Abstract
Cell adhesion molecules, such as N-cadherin (cdh2), are essential for normal neuronal development, and as such have been implicated in an array of processes including neuronal differentiation and migration, and axon growth and fasciculation. Cdh2 is expressed in neurons of the peripheral nervous system during development, but its role in these cells during this time is poorly understood. Using the transgenic zebrafish line, tg(p2xr3.2:eGFPsl1), we have examined the involvement of cdh2 in the formation of sensory circuits by the peripheral nervous system. The tg(p2xr3.2:eGFPsl1) fish allows visualization of neurons comprising gV, gVII, gIX and gX and their axons throughout development. Reduction of cdh2 in this line was achieved by either crosses to the cdh2-mutant strain, glass onion (glo) or injection of a cdh2 morpholino (MO) into single-cell embryos. Here we show that cdh2 function is required to alter the directional vectors of growing axons upon reaching intermediate targets. The central axons enter the hindbrain appropriately but fail to turn caudally towards their final targets. Similarly, the peripheral axons extend ventrally, but fail to turn and project along a rostral/caudal axis. Furthermore, by expressing dominant negative cdh2 constructs selectively within cranial sensory ganglia (CSG) neurons, we found that cdh2 function is necessary within the axons to elicit these stereotypic turns, thus demonstrating that cdh2 acts cell autonomously. Together, our in vivo data reveal a novel role for cdh2 in the establishment of circuits by peripheral sensory neurons.
Keywords: cadherin, axon, sensory, peripheral, ganglion, zebrafish
The cranial nerves connect the central nervous system (CNS) to the periphery, allowing vertebrates to sense and respond to their internal and external environment. Only four of the cranial nerves carry both sensory and motor information: the trigeminal (V), facial (VII), glossopharyngeal (IX), and vagal (X) nerves (Zhang and Ashwell, 2001). The sensory components of these four arise from the cranial sensory ganglia (CSG), which are clusters of neurons that reside outside the CNS in stereotypic positions. These ganglia transduce somatosensory, chemosensory and viscerosensory information to the brain from receptors in the head, throat, heart and viscera. While much is known about the anatomy and physiology of these ganglia in adult organisms, little is known about how these circuits are formed.
The process of peripheral sensory circuit formation is complex, requiring the interaction of multiple cues in the environment to position the neurons of each ganglia and guide the central and peripheral nerves to discrete targets [reviewed in (Tessier-Lavigne and Goodman, 1996)]. These cues may be diffusible or membrane-bound. Membrane-bound cues include not only receptors involved in bidirectional signaling such as the ephs/ephrins (Reber et al., 2007), but also proteins that participate in intercellular adhesion, such as the cadherins (Ranscht, 2000).
Cadherins are a family of calcium-dependent cell adhesion proteins. Their extracellular domains consist of cadherin repeats that selectively bind the extracellular domains of homotypic cadherins (i.e. cdh2 preferentially binds to other cdh2 proteins) (Ivanov et al., 2001). Type 1 cadherins, including N-, E- and R-cadherin have a single transmembrane and an intracellular domain that binds β- and α-catenin, to anchor the protein to the actin cytoskeleton (Ivanov et al., 2001). Additionally, the intracellular domain of cadherins interacts with many intracellular signaling paths affecting axon outgrowth, navigation and synaptogenesis [reviewed in (Nollet et al., 2000, Wheelock and Johnson, 2003)]. During neural circuit formation, axons expressing a set of cadherins will selectively contact neighboring cells or axons expressing the same cadherins (Redies et al., 1992, Ranscht, 2000). In such a way, cadherins can serve as molecular codes orchestrating connectivity of various peripheral sensory modalities to discrete targets in the CNS (Redies et al., 1997).
The expression pattern of N-cadherin (cadherin-2 or cdh2) has been studied extensively in mouse, chick, xenopus and zebrafish. During development cdh2 expression is detected in neural tissues, including subsets of the CSG (Simonneau et al., 1992, Bitzur et al., 1994, Redies, 1995, Liu et al., 2003). It has been shown that loss of cdh2 function in animals disturbs the formation of cranial ganglia (Kerstetter et al., 2004); however, these studies were unable to clearly distinguish effects on CSG afferents from those on efferents. Here we show that disruption of cdh2 function results in misguided CSG afferents to the periphery and the CNS. Furthermore, we show that these effects result from a cell autonomous role of cdh2 within the CSG neurons.
Materials and Methods
Maintenance of fish
Fish were kept on a 14-hr day, 10-hr night schedule at a constant 28.5 °C, with feeding done twice daily. All animal husbandry was carried out as described by Westerfield (Westerfield, 2000). Embryos were staged according to hours post-fertilization (hpf) and morphological criteria (Kimmel et al., 1995). Embryos used for microscopy were treated with 0.003% phenylthiourea to reduce pigmentation.
Injections into embryos
For injections, plasmid DNA or morpholino oligonucleotides (MO) were dissolved in 0.1 M KCl, 20 mM HEPES (pH 7.4) containing 0.01% Phenol Red and injected into single-cell embryos. 2-5 nanoliters were injected using a Picospritzer III (General Valve Corporation, Fairfield, NJ) attached to a broken glass capillary. The cdh2 morpholino oligonucleotide (MO) designed against basepairs -36 to -13 of N-cadherin cDNA was purchased from GeneTools (Open Biosystems, Huntsville, AL). 2-5ng of the morpholino was injected into 1-4-cell-stage embryos. Post injection, embryos were allowed to develop in fish water at 28.5 °C.
Zebrafish strains
Tg(UAS:kaede) and glass onion (glom117) embryos were obtained from the Zebrafish International Resource Center at the University of Oregon (Eugene, OR). Tg(p2xr3.2:eGFPsl1) embryos were described previously (Kucenas et al., 2006). glom117 heterozygotes were crossed to tg(p2xr3.2:eGFPsl1) fish to generate tg(p2xr3.2:eGFPsl1);glo heterozygotes. Such heterozygotes were in-crossed to create homozygous glom117 embryos containing the p2xr3.2:eGFP reporter. Tg(nkx2.2megfp) fish were a gift from Bruce Appel (Ng et al., 2005), tg(isl1:eGFP) were a gift of H. Okamota (Higashijima et al., 2000) and tg(PB4:gvp;UAS:kaede) are described below.
Generation of tg(PB4:gvp;UAS:kaede)
The PB4 promoter is composed of the 4kb of genomic sequence immediately upstream of the transcriptional start site of the p2xr3.2 gene (Kucenas et al., 2006), and was obtained by PCR and subcloned into the pEGFP-1 vector (Clontech) using Xho1/EcoR1 sites present in the PCR primers. Embryos injected with this construct expressed eGFP within the CSG in a pattern identical to the tg(p2xr3.2:eGFPsl1) line fish, but had no detectable eGFP expression in the hindbrain. This PB4 promoter fragment was excised via NheI and AgeI sites and inserted upstream of a gal4VP16 element in the tol2 transposon vector pCH mcs G4VP16 (a kind gift from Dr. Mike Nonet) yielding pCH.PB4:GVP. pCH.PB4:GVP (50 ng/μl) was coinjected with transposase RNA (30 ng/μl) (pCS-TP) into eggs from the AB genetic background. Transposase activity was verified by PCR analysis as described in (Kawakami et al., 1998). The pCH.PB4.GVP construct contains a second expression cassette that has the cardiac myosin light chain promoter in front of CFP, allowing us to identify carrier embryos by their cardiac CFP-expression. Carriers were raised to adulthood and intercrossed to identify founders. Approximately 10-15% of carriers were identified as founders. The tg(PB4:gvp) fish were crossed to tg(UAS:kaede) fish to generate embryos for the tg(PB4:gvp;UAS:kaede) line.
