μm final) were bound to glutathione-Sepharose 4B beads (Amersham Biosciences) and mixed with 2 μm full-length Srv2, Srv2-90, Srv2-91, or Srv2-94, in the presence and absence of 2 μm ADP-G-actin. In Fig. 5D, GST-N-Srv2 was immobilized on beads and incubated with soluble 1 μm Cof1 and/or 2 μm G-actin. Reactions were carried out in 5 mm Tris-HCl, pH 8.0, 0.2 mm CaCl2, 0.4 mm DTT, 0.2 mm ADP, 100 mm KCl as described (22). Data were analyzed using SigmaPlot 9.0 software (Systat Software).
Hydrodynamic Analysis of Srv2 Complexes—To estimate the molecular weights of complexes formed by purified Srv2 polypeptides, we measured Stokes radius by gel filtration on a Superose 6 column and sedimentation coefficient (S value) by sucrose gradient fractionation (10). From these values, we estimated the native molecular range of Srv2 using the following formula: M = (6πηNas)/(1 - νρ) (31), where M, molecular weight; η (buffer viscosity) = 1.002 × 10-2g/(cm·s); N, Avogadro's number; a, Stokes radius; s, sedimentation coefficient; ν (partial specific volume of an average protein) = 0.725 cm3/g; ρ (density of water) = 0.998g/cm3. In addition, sedimentation and diffusion coefficients were determined for full-length Srv2 and C-Srv2 by analytical ultracentrifugation at 20 °C using an Optima XL-I and An-50 Ti rotor (Beckman Coulter, Fullerton, CA). Cells with standard Epon two-channel centerpiece and sapphire windows were used. Absorbance (A280) scans were collected at 2-min intervals during sedimentation at 40,000 rpm. Partial specific volume of the protein, solvent density, and solvent viscosity were calculated from standard tables using the program SEDNTERP, version 1.08 (32). The resulting scans were analyzed using the continuous distribution (c(s)) analysis module in the program SEDFIT version 9.3 (33). Sedimentation coefficient increments of 100 were used, and the frictional coefficient was allowed to float during fitting. The weight average sedimentation coefficient was obtained by integrating the range of sedimentation coefficients in which the peaks were present. Complexes formed in vivo by full-length Srv2 and Srv2-ΔCC were analyzed by sedimentation velocity sucrose gradient fractionation of cell lysates and Western blotting with Srv2 antibodies (10).
Miscellaneous—Concentrations of purified Srv2, Cof1, and actin were determined by spectrophotometry using extinction coefficients of ε280 = 50,100 m-1 cm-1 for Srv2, ε280 = 15,930 m-1 cm-1 for Cof1, and ε290 = 26,600 m-1 cm-1 for actin. Concentrations of mutant Srv2 proteins were determined by comparing band intensities on Coomassie-stained gels against a standard curve of wild type Srv2 protein.
RESULTS
Reconstitution of Srv2-Actin Complex from Purified Components—Previously, we isolated native Srv2-actin complex from yeast cells and estimated its molecular mass at ∼600 kDa (10). However, using this purification approach we could isolate only very small quantities, precluding further biophysical analysis to more accurately define its oligomerization state and limiting efforts to dissect its structure and function. To bypass these obstacles, we developed a purification strategy for isolating large quantities of functional His6-tagged full-length S. cerevisiae Srv2 from E. coli (see “Experimental Procedures”).
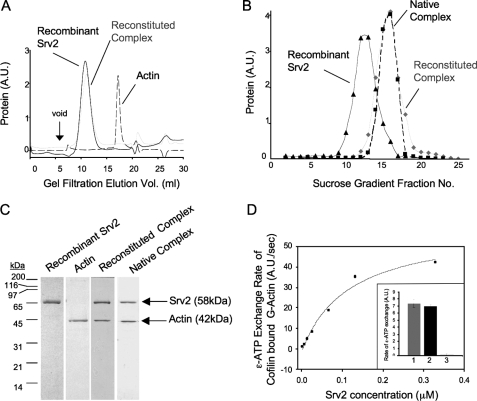
Recombinant full-length Srv2 alone and bound to G-actin was monodispersed by gel filtration (Fig. 1A) and sedimentation analysis (Fig. 1B). The Srv2-actin complex had a larger sedimentation coefficient than Srv2 alone, and the reconstituted complex showed a migration profile similar to native yeast Srv2-actin complex (Fig. 1B). Fig. 1C shows peak fractions from gel filtration analysis. Together, these data indicate that recombinant Srv2 stably associates with G-actin in a complex with similar hydrodynamic properties to native Srv2-actin complex (10). The reconstituted complex also promoted nucleotide exchange on cofilin-bound G-actin in a concentration-dependent manner (Fig. 1D), as described for native Srv2-actin complex (10), demonstrating it is active.
FIGURE 1.
Reconstitution of the Srv2-actin complex from purified proteins. A, gel filtration analysis of purified full-length recombinant Srv2, actin, and reconstituted Srv2-actin complex. B, sucrose gradient sedimentation analysis of recombinant full-length Srv2, reconstituted Srv2-actin complex, and native Srv2-actin complex. C, Coomassie-stained gel of peak fractions from gel filtration compared with native Srv2-actin complex isolated from yeast. D, recombinant Srv2 has concentration-dependent effects in relieving cofilin inhibition of ε-ATP nucleotide exchange on actin monomers. Inset, ε-ATP exchange rates for reactions containing 1 μm G-actin, 2 μm Cof1, and either 53 nm recombinant Srv2 (bar 1), 53 nm native Srv2 (bar 2), or no Srv2 (bar 3). A.U., arbitrary units.
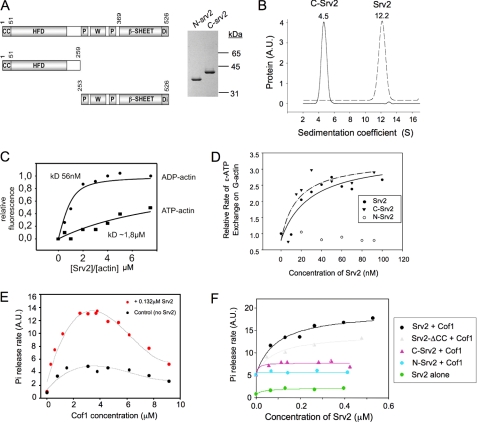
Purified Full-length Srv2 Forms Stable Oligomers—Next, we addressed Srv2 oligomerization. Previously, we estimated the mass of native S. cerevisiae Srv2-actin complex to be 587-645 kDa (10). Given the masses of Srv2 (57.5 kDa) and actin (41.7 kDa) and their apparent 1:1 molar ratio in the complex, this had suggested that an intact complex might consist of 5-6 molecules each of Srv2 and actin. Here, purification of recombinant Srv2 provided the quantities of protein required for an independent analysis of the oligomerization state by analytical ultracentrifugation (Fig. 2B). Purified Srv2 (without actin) behaved as a homogeneous species with a sedimentation coefficient of 12.2 S (Fig. 2B). This yielded an experimental molecular mass of 315 ± 30 kDa, which falls within the predicted range for an Srv2 oligomer comprised of 5-6 subunits (288-345 kDa). In contrast, the C-terminal half of Srv2 (C-Srv2) had a sedimentation coefficient of 4.5 S (Fig. 2B). This yielded an experimental mass of 63 ± 7 kDa, close to the predicted 60.6 kDa mass of a C-Srv2 dimer. These results are consistent with our earlier analyses on the native Srv2-actin complex, and demonstrate that Srv2 oligomerization is independent of actin association and requires the N-terminal half of Srv2.
FIGURE 2.
Domain requirements for Srv2 oligomerization, G-actin binding, catalysis of actin nucleotide exchange, and acceleration of actin turnover. A, schematic of Srv2 domain organization and Coomassie-stained gel of purified Srv2 fragments. CC, coiled coil domain; HFD, helical folded domain; P, polyproline-rich motifs; W, WASp-homology 2 (WH2) domain; Di, dimerization motif. B, analytical ultracentrifugation sedimentation coefficient distributions for purified full-length Srv2 (dotted line) and C-Srv2 (solid line). Sedimentation coefficients are shown above peaks. C, concentration-dependent binding of full-length Srv2 to 0.2 μm ATP-bound (squares) or ADP-bound (circles) NBD-labeled actin monomers. Values on the x axis are molar ratios of Srv2 to actin. Binding constants were calculated as described (24). D, concentration-dependent effects of full-length Srv2, N-Srv2, and C-Srv2 on rate of ε-ATP exchange on actin monomers. E, concentration-dependent effects of cofilin on rate of steady state turnover of F-actin (7.7 μm) measured by Pi release assay in the absence (black circles) and presence (red circles) of 0.132 μm full-length Srv2. F, concentration-dependent effects of different Srv2 proteins on rate of turnover of F-actin in the presence of a fixed concentration (3.8 μm) of cofilin. A.U., arbitrary units.
N-terminal Half of Srv2 Makes a Critical Contribution to Actin Turnover—To dissect the activities of the Srv2 complex, we compared purified full-length Srv2, N-Srv2, and C-Srv2 in assays for G-actin binding, effects on rate of nucleotide exchange on actin monomers, and acceleration of cofilin-dependent steady state turnover of F-actin. Previously, we reported that C-Srv2 binds to G-actin, whereas N-Srv2 does not (24). Here we found that full-length Srv2 binds to ADP-G-actin with high affinity (Kd = 56 nm) and ATP-G-actin with low affinity (Kd ∼2 μm) (Fig. 2C), very similar to the properties of C-Srv2. Full-length Srv2 and C-Srv2 also had similar concentration-dependent effects on rate of actin nucleotide exchange (Fig. 2D), further suggesting that their binding affinities for G-actin are similar. N-Srv2 had no appreciable effect on actin nucleotide exchange rate, consistent with its inability to bind actin (9). Taken together, these data indicate that most or all of the G-actin binding activity of Srv2 resides in its C-terminal half.
Next, we compared the effects of Srv2 constructs in an in vitro assay that measures rate of cofilin-mediated F-actin turnover by monitoring inorganic phosphate (Pi) release. This represents a more complete assay that includes multiple steps in actin turnover and is thus more reflective of cellular conditions. The assay contains both monomeric and polymeric actin (in a steady state equilibrium) and measures rate of flux of actin subunits between the monomer and polymer populations. In these reactions, the rate of ATP hydrolysis and Pi release (which occurs exclusively in polymer) is directly proportional to the rate of actin filament turnover.
As expected, cofilin increased the rate of actin turnover in a concentration-dependent manner, and at 3.8 μm cofilin reached a maximum effect of four times faster turnover than F-actin alone (Fig. 2E). This concentration of cofilin was used in all subsequent assays. Full-length Srv2 had no effect on rate of turnover in the absence of cofilin (Fig. 2F, green curve), but in the presence of 3.8 μm cofilin caused a concentration-dependent increase in turnover rate (black curve). The maximal rate of turnover achieved by combining cofilin and Srv2 was 14 times faster than F-actin alone. In contrast, C-Srv2 had only a minimal stimulatory effect on turnover in the presence of cofilin (Fig. 2F, light purple curve), and N-Srv2 had no detectable effect (light blue curve). Thus, both halves of Srv2 protein are required for its ability to accelerate cofilin-mediated actin turnover in vitro.
N-terminal HFD Domain Is Critical for SRV2 Function in Vivo—To better understand the essential contribution of the N-terminal half of Srv2 to actin turnover, we performed a mutational dissection of residues 50-259, which adopts a six helix bundled structure and dimerizes in antiparallel fashion (Fig. 3D) (18, 19). As mentioned above, we refer to this as the “helical folded domain” (HFD).