Dominant negative construction and injection
Full length zebrafish cdh2 (accession number: AF430842) was cloned from cDNA obtained from 4 days post fertilization (dpf) zebrafish by PCR (Phusion, New England Biolabs) and was verified by sequencing. The two dominant negative cdh2 were constructed as follows. The C-terminal deleted cdh2 (ΔC) was engineered by amplifying the coding region from the initiator ATG to amino acid E739, which was converted to a termination codon (TAG); this residue sits C-terminal to the transmembrane domain. The second dominant negative was a cdh2 molecule that did not contain the extracellular cadherin domains (ΔN) and was constructed in a two-step process. The first fragment spanned amino acid M1 through G30: this sequence contains the signal peptide and terminates at the start of the pro-domain (Malicki et al., 2003). The 3′-primer used to generate this fragment included an extra 15bp corresponding to 5 amino acids (S656-S660) situated 5′ of the transmembrane domain. A second PCR fragment was generated that began at these 5 amino acids and continued through the membrane spanning domain and ended at the native stop codon present in the 3′ primer. These two fragments were then used in an overlap PCR (Egan et al., 1998) such that the signal peptide was attached directly to a site 47 amino acids upstream of the transmembrane domain (Kintner, 1992). Both dominant negatives were then cloned into a UAS vector (a gift from Dr. R. Wong (Mumm et al., 2006)) at the Asp718/NotI sites. Primers used for PCR were as follows: full length – 5′-end: tataggtaccatcggtttctttatacagaac and 3′-end: tatagcggccgcctagtcgtcgttacctccgta; ΔC – 3′-end: tatagcggccgcctatttatcccgtctcttcat; ΔN – overlap forward: ccatgtcagcctggtagttacctggaaagt and overlap reverse: actttccaggtaactaccaggctgacatgg. All PCR was performed using the high fidelity DNA polymerase Phusion (New England Biolabs). PB4:GVP plasmid (50ng/μl) was coinjected with either UAS:ΔC or UAS:ΔN (25ng/μl).
Epifluorescent Microscopy
Embryos were anesthesized with 0.02% tricaine in fish water and transferred to a 96-well plate. Epifluorescent images were taken using a Nikon TE200 inverted microscope equipped with a CoolSnap HQ digital camera. Metamorph software (Universial Imaging Corp) was used to acquire and process images. Cropping and rotating of images was carried out using Adobe Photoshop. For timelapse imaging, embryos were embedded in 0.5% low-melting point agarose containing 0.02% tricaine. The embryos were maintained at 28-29°C using a heated plate, and images were taken every 5 minutes.
Confocal imaging of embryos
Live or fixed embryos were embedded in 0.5% low-melting point agarose containing 0.02% tricaine. Optical sections were taken using an Olympus FV1000 MPE and z-stacks were processed using Olympus Fluoview software and Adobe Photoshop. Kaede was photoconverted from red to green fluorescence under visual inspection by excitation with a 405nm laser.
Scoring of phenotype
Offspring from glom117; tg(p2xr3.2:eGFPsl1) adults were imaged under brightfield to identify homozygotes based on embryo morphology. At 4 days post fertilization (dpf), GFP-positive embryos glom117 embryos were imaged laterally under confocal microscopy as described above. The gIX central axon was not amenable to visualization due to its depth and the scattering of the epibranchial ganglia, so it was excluded from further analysis. The score of “no axon” was given only when no central gVII nerve (snVII) or gX nerve (snX) was visible. The score of “normally projecting” was given when the snVII axon terminated near its normal proximity in the hindbrain even when no plexus was formed, or when snX axons formed a plexus in the hindbrain, though this plexus was sometimes less defined. The score of “misrouted axons” was given when the snVII or snX was visible but followed a guidance path divergent from that seen in control embryos.
Results
Previous studies have shown that cdh2 is involved in the development of the CSG; however, these studies used traditional techniques to label axons (e.g., acetylated tubulin immunostaining) (Kerstetter et al., 2004) and as such were unable to identify all sensory axons, nor distinguish the labeled CSG afferents from the branchiomotor efferents that course alongside them. To overcome these problems and examine the role of cdh2 in the development of the CSG, we used zebrafish from the transgenic line tg(p2xr3.2:eGFPsl1). The utility of this line lies in the fact that the transgene reporter selectively labels the neurons and axons of the trigeminal (gV), facial (gVII), glossopharyngeal (gIX), and vagal (gX) ganglia (Kucenas et al., 2006) (Fig.1A,C). The soluble green fluorescent protein (eGFP) is detectable in CSG neurons as they migrate, cluster into ganglia, and extend axons to peripheral and central targets, thus allowing us to examine the formation of the central projections of the various CSG in intact, living animals.
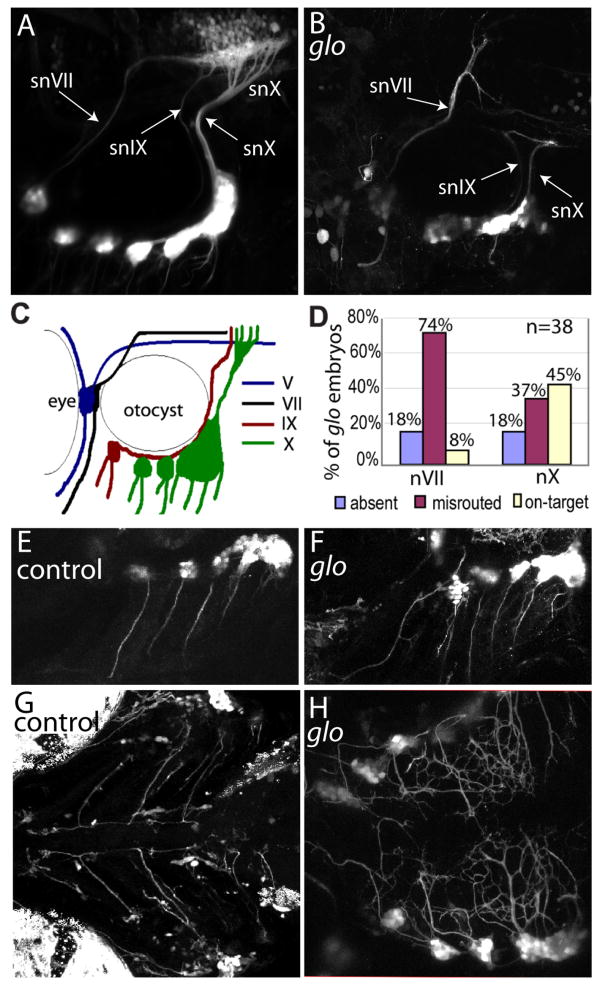
Figure 1. Absence of cdh2 function in glo mutant embryos disrupts axon pathfinding of the epibranchial ganglia.