FIGURE 3.
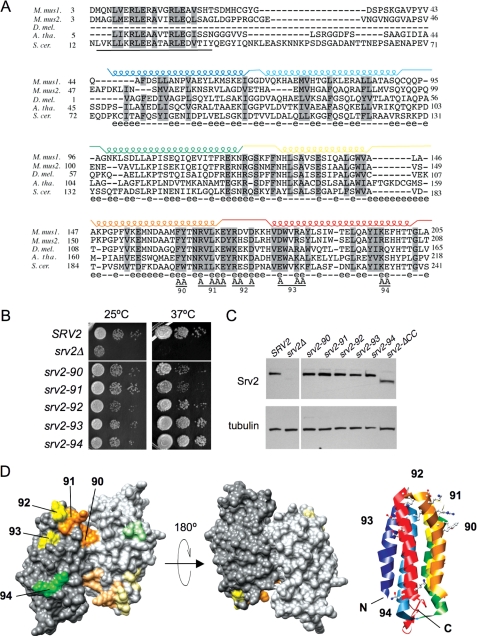
Mutational analysis of the Srv2 HFD domain. A, alignment of N-terminal sequences for diverse Srv2/CAP homologues using ClustalW. M. mus1, mouse CAP1; M. mus2, mouse CAP2; D. mel., Drosophila melanogaster CAP; A. tha, Arabidopsis thaliana CAP; and S. cer., S. cerevisiae Srv2. Residues 73-241 in S. cerevisiae Srv2 form the HFD domain (18). Each helix is indicated above the primary sequence and color-coded. Solvent-exposed residues are designated below the primary sequence (e indicates solvent-exposed; - indicates for solvent-inaccessible). Below the alignment, the predicted coiled coil domain (residues 14-34) is underlined, and residues changed to alanine are marked A for each srv2 allele (numbered 90-94). B, SRV2 and srv2 mutant strains were grown to log phase, serially diluted, and plated on YPD plates at 25 and 37 °C to compare cell growth. C, immunoblot of whole cell extracts from SRV2 and srv2 mutant strains probed with anti-Srv2 antibodies and tubulin antibodies as a loading control. D, srv2 mutations modeled on a rendered view of the crystal structure of the dimeric, anti-parallel HFD from Dictyostelium Srv2/CAP (Protein Data Bank code 1S0P). Residues mutated in each allele are color-coded by the relative severity of their cell growth phenotype (orange, severe; yellow, mild; green, pseudo-wild type). Shading is lighter on one Srv2 molecule in the dimer. A ribbon structure is also shown for one molecule in the dimer, with helices color-coded to match those in A. The ribbon view has the same orientation as the rendered structure on the far left.
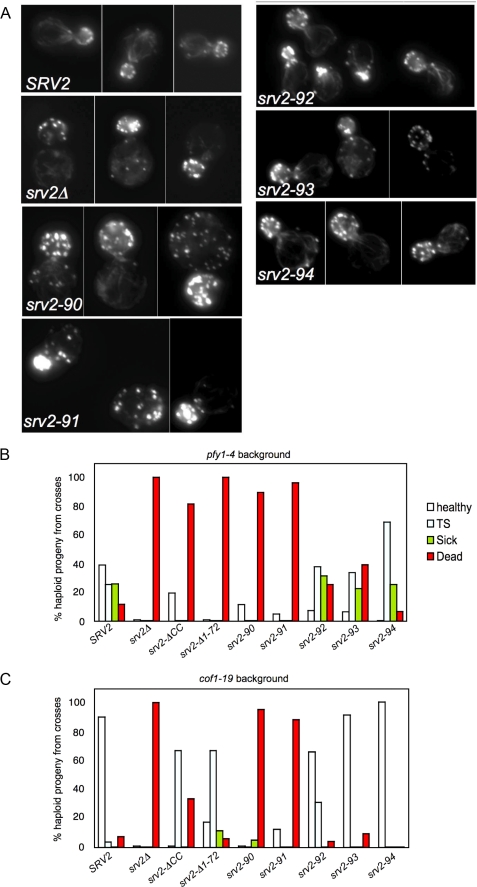
Using site-directed mutagenesis, we introduced five srv2 alleles, each containing 2-3 alanine substitutions at conserved solvent-exposed residues (srv2-90 to srv2-94, Fig. 3A). Each allele was integrated, replacing wild type SRV2, and analyzed for defects in cell growth (Fig. 3B). Two alleles (srv2-90 and srv2-91) had impaired cell growth at 37 °C, whereas the remaining three alleles (srv2-92, srv2-93, and srv2-94) had no obvious defects in growth compared with the wild type SRV2 strain. Immunoblotting confirmed that the mutant Srv2 proteins are expressed at normal levels (Fig. 3C), and immunofluorescence microscopy verified that each mutant protein localized to patch-like structures that co-stained with actin similar to wild type Srv2 protein (data not shown). The positions of the surface residues mutated in each srv2 allele are modeled on the HFD crystal structure, with alleles color-coded by severity of growth phenotype (Fig. 3D).
We also compared the srv2 alleles for defects in cell morphology and actin organization (Fig. 4A). The two alleles with obvious defects in cell growth (srv2-90 and srv2-91) had abnormally large and rounded cell morphologies, diminished actin cable staining, and depolarization of actin patches. The remaining three alleles that showed no obvious defects in cell growth (srv2-92, srv2-93, and srv2-94) were pseudo-wild type for morphology and actin organization.
FIGURE 4.