Confocal Z-stacks comparing control CSG (A,E,G) and glo CSG (B,F,H) at 4dpf. In all panels, rostral is to the left. A, B, Lateral views of 4dpf control (A) and homozygous glom117;tg(p2xr3.2:eGFPsl1) (B) embryos showing defects in the gVII, gIX and gX central axonal projections (labeled snVII, snIX and snX, respectively). C, Schematic illustration of GFP-labeled nerves in A. D, Graph representing the percentage of glo mutant embryos displaying normally projecting, aberrantly projecting or absent gVII or gX axons. 38 homozygous glo m117;tg(p2xr3.2:eGFPsl1) embryos from two clutches were examined under confocal microscopy and their central projections were scored. E and F, Lateral views of tg(p2xr3.2:eGFP) control (E) and glo (F) embryos showing bundles of axons extending ventrally from each epibranchial ganglia in both control and glo mutant embryos. G and H, Ventral views of control (G) and glo; tg(p2xr3.2:eGFPsl1) mutant (H) embryos showing the bilateral ganglia extending peripheral processes to the ventral midline. In glo embryos the processes defasciulate as they reach the midline and fail to turn and extend along the rostral-caudal axis. snVII, snIX and snX are abbreviations for gVII, gIX and gX nerves, respectively.
cdh2 knockdown disrupts axon pathfinding of the epibranchial ganglia
Our first step in determining the role of cdh2 in the formation of the ganglionic circuits was to examine the effects of cdh2 disruption on ganglion formation and axon guidance. This was accomplished in two ways: injecting a cdh2 morpholino oligonucleotide (MO) into tg(p2xr3.2:eGFPsl1) embryos to block translation of the cognate mRNA, or by crossing the tg(p2xr3.2:eGFPsl1) line fish into the cdh2 mutant, glass onionm117 (glom117). The glom117 mutant contains a point mutation in the extracellular domain of cdh2 that changes a tryptophan to a glycine (Malicki et al., 2003), resulting in a protein that is unable to oligomerize and thus cannot participate in adhesion. The CSG in living morphant or mutant embryos were analyzed using confocal microscopy, with both sets of embryos yielding identical results.
The most anterior CSG, gV, was only slightly affected as the neurons forming these ganglia were not as compacted as in control animals (Fig. 1B) [see also (Kerstetter et al., 2004)]. The central gV afferent nerve (snV) was slightly defasciculated, but properly routed (data not shown).
The epibranchial ganglia (gVII, gIX and gX) were affected to differing degrees. The most obvious defect seen was the misrouting of the gVII central nerve (snVII) in the hindbrain (Fig. 1B). The gVII sits just rostro-medial to the ear. In control transgenic embryos, the central afferent nerve a tightly fasciculated bundle of axons that projects dorsally along the rostral edge of the ear. It enters the hindbrain at rhombomere 4, grows medially, forms a genu and extends caudally through the hindbrain to join the gIX and gX central projections (snIX and snX) at a plexus in the hindbrain (Fig. 1A). In both mutants and morphants, the snVII did not form the genu and failed to extend caudally to the hindbrain plexus (Fig. 1B). In most mutants (28 out of 38 mutant glo embryos), the snVII continued its dorsal path after entering the hindbrain and projected aberrantly in rhombomere 4 or 5 (Fig. 1B,D) However, in some mutants (7 out of 38 mutant glo embryos), the gVII failed to extend a central axon at all. In a few mutant embryos (3 out of 38 mutant glo embryos) snVII did extend caudally as in control (Fig. 1D).
The other two epibranchial ganglia, the gIX and gX, also had disrupted central projections in mutants and morphants. In control embryos, gIX and gX are discrete neuronal clusters that lie ventral to the otocyst. gIX sits at the anterioventral corner of the otocyst and snIX extends along the ventral edge of the otocyst until, at the posterior edge, it turns dorsally and enters the hindbrain at rhombomere 6 (Fig. 1A,C). gX consists of several neuronal clusters ventral and caudal to the otocyst (Fig. 1A,C). The snX axons course ventrally around the ear and along the posterior margin until they meet at the dorsal edge of gX, where they form a bundle, course dorsally and project into the hindbrain at rhombomere 7 (Fig. 1A,C). The snIX and snX project along distinct paths, but meet with the snVII at a characteristic plexus in the hindbrain (Fig.1A).
In both mutants and morphants, gIX neurons were less tightly compacted in the ganglion and snIX failed to project to the hindbrain plexus. In some embryos (1 out of 38), snIX curved around the ear, projected dorsally but missed the plexus and extended anteriorly into the hindbrain. In other individuals (2 out of 38), snIX extended around the ear to the location of the plexus even though the plexus was absent. However, in the majority of embryos (35 out of 38), snIX was not clearly visible. This could be because it was either absent, bundled with snX, or its thin fibers were obscured by the gX neurons that were scattered in the glom117 embryos.
In nearly half of glo mutants (17 out of 38), snX did reach the hindbrain; however, there was little or no branching and as such the plexus was absent or weakly defined (Fig. 1D). For other mutants (14 out of 38), an axon bundle grew dorsally but was misdirected, extending aberrantly into the hindbrain or turning and projecting back towards the ganglia. For still others (7 out of 38 glo mutant embryos), the gX failed to extend an axon dorsally at all.
The peripheral processes of these ganglia were also disrupted in cdh2 mutants. In control embryos, the peripheral processes extend from each ganglia into the branchial arches as two bundles of ventrally oriented fascicles. The bundles follow the gill arches to near the ventral midline, at which point they turn and join a fiber bundle oriented in the rostral-caudal direction (Fig. 1E,G). The severely disrupted head and jaws of both mutants made visualization of the ventral processes difficult, but axons could be seen projecting ventrally from each epibranchial ganglia in mutants as in controls (Fig. 1E,F). However, these peripheral processes were defasciculated and failed to turn at the midline (Fig. 1H). All of these results support a role for cdh2 in normal CSG circuit formation, both at the level of ganglia formation and axon guidance.
Timelapse imaging reveals a pause in gVII elongation between 50-60hpf
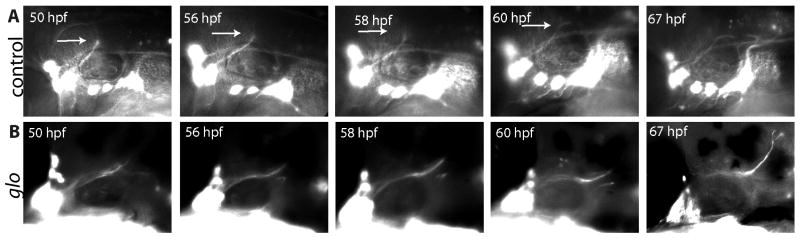
To further elucidate the role that cdh2 plays in afferent axon guidance to the CNS, we carried out in vivo timelapse imaging of snVII as it grows to its central targets. In tg(p2xr3.2:eGFPsl1) embryos, gVII first becomes apparent at 33hpf. A bundle of axons then grows dorsally from the ganglia and then medially into rhombomere 4 at 50 hpf. From 50-60 hpf, small projections ending in growth cones can be seen extending and retracting in nearly all directions from the end of the fascicle, but the extensions are transient and no elongation of the axon is detected. The extension of transient neurites in multiple directions indicates that at the level of rhombomere 4 in control embryos snVII axons are detecting and responding to cues in the environment. Around 60 hpf, the protrusions begin favoring the caudal direction and the snVII forms a genu as it grows to rhombomere 7 and the caudal hindbrain, where it meets snIX and snX at the hindbrain plexus (Fig. 2A).
Figure 2. Timelaspe imaging reveals a pause in snVII growth accompanied by transient axonal extension and retraction in control, but not in glom117 embryos as snVII turns from a dorsal to a caudal vector.
Row A shows frames from a tg(p2xr3.2:eGFPsl1) control timelapse movie. In control embryos, elongation of snVII pauses between 50h and 60h. Arrows in row A indicate the axon fascicles that are extended and retracted before snVII turns to grow caudally and meet snIX and snX at the hindbrain plexus. Row B shows frames from a timelapse movie of a tg(p2xr3.2:eGFPsl1);glo m117 mutant embryo. In the mutant, snVII continues growing in the dorsal direction and the transient axon extensions and retractions are absent. Age of embryos is shown in upper left corner of each image. In all images rostral is to the left, dorsal to the top.
Initially in glom117;tg(p2xr3.2:eGFPsl1) embryos, gVII formed similarly to that of control transgenic embryos. gVII neurons became apparent around 33 hpf and central bundles began growing dorsally. However, between 50 and 60 hpf snVII did not pause and extend growth cones in various directions as seen in controls, but instead continued to grow in the dorsal direction (Fig. 2B) and thus failed to contribute to the formation of the hindbrain plexus. The misdirected nerve also showed signs of defasciculation in the hindbrain around 72hpf. The snIX and snX bundles follow the same time course in the mutants as they do in the controls, exhibiting their defects only after reaching the area where the plexus normally forms (data not shown).
These findings illustrate one major way that cdh2 mutant embryos differ from controls is through the absence of an snVII pause and genu formation at the level of rhombomere 4. It is interesting that up to this caudal turn, snVII courses in a path distinct from, but parallel to, the efferent branchiomotor VII nerve (mnVII), raising the possibility that mnVII is influencing snVII axon guidance. We address this possibility by looking at efferent fibers in cdh2-deficient embryos.
Efferent fibers are not affected by cdh2 knockdown
The branchiomotor nuclei in the hindbrain arise earlier than the CSG and their efferents begin pathfinding ventrally from the branchiomotor neurons to peripheral targets, passing alongside the various CSG, before the central processes of the CSG begin growing towards the hindbrain (Higashijima et al., 2000). This supports the hypothesis that the pathfinding of the efferents may influence the guidance of the afferents to the hindbrain and central targets. If this were the case, and given the defects seen in the sensory circuits, we would expect that branchiomotor nerves would also be misguided in cdh2 morphant embryos.
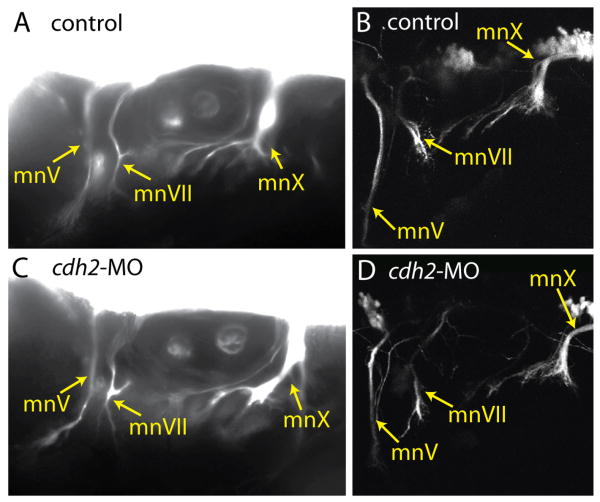
We tested this possibility by first examining the effect of cdh2 disruption on the peripheral motor nerves using both tg(Isl1:eGFP) and tg(nkx2.2:megfp) embryos. In the tg(Isl1:eGFP) fish, soluble GFP is expressed by the cranial motor neurons and some neurons of the CSG (except gV) (Higashijima et al., 2000), allowing visualization of both cell bodies and axons. In the tg(nkx2.2:megfp) fish, membrane targeted eGFP is expressed in the neural tube and branchial arches (Ng et al., 2005); the only labeled peripheral axons in the head are the cranial motor nerves. We injected cdh2 MO into embryos of these lines and visualized the efferent fibers at 3 days post fertilization (dpf) by epifluorescent and confocal microscopy. We found that in morphant embryos the Vth, VIIth and Xth branchiomotor nerves (mnV, mnVII and mnX, respectively) projected through the periphery (Fig. 3C,D) in manners similar to those in control transgenic embryos (Fig. 3A,B). These findings indicate that the peripheral defects seen in the sensory limb do not arise from misrouted motor nerves.
Figure 3. Efferent fibers are not affected by loss of cdh2.
Comparison of control (A,B,C,D) and cdh2-MO injected (E,F,G,H) motor projections using tg(nkx2.2megfp) (A,E) and tg(Isl1:eGFP) (B,C,D,F,G,H). A,B,E,F are epifuorescent images of lateral views of 72 hpf embryos; C and G are lateral confocal images from 48 hpf embryos; D and H are 96hpf dorsal confocal images. In all images rostral is left. mnV, mnVII and mnX are abbreviations for respective motor nerves, whereas mV, mVII and mX are abbreviations for the respective motor nuclei.
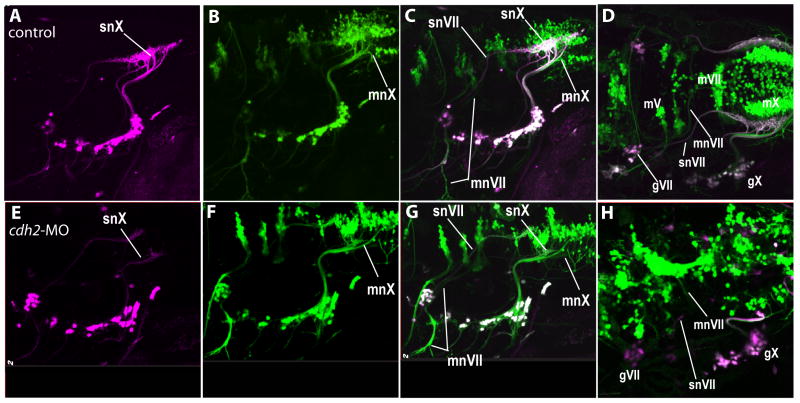
Our next step was to examine the effects of cdh2 disruption on the CNS portions of the efferent nerves. We designed an experimental approach, using dual transgenic animals that would enable us to simultaneously visualize and distinguish between the motor and sensory fibers. This was necessary, as there was a wide diversity of afferent nerve defects that had been observed in both morphant and mutant fish and it was essential to compare both afferent and efferent nerves in the same animal in order to reach valid conclusions about the role(s) of the efferents. To accomplish this goal, we first identified a p2xr3.2 promoter fragment that allowed us to selectively express proteins within the peripheral sensory neurons, but not in hindbrain neurons. We achieved this by cloning consecutively smaller portions of the 10kb p2xr3.2 promoter used to generate the tg(p2rx3.2:eGFPsl1) into an eGFP reporter plasmid. This led to the identification of a 4kb fragment, referred to as PB4, which gave cranial expression in only the gV, gVII, gIX and gX sensory neurons. This promoter was used to engineer tg(PB4:GVP;UAS:kaede) (as described in the methods section), which drives the photoconvertible protein kaede specifically within the CSG. By exposing tg(PB4:GVP;UAS:kaede) embryos to UV light, kaede expressed in sensory neurons and their axons was converted from a green to a red fluorescent protein (Hatta et al., 2006). Thus, photoconversion of 4dpf embryos from a cross of tg(PB4:GVP;UAS:kaede) and tg(Isl1:eGFP) yielded CSG neurons and axons that were detected in the red channel (shown as false-color magenta in Fig. 4A,E), whereas the motor neurons and their axons emit only green fluorescence (Fig. 4B,F). This differential reporter expression allows us to distinguish the afferent and efferent fibers simultaneously (merged images, Fig. 4C,G). Cdh2-MO was injected into eggs derived from these crosses and the effects of cdh2 knock-down on the afferent and efferent fibers were examined simultaneously (Fig. 4E-H).