Cellular actin organization and genetic interactions of srv2 mutants. A, wild type SRV2 and srv2 mutant strains were grown to log phase, fixed, and stained with Alexa488-phalloidin to visualize filamentous actin. B and C, haploid yeast strains carrying integrated srv2 alleles were crossed to haploid pfy1-4 (B) and cof1-19 (C) strains. Diploids were sporulated and tetrads dissected (minimum 20 tetrads, 80 spores), and all haploid progeny were compared for cell growth on YPD plates at 25, 30, 34, and 37 °C. For each cross, we determined the percentage of haploid progeny that grew normally at all temperatures compared with a wild type strain (healthy), exhibited impaired growth at 37 °C (TS), exhibited impaired growth at all temperatures (sick), or were dead at 25 °C (dead).
As an independent test of in vivo function, we crossed each of the srv2 alleles to pfy1-4 and cof1-19 (Fig. 4, B and C), mutations that are synthetic lethal with srv2Δ (10, 34). Single and double mutant progeny were compared for cell growth defects. The pfy1-4 background is more sensitive than cof1-19 and therefore causes more severe defects in cell growth when combined with partial loss of function srv2 alleles. srv2-90 and srv2-91 displayed strong synthetic lethality in both backgrounds; srv2-92, srv2-93, and srv2-94 were pseudo-wild type. The severity of srv2 genetic interactions with pfy1-4 and cof1-19 correlated closely with the severity of their cell growth, morphology, and actin organization defects.
Taken together, the data in Figs. 3 and 4 pinpoint a conserved surface on one face of the dimeric HFD as being essential for SRV2 function in vivo (Fig. 3D). This also means that there are two functional sites per HFD dimer, which are positioned at opposite ends of its conserved face (Fig. 3D, orange surfaces). We note however that our mutagenesis of this domain was not comprehensive, and thus leaves open the possibility that additional surfaces might contribute to function.
Loss of Actin Turnover Activity for srv2 Mutants Correlates with Loss of in Vivo Function—To dissect the mechanism underlying the critical in vivo contributions made by the HFD, we purified from E. coli full-length Srv2 proteins containing the HFD point mutations (Fig. 5A). Urea denaturation experiments verified that the mutant proteins were properly folded (data not shown), and gel filtration analysis showed that each mutant protein was monodispersed and had an oligomerization state similar to wild type Srv2 (Table 1). We then compared wild type and mutant Srv2 proteins for concentration-dependent effects on cofilin-mediated actin turnover in the Pi release assay (Fig. 5B). The two mutants with strong defects in vivo (Srv2-90 and Srv2-91) were severely compromised in promoting actin turnover. Furthermore, their activity profiles were similar to C-Srv2 alone, which suggests that these mutations (srv2-90 and srv2-91) abolish the functional contributions of the N-terminal half of Srv2. In contrast, two mutants that were pseudo-wild type in vivo (Srv2-93 and Srv2-94) had similar activities to wild type Srv2 protein in the actin turnover assay. Furthermore, wild type and mutant Srv2 polypeptides showed similar effects on Cof1-mediated actin turnover in assays using rabbit muscle actin versus yeast actin (Fig. 5C). From these data, we conclude that there is a strong correlation between mutant loss of function in vivo and loss of actin turnover activity in vitro.
TABLE 1.
Hydrodynamic properties of purified Srv2 polypeptides
| Construct name | Residues | Molecular massa | Sedimentation coefficientb | Stokes radiusc | Native massd | Oligomerization statee |
|---|---|---|---|---|---|---|
| kDa | S | nm | kDa | |||
| Srv2:actin | 1-526 | 100.8 | 16.1 | 9.9 | 660 | 6.5 |
| C-Srv2 | 253-526 | 30.3 | 2.4 | 5.8 | 57 | 1.9 |
| Srv2ΔCC | 51-526 | 53.0 | 3.4 | 6.7 | 93 | 1.8 |
| Srv2-90 | 1-526 | 58.6 | 9.8 | |||
| Srv2-91 | 1-526 | 58.5 | 9.7 | |||
| Srv2-92 | 1-526 | 58.6 | 9.8 | |||
| Srv2-93 | 1-526 | 58.7 | 9.8 | |||
| Srv2-94 | 1-526 | 58.7 | 9.9 |
Molecular mass of a monomeric polypeptide is shown, inclusive of its His6 tag.
Data were determined by sucrose gradient sedimentation velocity analysis.
Data were determined by gel filtration.
Data were calculated from Stokes radius and S value (see “Experimental Procedures”).
Ratio of native molecular mass to monomeric molecular mass is shown.
Srv2 HFD Domain Binds Cofilin-Actin Complexes—One earlier report showed that the N-terminal half of human CAP1 binds to cofilin-actin complexes (9). This led us to test whether the interaction is conserved for yeast Srv2, and whether the conserved functional surface we identified on the Srv2 HFD domain might be the binding site for cofilin-actin.
To test this model, we assessed the ability of full-length Srv2 to interact with cofilin-actin (immobilized on beads) in a supernatant depletion assay (Fig. 5D). In the presence of ADP-G-actin, Srv2 was depleted from the supernatant by GST-cofilin beads but not by control GST beads. However, in the absence of G-actin, Srv2 did not display detectable binding to GST-cofilin. Similar results were obtained when this assay was carried out using GST-N-Srv2 instead of full-length Srv2 (Fig. 5E). These data confirm that the N-terminal half of yeast Srv2 binds to cofilin-actin, consistent with the observations of human CAP from Moriyama and Yahara (9). Thus, the interaction is conserved across distant species lines in the Srv2/CAP family.
We next used this assay to compare the abilities of wild type Srv2 and three mutant Srv2 proteins (Srv2-90, Srv2-91, and Srv2-94) to bind cofilin-actin (Fig. 5F). Wild type Srv2 and pseudo-wild type Srv2-94 showed a similar ability to bind cofilin/actin, whereas Srv2-90 and Srv2-91 were defective. These data suggest that the conserved residues on the HFD domain of Srv2 that are critical for its in vivo functions and in vitro actin turnover activity are also crucial for binding cofilin-actin.