Figure 4. The effects of cdh2 knock-down are specific to the afferent fibers.
A cross between tg(PB4:GVP;UAS:kaede) and tg(Isl1:eGFP) fish show that snVII central processes project aberrantly even though mnVII projects normally in cdh2-MO injected embryos. A-D depict a control embryo, E-H depict a cdh2-MO injected embryo. Note also, neurons in the CSG appear scattered in cdh2-MO injected embryos (E-G). D and H are dorsolateral views showing the positioning of the motor nuclei. In all images, the sensory circuits are labeled with photoconverted kaede, shown as magenta, and all motor circuits (and some sensory neurons) contain GFP labeling that is shown in green. Cells expressing both proteins are seen as white. Images are from 96hpf embryos. Rostral is left, dorsal is top.
At 4dpf the VII branchiomotor nuclei (mVII) of control embryos sit near the midline in rhombomeres 5 and 6 (Fig. 4D). The mVII efferents (mnVII) project anteriorly, then turn laterally, exit the hindbrain, and course ventrally between the gVII and the otocyst (Fig. 4C,D). The snVII takes a distinct path from mnVII (Higashijima et al., 2000) (Fig 4C,D). During their ascent, snVII afferent fibers follow a course lateral to the descending mnVII efferents. The snVII then extends medially into the CNS at a point just caudal to the exit point of the mnVII. However, just past its entry point into the CNS, the snVII diverges from its path alongside mnVII, forms a genu, and grows caudally to the plexus in the hindbrain. In cdh2-MO injected embryos where the snVII failed to make its caudal turn and project to the hindbrain, the axons forming the mnVII were still able to leave the branchial motor nuclei as a fascicle, make a lateral turn and exit the hindbrain into the periphery as in control embryos. This occurred even though the afferent limb was misrouted (Fig. 4G,H). Similar findings were observed for the vagal system, where in animals without a properly formed plexus the mnX axons became fasciculated, grew anteriorly and exited the hindbrain as normal (Fig. 4G,H). Together, these results showed that the misrouting of the afferents in the morphants was not due to defects in the branchiomotor efferent projections.
However, the positioning of the branchiomotor nuclei in the brain was irregular in many morphants, with neurons of the branchiomotor nuclei crossing the midline and spreading across rhombomeric borders (Fig. 4H; see also (Lele et al., 2002)), confirming that hindbrain organization is affected by loss of cdh2 function. To determine if this disorganization was responsible for the misrouting of the CSG afferents, we next examined the pathfinding of cdh2-deficient neurons in a wild-type background.
Impairment of cdh2 function selectively within CSG neurons mimics the affects of global cdh2 knockdown on snVII and snX
Loss of cdh2 throughout an embryo results in animals with curved trunks, smaller eyes, an indistinct midbrain-hindbrain boundary (Fig. 5A,B), a dorsally-broadened neural tube, and neuroepithelial cells, motoneurons and hindbrain interneurons that are abnormally arranged (Fig. 4 and (Lele et al., 2002)). Incorrect CSG afferent pathfinding could therefore arise due to disruption of hindbrain environment, loss of cdh2 in the environment and/or loss of cdh2 in CSG. To distinguish among these possibilities, we specifically interfered with cdh2 function in only CSG neurons by expressing dominant negative cdh2 constructs within these cells.
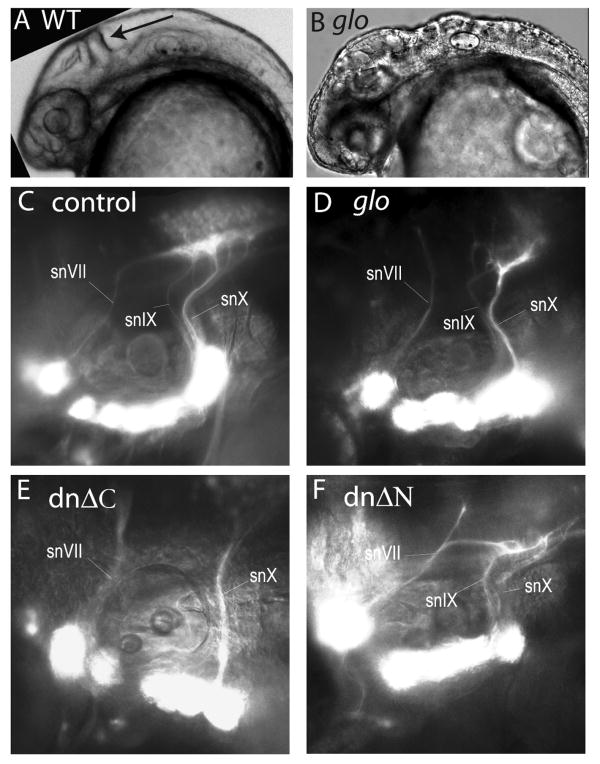
Figure 5. Cdh2 is necessary within CSG neurons for proper pathfinding of gVII, gIX, and gX central axonal bundles.
Heads of control (A) and glo mutant (B) embryos at 30hpf are shown as brightfield images. The midbrain-hindbrain is indicated with arrow in A. Epifluorescent images in C-F are lateral views of 4dpf tg(p2xr3.2:eGFPsl1) embryos which are uninjected (C), cdh2-MO injected (D) or co-injected with PB4:GVP and a UAS:ΔC (E) or UAS:ΔN (F). Arrowhead indicates snVII, which projects aberrantly in D, E, and F. Rostral is left; dorsal is up.
Overexpression of these constructs within the CSG neurons was achieved using the PB4 promoter and the binary Gal4-VP16/UAS system (Halpern et al., 2008). Gal4-VP16b (GVP) is a potent transactivator that drives overexpression by binding at multiple, tandem UAS sites (Halpern et al., 2008). Thus, a dominant negative construct under the control of a UAS/basal promoter combination would be expected to achieve the high levels of dominant negative protein needed to disrupt the function of the endogenous proteins.
The first dominant negative we tested was cdh2 without the intracellular domain. Deletions of this cytoplasmic domain (ΔC), which anchors the cadherin protein to the cytoskeleton, have been shown to interfere with cdh-mediated adhesion in xenopus, zebrafish, chick and mice (Kintner, 1992, Levine et al., 1994, Nakagawa and Takeichi, 1998, Castro et al., 2004, Babb et al., 2005). This construct was engineered by inserting a stop codon just downstream of the transmembrane domain. Tg(p2xr3.2:eGFPsl1) embryos were then coinjected with PB4:GVP and UAS:cdh2ΔC in order to achieve overexpression of cdh2ΔC in the CSG neurons. Unlike control embryos (either uninjected or co-injected with PB4:GVP and UAS:mYFP, Fig. 5C) and similar to morphants (Fig. 5D), embryos injected with cdh2ΔC showed misrouting of CSG afferents (Fig. 5E). The animals exhibiting these defects had normal heads and bodies, suggesting minimal to no ectopic expression of cdh2ΔC. We found that transient expression frequently yielded mosaic expression patterns in the CSG (data not shown). Even so, we found snVII, snIX or snX to be misrouted or absent in 21.4% (n=537) of embryos coinjected with PB4:GVP and UAS:cdh2ΔC, compared to less than 1% of control animals.
The transmembrane and the extracellular domain are still present in the cdh2ΔC construct. To determine if the extracellular domain of cdh2, which contains the adhesion motifs, was essential in afferent pathfinding, we tested the effects of a second dominant negative (cdh2ΔN). This cdh2 construct lacks the 5 extracellular cadherin repeats, but still retains the complete intracellular domain. As with the cdh2ΔC construct, we found defects in the routing of snVII, snIX and snX in 12.0% (n=279) of embryos injected with PB4:GVP and UAS:cdh2ΔN (Fig. 5F). Previous supports suggest that cdh2ΔN acts as a pan-cadherin dominant negative (Jontes et al., 2004). The fact that the phenotypes exhibited in cdh2ΔC- and cdh2ΔN-expressing embryos are identical would suggest that cdh2 function predominates in formation of these circuits. The defects in axon guidance produced by expression of either the ΔC or ΔN construct indicate that even in a control environment, gVII, gIX and gX axons which lack cdh2 function are unable to find their central targets.
A common test for cell autonomy is to test the effects of the wild-type molecule within mutant cells. To this end, a full-length zebrafish cdh2 was expressed specifically within the CSG neurons of glo embryos using the PB4:GVP/UAS system. In these experiments, we detected no difference in the percentage of embryos exhibiting defective axon pathfinding between groups of injected and uninjected glo;tg(p2xr3.2:eGFP) embryos (data not shown). These results can be due to a number of potential mechanisms. One possibility is that we were unable to achieve high enough levels of cdh2FL in sufficient numbers of neurons within a ganglion to rescue the phenotype. Alternatively, it is possible that cdh2-expression within CSG neurons is unable to rescue afferents in cdh2-deficient embryos because cdh2 on axon membranes must interact with cdh2 protein on neighboring cells, such as glial cells, to pathfind properly. Another possibility is that the physical disruptions throughout the hindbrain and neural tube of cdh2-deficeint embryos might be sufficient to disrupt afferent pathfinding regardless of whether functional cdh2 is present within the CSG neurons or not. It is not possible to definitively conclude from our results which of these mechanisms is operative.
Discussion
N-cadherin (cdh2) has been implicated in morphogenesis of neural systems across species (Bronner-Fraser et al., 1992, Radice et al., 1997, Luo et al., 2001, Lele et al., 2002). Sensory neurons express cdh2 during their development, but little is known about its function in this process. In this study we investigated the role of cdh2 in the formation of sensory circuits by the CSG. We demonstrate that cdh2 activity is a requirement for the proper pathfinding of both the central and peripheral projections of the epibranchial CSG, and that this action is dependent on the expression of functional cdh2 in CSG neurons themselves. These findings highlight a novel in vivo role of cdh2 in neural circuit formation.
There are multiple hindbrain defects seen in zebrafish cdh2 mutants (Lele et al., 2002). Considering these abnormalities, it would not be unexpected that peripheral sensory neural circuits would also be affected. Indeed, we found this to be the case in both mutant (glom117) and cdh2 morpholino-injected embryos. In these animals, the CSG were found in their appropriate positions near the eye (gV) or ventrolateral to the ear (gVII, gIX and gX), ruling out a role for cdh2 in anatomical positioning. However, these embryos did exhibit a spectrum of defects in gVII, gIX or gX circuit formation, ranging from axonal misrouting (seen in the majority of cases) to absence of a central projection from single ganglia. This lack of a central projection was not due to loss of neurons, as all ganglia were present in all animals, and each of the ganglia produced proximally fasciculated peripheral axons. These findings suggest that either the neurons were unable to generate a viable central axon branch or that both the central and peripheral branches were bundled together, with the peripheral branch dominating the pathfinding process. Future studies will be needed to determine which of these mechanisms is responsible. As for embryos exhibiting misrouting of projections, even in the most severe cases axons always exited and were maintained for some distance from the ganglion as tightly fasciculated bundles in a manner similar to control animals, indicating that cdh2 is not required for the initial fasciculation of CSG axons.
In the misrouting cases neither peripheral nor central projections reached their final targets. While the CSG central projections entered the hindbrain correctly, the snVII, snIX, and/or snX failed to make stereotypical caudal turns or branches leading to their final target zone in the hindbrain. For the peripheral processes, fascicles from each ganglion coursed ventrally along the gill arches, became defasciculated, and failed to make the typical orthogonal turn to project along the midline upon reaching their ventral limit. Thus for both sets of projections, it appeared that loss of cdh2 function resulted in an inability for axons to undergo a change in direction at particular, stereotypical points which we refer to as ‘choice points’. These behaviors lead us to propose that cdh2 permits directional changes during axonal growth in response to environmental cues.
One difficulty in interpreting the results from mutant and morphant experiments is that global disruption of cdh2 function impacts numerous neurodevelopmental processes. Thus, we could not discern whether the defective CSG circuits were due to loss of cdh2 activity within the CSG axons, to loss of activity within the axonal environment, or a combination of both mechanisms. By selectively over-expressing two different dominant negative cdh2 constructs in the sensory neurons of wild-type fish, we were able to demonstrate that cdh2 has an essential role within the CSG axons.
So how does cdh2 function within CSG neurons mediate axon guidance at choice points? We propose two hypotheses to explain the defects in central pathfinding we observed. First, as a homotypic cell adhesion protein cdh2 could selectively bind and adhere the growing axon to cdh2 proteins on neighboring cells. This has been observed in in vitro experiments where cdh2-positive sensory axons preferentially grew along the cell borders of cdh2-expressing cells (Redies et al., 1992). In zebrafish, a trail of cdh2-bearing cells could act to guide the cdh2-expressing snVII, snIX or snX nerves through the hindbrain to their final destinations. Therefore, when homotypic cdh2-mediated adhesion occurred, it would act to trigger further axonal growth along this cellular tract. The pause in snVII axon growth seen from 50-60 hpf could be explained by the need for cdh2-positive cells, such as glia (Tomaselli et al., 1988), to migrate to this choice point where they contact and stimulate the snVII axons to continue on in a new direction. When no homotypic adhesion occurs, as with cdh2 disruption, the axons do not pause but instead continue growing in an incorrect direction due to inappropriate integration of surrounding inputs. Thus, lack of cdh2 function in CSG leads to loss of adhesion resulting in altered axon outgrowth.