Identification of an Srv2-binding Site on Cofilin—We next attempted to identify the cognate Srv2-binding surface on cofilin. Cofilin is a small (18 kDa) globular protein with a well defined actin-binding surface. A systematic mutational analysis of conserved charged surface residues on yeast cofilin previously identified sites important for its function in vivo (35). Most of the cof1 alleles that showed defects in cell growth and actin organization in vivo were found to be impaired in binding G-actin and/or F-actin. However, two alleles with strong growth defects in vivo, cof1-5 (temperature-sensitive) and cof1-9 (lethal), had normal biochemical interactions with G-actin and F-actin. This led the authors to conclude, “It is possible that the phenotypes of these two mutants result from a currently unrecognized activity of cofilin” (35). We purified Cof1-5 and Cof1-9 and compared them with wild type Cof1 for the ability to bind N-Srv2 in the presence and absence of ADP-G-actin (Fig. 5G). These data show that Cof1-5 and Cof1-9, in complex with actin, are defective in binding Srv2.
Together, the data demonstrate that the conserved surface on cofilin mutated in cof1-5 and cof1-9 is critical for binding interactions with the HFD domain of Srv2. Importantly, this Srv2-binding surface on cofilin is nonoverlapping with its actin-binding surface (Fig. 5H).
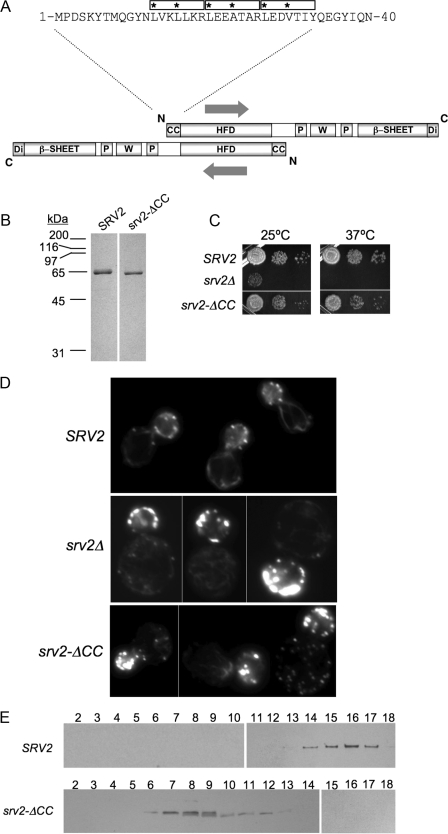
Srv2 Oligomerization Is Not Critical for but Optimizes Actin Turnover Function—We next addressed the importance of Srv2 oligomerization for its actin turnover function. In all species examined, Srv2 oligomerizes to form a high molecular weight complex, yet the functional significance of oligomerization has not been addressed. Our hydrodynamic comparison of full-length Srv2 and C-Srv2 demonstrated that the N-terminal half of Srv2 is required for higher order oligomerization (Fig. 2B). The predicted coiled coil domain found at the N terminus of Srv2 (Fig. 6A) represented a strong candidate for mediating oligomerization, in particular because it is positioned at opposite ends of the dimeric anti-parallel HFD domain. Thus, the CC domain is unlikely to mediate intra-dimer associations and more likely to mediate inter-dimer associations leading to the formation of higher order Srv2 oligomers.
FIGURE 6.
Biochemical and in vivo analysis of a coiled coil domain Srv2 mutant. A, schematic of an anti-parallel Srv2 dimer (same domain abbreviations as in Fig. 2A). The HFD is an anti-parallel dimer, which positions the two CC domains on opposite ends of the dimer such that they are unlikely to form an intra-dimer association, and instead they may be used to form inter-dimer associations leading to oligomerization into higher order complexes. Shown above the schematic are residues 1-40 of S. cerevisiae Srv2, highlighting three predicted heptad repeats in its coiled coil domain (boxed, asterisks at positions 1 and 4 of each heptad). B, Coomassie-stained gels of purified full-length Srv2 and Srv2-ΔCC. C, SRV2, srv2Δ, and srv2-ΔCC strains were grown to log phase, serially diluted, plated on YPD plates, and compared for growth at 25 and 37 °C. D, same strains were examined for actin organization by staining fixed cells with Alexa488-phalloidin. E, sedimentation velocity profiles for full-length Srv2 and Srv2-ΔCC. Lysates from SRV2 and srv2-ΔCC strains were fractionated on sucrose gradients. Blotted fractions were probed with anti-Srv2 antibodies.
To test this model, we purified a truncation mutant lacking the CC domain (Fig. 6B, Srv2-ΔCC). The oligomerization state of Srv2-ΔCC was assessed by gel filtration and sucrose gradient fractionation (Table 1); the data strongly indicate that it dimerizes. This demonstrates that the CC domain of Srv2 is required for higher order oligomerization. Over a range of concentrations, purified Srv2-ΔCC promoted cofilin-mediated actin turnover in the Pi release assay (Fig. 2F, gray curve), albeit somewhat less efficiently than full-length Srv2. We conclude that oligomerization is not critical for the actin turnover effects of Srv2 in vitro, but it helps optimize the activity.
To address the in vivo significance of Srv2 oligomerization, we integrated the srv2-ΔCC allele. We found that it caused partial defects in cell growth (Fig. 6C) and in cell morphology and actin organization (Fig. 6D). srv2-ΔCC cells were slightly larger and more rounded than wild type cells and showed decreased actin cable staining. These defects were much less pronounced than the defects of srv2Δ cells, indicating that srv2-ΔCC causes only a partial loss of function. This view is supported further by genetic crosses between srv2-ΔCC and pyf1-4 (Fig. 4B) and cof1-19 (Fig. 4C). These data show that srv2-ΔCC is more defective than srv2-92 but less defective than srv2-90 or srv2-91. A second truncation allele that we generated, with a more extensive N-terminal deletion (srv2Δ1-72), had cell defects (not shown) and genetic interactions similar to those of srv2-ΔCC (Fig. 4, B and C). Sucrose gradient fractionation experiments verified that Srv2-ΔCC disrupted higher order Srv2 oligomerization in vivo (Fig. 6E). Taken together, these data suggest that deletion of the CC domain leads to a partial disruption of Srv2 function in vivo, consistent with the partial defects in turnover activity observed for purified Srv2-ΔCC.
DISCUSSION
The regulation of actin turnover is a conserved process, governed centrally by actin depolymerizing factor/cofilin acting in concert with several other highly conserved actin-binding proteins, including profilin, Aip1, coronin, and Srv2/CAP (10, 36-39). Srv2 plays a central and conserved role in driving in vivo actin turnover by catalyzing the recycling of cofilin and actin monomers, which fuels cell movement, cell division, and cell morphogenesis.
Efforts to unravel the Srv2 mechanism of action have been hindered by the large size of the native complex it forms and by the multiplicity of Srv2 domains and interactions. Here we have made key advances in elucidating the Srv2 mechanism as follows: 1) by reconstituting the 600-kDa Srv2-actin complex from purified components; 2) by identifying a critical (and conserved) functional surface on its N-terminal HFD domain that binds cofilin-actin; 3) by identifying a cognate and conserved Srv2-binding site on cofilin; 4) by demonstrating that interactions with cofilin-actin are required for Srv2 activity and in vivo function in promoting actin turnover; 5) by showing that the coiled coil domain of Srv2 is required for higher order oligomerization; and 6) by demonstrating that oligomerization contributes to Srv2 activity in vitro and in vivo. Based on these findings, we propose a revised mechanism for how Srv2 catalyzes cofilin-dependent actin turnover. This model has important implications for understanding actin turnover in all eukaryotes, because Srv2/CAP and cofilin are ubiquitous, and the functional sites on Srv2 and cofilin that we identify here are conserved.
Revised Model for the Srv2 Mechanism of Action—Models to date for Srv2 function have focused almost exclusively on the role of its C-terminal actin-binding domain and have depicted this domain as being sufficient for driving cofilin-mediated actin turnover. However, a pioneering study by Moriyama and Yahara (9) on human Srv2/CAP found that both halves of the protein were required for promoting cofilin-mediated actin turnover in vitro. Our data for yeast Srv2 are in close agreement. We found that C-Srv2 had activities similar to full-length Srv2 in promoting nucleotide exchange on ADP-actin monomers, whereas both halves (N-Srv2 and C-Srv2) were required for accelerating cofilin-mediated actin turnover in the Pi release assay. The latter assay more closely mimics in vivo conditions for actin turnover, because it depends equally on filament disassembly, monomer processing, and growth of filaments. Furthermore, the in vitro data from this assay are highly consistent with our in vivo observations showing that the HFD domain of Srv2 makes a critical contribution to SRV2 cellular function. Thus, our data closely agree with those of Moriyama and Yahara (9) and together with their data indicate that the activity of N-Srv2 is conserved from yeast to humans. Furthermore, we have extended the analyses of this domain by identifying the binding site for cofilin-actin, and by using point mutations at this site to probe the importance of these interactions for Srv2 function in vivo.
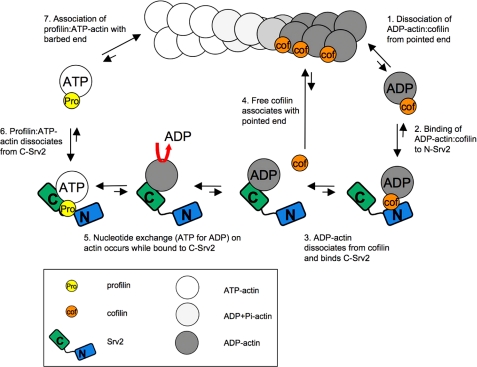
Incorporating our new findings with data from previous studies, we have built a substantially revised model for the Srv2 mechanism (Fig. 7). The steps in our new model are as follows. 1) Cofilin-bound ADP-actin monomers dissociate from the pointed end of a filament. 2) Free cofilin-ADP-G-actin complexes bind to N-Srv2. Preferred binding of cofilin-ADP-G-actin complexes to N-Srv2, as opposed to C-Srv2, is consistent with competition between cofilin and C-Srv2 for binding ADP-G-actin (24) and observations that N-Srv2 does not compete with cofilin for G-actin binding (data not shown). 3) Cofilin dissociates from ADP-G-actin (KD ∼100 nm), which enables C-Srv2 to bind ADP-G-actin with high affinity (KD ∼20 nm). Free cofilin has no detectable affinity for N-Srv2, so it freely dissociates from the complex. 4) Free cofilin re-associates with ADP-rich regions of actin filaments to promote further severing and depolymerization. 5) C-Srv2 binding to ADP-G-actin induces rapid exchange of nucleotide (ATP for ADP). This step may also be facilitated by profilin, which binds to the first proline-rich motif of Srv2 and genetically contributes to Srv2 function in actin turnover (22). It remains to be determined whether profilin interactions accelerate C-Srv2-catalyzed nucleotide exchange on actin monomers and/or catalyze dissociation of ATP-G-actin from C-Srv2. 6) ATP-G-actin dissociates from C-Srv2, as a result of 100-fold reduced binding affinity of C-Srv2 for actin in the ATP-bound state (KD ∼2 μm), and associates with profilin (24). 7) Profilin-bound ATP-G-actin readily associates with the barbed end of a growing filament.
FIGURE 7.
Model for Srv2 mechanism in actin turnover. In sequential steps, the activities of the N- and C-terminal halves of Srv2 (shaded blue and green, respectively) are coordinated to accelerate conversion of ADP-actin monomers to ATP-actin monomers and recycle cofilin for new rounds of filament severing (see “Discussion” for details).