An alternative hypothesis is that it is the intracellular signaling mediated and/or modulated by the C-terminus of cdh2, rather than adhesion, which is critical. The C-terminus of cdh2 interacts with a number of proteins that modulate the adhesive properties of cdh2 and/or activate downstream signaling pathways. Molecules that can interact with the intracellular domain of cdh2, include tyrosine kinases, phosphotases, the Rho family of GTPases and effectors involved in Wnt signaling (Wheelock and Johnson, 2003, Bamji, 2005, Bruses, 2006). These molecules are implicated in the regulation of cadherin-mediated cell adhesion at the level of gene transcription, post translational modifications, or by changes in extracellular calcium concentration. Furthermore, cdh2 influences signaling cascades affecting actin dynamics or synapse formation by interacting with these molecules (Ranscht, 2000). Thus, this domain could be critical for the proper integration of signaling by multiple guidance cues acting upon the axons, and without this action of cdh2, even in the presence of adhesion, the axon cannot generate the proper cue response and instead grows in an inappropriate direction. Distinguishing between the adhesive and signaling aspects of cdh2 function has been proposed to be problematic (Kintner, 1992, Bruses, 2006); we also were unable to differentiate between these two properties in our dominant negative experiments.
There are notable differences between pathfinding errors of different CSG afferents in embryos with defective cdh2 function. For gVII, the central afferent failed to turn caudally and instead extended in an aberrant fashion. In contrast, snIX and snX did make their caudal turns, but then failed to branch and form the plexus. One explanation for these differences could lie in reports that CSG neurons express multiple cadherin isoforms; in zebrafish, cdh2 is expressed in gV, gVII and gX neurons (Liu et al., 2003), cdh1 (E-cadherin) in gV and gVII, cdh4 (R-cadherin) and cdh6 within gV and gX (Liu et al., 2003, Liu et al., 2006b), and cdh10 in gV, gVII and gX (Liu et al., 2006a). This differential expression of cadherins may provide differing degrees of redundancy with respect to cdh2 function and thus cause the observed differences.
Another functional outcome attributed to this combinatorial cadherin expression is the creation of a molecular code to guide axons from neurons that share common functional features or neuroanatomical connections (Redies et al., 1992, Redies and Takeichi, 1993, Redies et al., 1997, Ranscht, 2000). Cadherins have been shown to be expressed differentially in regions of the floor plate (Redies et al., 1997) and thus may act to selectively mark intermediate targets for the different CSG central nerves. The afferents of cdh2-deficient embryos become misguided after reaching these intermediate targets, suggesting that cdh2 is not involved in that stage of guidance, but instead that it modifies the response of the afferents to environmental cues after they pass these target zones. For instance, cdh2 expression at the floor plate may trigger snIX and snX to defasciculate, form branches to and establish synapses in the hindbrain plexus similar to what has been reported for cadherins in the tectofugal tract in the chick hindbrain (Redies et al., 1993, Redies et al., 1997). One note of interest is that while our studies indicate disruption of cdh2 in the ganglia affects pathfinding of snIX, previous studies did not report cdh2 expression within gIX (Liu et al., 2003). However, a low copy number of transcripts or proteins would make it difficult to reliably detect cdh2 expression in gIX, given the small number of neurons composing it, together with its medial location.
Our results show for the first time that cdh2 expression is required within the CSG neurons to allow discrete turning or branching of their nerves in the hindbrain. The mechanism responsible for this function of cdh2 within the CSG neurons remains to be elucidated; however, our results clearly show that cdh2 function is a requirement at these “choice point(s)”, where it enables a change in direction of axonal outgrowth. Additionally, we have demonstrated the utility of the tg(p2xr3.2:eGFPsl1) and the PB4 promoter for assessing the role of genes in CSG development. These tools should prove valuable in future studies involving genes critical to the development of the CSG.
Acknowledgments
We would like to thank Jasmina Mandzukic for help with fish husbandry, Dr. Jane Cox for suggestions and review of the manuscript. The zn8 antibody developed by Dr. W. Trevarrow was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. This work was supported by NIH grant NS060074.
Abbreviations
- CSG
cranial sensory ganglia
- CNS
central nervous system
- glo
glass onion mutant
- gV
trigeminal ganglia
- snV
trigeminal afferent
- mV
trigeminal nuclei
- mnV
trigeminal efferent
- gVII
facial ganglia
- snVII
facial afferent
- mVII
facial nuclei
- mnVII
facial efferent
- gIX
glossopharyngeal ganglia
- snIX
glossopharyngeal afferent
- mIX
glossopharyngeal nuclei
- mnIX
glossopharyngeal efferent
- gX
vagal ganglia
- snX
vagal afferent
- mX
vagal nuclei
- mnX
vagal efferent
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babb SG, Kotradi SM, Shah B, Chiappini-Williamson C, Bell LN, Schmeiser G, Chen E, Liu Q, Marrs JA. Zebrafish R-cadherin (Cdh4) controls visual system development and differentiation. Dev Dyn. 2005;233:930–945. doi: 10.1002/dvdy.20431. [DOI] [PubMed] [Google Scholar]
- Bamji SX. Cadherins: actin with the cytoskeleton to form synapses. Neuron. 2005;47:175–178. doi: 10.1016/j.neuron.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Bitzur S, Kam Z, Geiger B. Structure and distribution of N-cadherin in developing zebrafish embryos: morphogenetic effects of ectopic overexpression. Dev Dyn. 1994;201:121–136. doi: 10.1002/aja.1002010204. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, Wolf JJ, Murray BA. Effects of antibodies against N-cadherin and N-CAM on the cranial neural crest and neural tube. Developmental biology. 1992;153:291–301. doi: 10.1016/0012-1606(92)90114-v. [DOI] [PubMed] [Google Scholar]
- Bruses JL. N-cadherin signaling in synapse formation and neuronal physiology. Molecular neurobiology. 2006;33:237–252. doi: 10.1385/MN:33:3:237. [DOI] [PubMed] [Google Scholar]
- Castro CH, Shin CS, Stains JP, Cheng SL, Sheikh S, Mbalaviele G, Szejnfeld VL, Civitelli R. Targeted expression of a dominant-negative N-cadherin in vivo delays peak bone mass and increases adipogenesis. Journal of cell science. 2004;117:2853–2864. doi: 10.1242/jcs.01133. [DOI] [PubMed] [Google Scholar]