By this mechanism, we propose that the two halves of Srv2 act sequentially to drive distinct steps in actin and cofilin recycling. We also considered the thermodynamic favorability of each step, depicted with bidirectional arrows. Only steps 1 and 6 are predicted to be energetically unfavorable. We speculate that these steps move forward rapidly because of the more favorable subsequent steps driving the reaction. Importantly, these effects of Srv2 also appear to be catalytic, occurring at low stoichiometries of Srv2 to cofilin-actin, suggesting that Srv2 may be able to promote actin turnover even when its cellular expression levels are low.
Roles of Srv2 and Profilin in Promoting Nucleotide Exchange on Actin Monomers—Our model above emphasizes the central role of Srv2 in promoting nucleotide exchange on monomeric actin. Although this role is typically assigned to profilin, it can be argued that there is stronger evidence for Srv2 holding this function. Specifically, the role of profilin in promoting nucleotide exchange on cofilin-bound ADP-G-actin (rather than ATP-G-actin) remains in question. Profilin has a strong binding preference for ATP-G-actin over ADP-G-actin (40), and as such does not effectively compete with cofilin for binding ADP-G-actin. Indeed, direct tests show that profilin is ineffective in catalyzing nucleotide exchange on cofilin-bound ADP-G-actin (10). By comparison, Srv2 strongly catalyzes nucleotide exchange on cofilin-bound ADP-G-actin. This effect can be attributed to the 100-fold stronger binding affinity of C-Srv2 for ADP-G-actin compared with ATP-G-actin, the opposite of the binding preference of profilin (24), and the ability of N-Srv2 to bind cofilin-actin, which may promote cofilin displacement. Furthermore, this function appears to be widely conserved in the Srv2/CAP protein family, as our results showing that yeast C-Srv2 promotes nucleotide exchange on G-actin (Fig. 2D) agree with the activities of plant and human Srv2/CAP (9, 16). Together, these observations suggest that the widely accepted role of profilin in catalyzing nucleotide exchange on actin monomers under physiological conditions (i.e. on cofilin-bound ADP-G-actin) requires reevaluation.
Functional Role(s) of Srv2 Oligomerization—One of the most intriguing properties of Srv2/CAP is its ability to homo-oligomerize into a very large multimeric complex. Until now the functional importance of oligomerization has not been tested. We report that oligomerization requires the coiled coil domain of Srv2. Constructs lacking this domain (C-Srv2 and Srv2-ΔCC) were dimeric (Fig. 2B and Table 1), presumably maintained by dimerization of the HFD domain (18) and/or dimerization of the C-terminal actin-binding domain (21). We also observed that full-length Srv2 forms high order oligomers both in the presence and absence of G-actin, and that oligomerization is not critical for but optimizes actin turnover activity and cellular functions. Therefore, it is possible that oligomerization has evolved to fine-tune Srv2 function in actin turnover. Alternatively, oligomerization may be more relevant to the genetically separate function of Srv2 in RAS-mediated cAMP signaling (41-43). This latter function depends on an association between the coiled coil domain of Srv2 and adenylyl cyclase. Thus, Srv2 oligomerization could play a more fundamental role in facilitating these interactions, especially given that adenylyl cyclase itself hexamerizes. Determination of the structure of the Srv2 complex will reveal how the different functional domains of Srv2 are spatially arranged in the complex and should clarify the Srv2 mechanism in both actin turnover and signal transduction.
Acknowledgments
We are grateful to J. Gelles, A. G. DuPage, A. Rodal, and E. Yu for intellectual and technical advice and editing of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM63691 (to B. G.). This work was also supported by grants from the Academy of Finland and Sigrid Juselius Foundation (to P. L.) and from the American Cancer Society.
Footnotes
The abbreviations used are: CAP, cyclase-associated protein; HFD, helical folded domain; DTT, dithiothreitol; GST, glutathione S-transferase; CC, coiled coil; NBD, 12-(N-methyl-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)).
References
- 1.Pollard, T. D. (2007) Annu. Rev. Biophys. Biomol. Struct. 36 451-477 [DOI] [PubMed] [Google Scholar]
- 2.Goode, B. L., and Eck, M. J. (2007) Annu. Rev. Biochem. 76 593-627 [DOI] [PubMed] [Google Scholar]
- 3.Pollard, T. D. (1986) J. Cell Biol. 103 2747-2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mabuchi, I. (1983) J. Cell Biol. 97 1612-1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maciver, S. K., Zot, H. G., and Pollard, T. D. (1991) J. Cell Biol. 115 1611-1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishida, E., Maekawa, S., and Sakai, H. (1984) Biochemistry 23 5307-5313 [DOI] [PubMed] [Google Scholar]
- 7.Carlier, M.-F., Laurent, V., Santolini, J., Melki, R., Didry, D., Xia, G.-X., Hong, Y., Chua, N.-H., and Pantaloni, D. (1997) J. Cell Biol. 136 1307-1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishida, E. (1985) Biochemistry 24 1160-1164 [DOI] [PubMed] [Google Scholar]
- 9.Moriyama, K., and Yahara, I. (2002) J. Cell Sci. 115 1591-1601 [DOI] [PubMed] [Google Scholar]
- 10.Balcer, H. I., Goodman, A. L., Rodal, A. A., Smith, E., Kugler, J., Heuser, J. E., and Goode, B. L. (2003) Curr. Biol. 13 2159-2169 [DOI] [PubMed] [Google Scholar]
- 11.Hubberstey, A. V., and Mottillo, E. P. (2002) FASEB J. 16 487-499 [DOI] [PubMed] [Google Scholar]
- 12.Gieselmann, R., and Mann, K. (1992) FEBS Lett. 298 149-153 [DOI] [PubMed] [Google Scholar]
- 13.Freeman, N. L., Chen, Z., Horenstein, J., Weber, A., and Field, J. (1995) J. Biol. Chem. 270 5680-5685 [DOI] [PubMed] [Google Scholar]
- 14.Hubberstey, A., Yu, G., Loewith, R., Lakusta, C., and Young, D. (1996) J. Cell. Biochem. 60 459-466 [DOI] [PubMed] [Google Scholar]
- 15.Zelicof, A., Protopopov, V., David, D., Lin, X. Y., Lustgarten, V., and Gerst, J. E. (1996) J. Biol. Chem. 271 18243-18252 [DOI] [PubMed] [Google Scholar]
- 16.Chaudhry, F., Guérin, C., von Witsch, M., Blanchoin, L., and Staiger, C. (2007) Mol. Biol. Cell 18 3002-3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang, J., Suzuki, N., and Kataoka, T. (1992) Mol. Cell. Biol. 12 4937-4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ksiazek, D., Bandstetter, H., Israel, L., Bourenkov, G. P., Katchalova, G., Janssen, K. P., Bartunik, H. D., Noegel, A. A., Schleicher, M., and Holak, T. (2003) Structure (Lond.) 11 1171-1178 [DOI] [PubMed] [Google Scholar]
- 19.Mavoungou, C., Israel, L., Rehm, T., Ksiazek, D., Krajewski, M., Popowicz, G., Noegel, A., Schleicher, M., and Holak, T. (2004) J. Biomol. NMR 29 73-84 [DOI] [PubMed] [Google Scholar]
- 20.Yusof, A., Hu, N.-J., Wlodawer, A., and Hofmann, A. (2005) Proteins Struct. Funct. Bioinformatics 58 255-262 [DOI] [PubMed] [Google Scholar]
- 21.Dodatko, T., Fedorov, A. A., Grynberg, M., Patskovsky, Y., Rozwarski, D., Jaroszewski, L., Aronoff-Spencer, E., Kondraskina, E., Irving, T., Godzik, A., and Almo, S. (2004) Biochemistry 43 10628-10641 [DOI] [PubMed] [Google Scholar]
- 22.Bertling, E., Quintero-Monzon, O., Mattila, P. K., Goode, B. L., and Lappalainen, P. (2007) J. Cell Sci. 120 1225-1234 [DOI] [PubMed] [Google Scholar]
- 23.Lila, T., and Drubin, D. (1997) Mol. Biol. Cell 8 367-385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattila, P. K., Quintero-Monzon, O., Kugler, J., Moseley, J. B., Almo, S. C., Lappalainen, P., and Goode, B. L. (2004) Mol. Biol. Cell 15 5158-5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quintero-Monzon, O., Rodal, A. A., Strokopytov, B., Almo, S. C., and Goode, B. L. (2005) Mol. Biol. Cell 16 3128-3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spudich, J. A., and Watt, S. (1971) J. Biol. Chem. 246 4866-4871 [PubMed] [Google Scholar]
- 27.Vartiainen, M., Sarkkinen, E. M., Matilainen, T., Salminen, M., and Lappalainen, P. (2003) J. Biol. Chem. 278 34347-34355 [DOI] [PubMed] [Google Scholar]
- 28.Pringle, J., Adams, A., Drubin, D., and Haarer, B. K. (1991) Methods Enzymol. 194 565-602 [DOI] [PubMed] [Google Scholar]
- 29.Kushnirov, V. V. (2000) Yeast 16 857-860 [DOI] [PubMed] [Google Scholar]
- 30.Harlow, E., and Lane, D. (1988) Using Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 31.Siegel, L. M., and Monty, K. J. (1966) Biochim. Biophys. Acta 112 346-362 [DOI] [PubMed] [Google Scholar]
- 32.Laue, T. M., Shah, B., Ridgeway, T., and Pelletier, S. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding, S. E., ed) Royal Society of Chemistry, Cambridge, UK
- 33.Schuck, P., Perugini, M. S., Gonzale, N. R., Howlett, G. J., and Schubert, D. (2002) Biophys. J. 82 1096-1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolven, A., Belmont, L., Mahoney, N., Almo, S., and Drubin, D. (2000) J. Cell Biol. 150 895-903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lappalainen, P., Fedorov, E. V., Fedorov, A. A., Almo, S. C., and Drubin, D. G. (1997) EMBO J. 16 5520-5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertling, E., Hotulainen, P., Mattila, P., Matilainen, T., Salminen, M., and Lappalainen, P. (2004) Mol. Biol. Cell 15 2324-2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brieher, W. M., Kueh, H. Y., Ballif, B. A., and Mitchison, T. J. (2006) J. Cell Biol. 175 315-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada, K., Ravi, H., Smith, E. M., and Goode, B. L. (2006) Mol. Biol. Cell 17 2855-2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai, L., Makhov, A. M., and Bear, J. E. (2007) J. Cell Sci. 120 1779-1790 [DOI] [PubMed] [Google Scholar]
- 40.Vinson, V. K., Delacruz, E. M., Higgs, H. N., and Pollard, T. D. (1998) Biochemistry 37 10871-10880 [DOI] [PubMed] [Google Scholar]
- 41.Gerst, J., Ferguson, K., Vojtek, A., Wigler, M., and Field, J. (1991) Mol. Biol. Cell 11 1248-1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, J., Wang, C., Palmieri, S. J., Haarer, B. K., and Field, J. (1999) J. Biol. Chem. 274 19985-19991 [DOI] [PubMed] [Google Scholar]
- 43.Nishida, Y., Shima, F., Sen, H., Tanaka, Y., Yanagihara, C., Yamawaki-Kataoka, Y., Kariya, K., and Kataoka, T. (1998) J. Biol. Chem. 273 28019-28024 [DOI] [PubMed] [Google Scholar]
- 44.Wriggers, W., Tang, J. X., Azuma, T., Marks, P. W., and Janmey, P. A. (1998) J. Mol. Biol. 282 921-932 [DOI] [PubMed] [Google Scholar]