- Egan TM, Haines WR, Voigt MM. A domain contributing to the ion channel of ATP-gated P2X2 receptors identified by the substituted cysteine accessibility method. J Neurosci. 1998;18:2350–2359. doi: 10.1523/JNEUROSCI.18-07-02350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern ME, Rhee J, Goll MG, Akitake CM, Parsons M, Leach SD. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish. 2008;5:97–110. doi: 10.1089/zeb.2008.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta K, Tsujii H, Omura T. Cell tracking using a photoconvertible fluorescent protein. Nature protocols. 2006;1:960–967. doi: 10.1038/nprot.2006.96. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov DB, Philippova MP, Tkachuk VA. Structure and functions of classical cadherins. Biochemistry. 2001;66:1174–1186. doi: 10.1023/a:1012445316415. [DOI] [PubMed] [Google Scholar]
- Jontes JD, Emond MR, Smith SJ. In vivo trafficking and targeting of N-cadherin to nascent presynaptic terminals. J Neurosci. 2004;24:9027–9034. doi: 10.1523/JNEUROSCI.5399-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Koga A, Hori H, Shima A. Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene. 1998;225:17–22. doi: 10.1016/s0378-1119(98)00537-x. [DOI] [PubMed] [Google Scholar]
- Kerstetter AE, Azodi E, Marrs JA, Liu Q. Cadherin-2 function in the cranial ganglia and lateral line system of developing zebrafish. Dev Dyn. 2004;230:137–143. doi: 10.1002/dvdy.20021. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kintner C. Regulation of embryonic cell adhesion by the cadherin cytoplasmic domain. Cell. 1992;69:225–236. doi: 10.1016/0092-8674(92)90404-z. [DOI] [PubMed] [Google Scholar]
- Kucenas S, Soto F, Cox JA, Voigt MM. Selective labeling of central and peripheral sensory neurons in the developing zebrafish using P2X(3) receptor subunit transgenes. Neuroscience. 2006;138:641–652. doi: 10.1016/j.neuroscience.2005.11.058. [DOI] [PubMed] [Google Scholar]
- Lele Z, Folchert A, Concha M, Rauch GJ, Geisler R, Rosa F, Wilson SW, Hammerschmidt M, Bally-Cuif L. parachute/n-cadherin is required for morphogenesis and maintained integrity of the zebrafish neural tube. Development (Cambridge, England) 2002;129:3281–3294. doi: 10.1242/dev.129.14.3281. [DOI] [PubMed] [Google Scholar]
- Levine E, Lee CH, Kintner C, Gumbiner BM. Selective disruption of E-cadherin function in early Xenopus embryos by a dominant negative mutant. Development (Cambridge, England) 1994;120:901–909. doi: 10.1242/dev.120.4.901. [DOI] [PubMed] [Google Scholar]
- Liu Q, Duff RJ, Liu B, Wilson AL, Babb-Clendenon SG, Francl J, Marrs JA. Expression of cadherin10, a type II classic cadherin gene, in the nervous system of the embryonic zebrafish. Gene Expr Patterns. 2006a;6:703–710. doi: 10.1016/j.modgep.2005.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Ensign RD, Azodi E. Cadherin-1, -2 and -4 expression in the cranial ganglia and lateral line system of developing zebrafish. Gene Expr Patterns. 2003;3:653–658. doi: 10.1016/s1567-133x(03)00109-1. [DOI] [PubMed] [Google Scholar]
- Liu Q, Liu B, Wilson AL, Rostedt J. cadherin-6 message expression in the nervous system of developing zebrafish. Dev Dyn. 2006b;235:272–278. doi: 10.1002/dvdy.20607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Ferreira-Cornwell M, Baldwin H, Kostetskii I, Lenox J, Lieberman M, Radice G. Rescuing the N-cadherin knockout by cardiac-specific expression of N- or E-cadherin. Development (Cambridge, England) 2001;128:459–469. doi: 10.1242/dev.128.4.459. [DOI] [PubMed] [Google Scholar]
- Malicki J, Jo H, Pujic Z. Zebrafish N-cadherin, encoded by the glass onion locus, plays an essential role in retinal patterning. Developmental biology. 2003;259:95–108. doi: 10.1016/s0012-1606(03)00181-7. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Williams PR, Godinho L, Koerber A, Pittman AJ, Roeser T, Chien CB, Baier H, Wong RO. In vivo imaging reveals dendritic targeting of laminated afferents by zebrafish retinal ganglion cells. Neuron. 2006;52:609–621. doi: 10.1016/j.neuron.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Takeichi M. Neural crest emigration from the neural tube depends on regulated cadherin expression. Development (Cambridge, England) 1998;125:2963–2971. doi: 10.1242/dev.125.15.2963. [DOI] [PubMed] [Google Scholar]
- Ng AN, de Jong-Curtain TA, Mawdsley DJ, White SJ, Shin J, Appel B, Dong PD, Stainier DY, Heath JK. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Developmental biology. 2005;286:114–135. doi: 10.1016/j.ydbio.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. Journal of molecular biology. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- Radice GL, Rayburn H, Matsunami H, Knudsen KA, Takeichi M, Hynes RO. Developmental defects in mouse embryos lacking N-cadherin. Developmental biology. 1997;181:64–78. doi: 10.1006/dbio.1996.8443. [DOI] [PubMed] [Google Scholar]
- Ranscht B. Cadherins: molecular codes for axon guidance and synapse formation. Int J Dev Neurosci. 2000;18:643–651. doi: 10.1016/s0736-5748(00)00030-7. [DOI] [PubMed] [Google Scholar]
- Reber M, Hindges R, Lemke G. Eph receptors and ephrin ligands in axon guidance. Advances in experimental medicine and biology. 2007;621:32–49. doi: 10.1007/978-0-387-76715-4_3. [DOI] [PubMed] [Google Scholar]
- Redies C. Cadherin expression in the developing vertebrate CNS: from neuromeres to brain nuclei and neural circuits. Experimental cell research. 1995;220:243–256. doi: 10.1006/excr.1995.1313. [DOI] [PubMed] [Google Scholar]
- Redies C, Arndt K, Ast M. Expression of the cell adhesion molecule axonin-1 in neuromeres of the chicken diencephalon. The Journal of comparative neurology. 1997;381:230–252. doi: 10.1002/(sici)1096-9861(19970505)381:2<230::aid-cne9>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Redies C, Engelhart K, Takeichi M. Differential expression of N- and R-cadherin in functional neuronal systems and other structures of the developing chicken brain. The Journal of comparative neurology. 1993;333:398–416. doi: 10.1002/cne.903330307. [DOI] [PubMed] [Google Scholar]
- Redies C, Inuzuka H, Takeichi M. Restricted expression of N- and R-cadherin on neurites of the developing chicken CNS. J Neurosci. 1992;12:3525–3534. doi: 10.1523/JNEUROSCI.12-09-03525.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redies C, Takeichi M. Expression of N-cadherin mRNA during development of the mouse brain. Dev Dyn. 1993;197:26–39. doi: 10.1002/aja.1001970104. [DOI] [PubMed] [Google Scholar]
- Simonneau L, Broders F, Thiery JP. N-cadherin transcripts in Xenopus laevis from early tailbud to tadpole. Dev Dyn. 1992;194:247–260. doi: 10.1002/aja.1001940402. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science (New York, NY. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- Tomaselli KJ, Neugebauer KM, Bixby JL, Lilien J, Reichardt LF. N-cadherin and integrins: two receptor systems that mediate neuronal process outgrowth on astrocyte surfaces. Neuron. 1988;1:33–43. doi: 10.1016/0896-6273(88)90207-3. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. Eugene, Oregon: University of Oregon; 2000. [Google Scholar]
- Wheelock MJ, Johnson KR. Cadherin-mediated cellular signaling. Current opinion in cell biology. 2003;15:509–514. doi: 10.1016/s0955-0674(03)00101-7. [DOI] [PubMed] [Google Scholar]
- Zhang LL, Ashwell KW. The development of cranial nerve and visceral afferents to the nucleus of the solitary tract in the rat. Anatomy and embryology. 2001;204:135–151. doi: 10.1007/s004290100185. [DOI] [PubMed] [Google Scholar]