Abstract
New antibiotics to combat the emerging pandemic of drug-resistant strains of Mycobacterium tuberculosis are urgently needed. We have investigated the effects on M. tuberculosis of phosphorothioate-modified antisense oligodeoxyribonucleotides (PS-ODNs) against the mRNA of glutamine synthetase, an enzyme whose export is associated with pathogenicity and with the formation of a poly-l-glutamate/glutamine cell wall structure. Treatment of virulent M. tuberculosis with 10 μM antisense PS-ODNs reduced glutamine synthetase activity and expression by 25–50% depending on whether one, two, or three different PS-ODNs were used and the PS-ODNs' specific target sites on the mRNA. Treatment with PS-ODNs of a recombinant strain of Mycobacterium smegmatis expressing M. tuberculosis glutamine synthetase selectively inhibited the recombinant enzyme but not the endogenous enzyme for which the mRNA transcript was mismatched by 2–4 nt. Treatment of M. tuberculosis with the antisense PS-ODNs also reduced the amount of poly-l-glutamate/glutamine in the cell wall by 24%. Finally, treatment with antisense PS-ODNs reduced M. tuberculosis growth by 0.7 logs (1 PS-ODN) to 1.25 logs (3 PS-ODNs) but had no effect on the growth of M. smegmatis, which does not export glutamine synthetase nor possess the poly-l-glutamate/glutamine (P-l-glx) cell wall structure. The experiments indicate that the antisense PS-ODNs enter the cytoplasm of M. tuberculosis and bind to their cognate targets. Although more potent ODN technology is needed, this study demonstrates the feasibility of using antisense ODNs in the antibiotic armamentarium against M. tuberculosis.
Mycobacterium tuberculosis remains one of the world's most important pathogens. Responsible for millions of new cases of tuberculosis annually (1), it is the leading cause of death from a single infectious agent. The emergence of multidrug-resistant M. tuberculosis underscores the need for new approaches to combat this pathogen (1).
We recently have identified glutamine synthetase [l-glutamate:ammonia ligase (ADP-forming); EC 6.3.1.2] as an important determinant of M. tuberculosis pathogenesis (2–4). An enzyme that plays a central role in nitrogen metabolism in every cell, glutamine synthetase is 1 of 10 proteins released in large quantity into the bacterium's extracellular milieu, whether the bacterium is growing axenically or intraphagosomally in human mononuclear phagocytes, the primary host cells (2). Of great interest, extracellular release of glutamine synthetase is unique to pathogenic mycobacteria and correlated with the presence of a poly-l-glutamate/glutamine (P-l-glx) component in the cell wall of these organisms, suggesting that not only is the enzyme involved in the synthesis of this heteropolymer but also that its presence is significant to virulence (4). Treatment of bacteria with l-methionine-S-sulfoximine, an irreversible inhibitor of glutamine synthetase, results in a decrease in extracellular glutamine synthetase activity, a marked reduction in the amount of cell wall P-l-glx, and inhibition of bacterial growth both in broth culture and in human macrophages (4).
Antisense oligodeoxyribonucleotides (ODNs), which can base pair with a gene's transcript, constitute a new technology for the control of gene expression in prokaryotes and eukaryotes, including mammalian cells (5, 6). In addition, this technology shows promise as a means for developing new chemotherapeutic agents against human diseases, including antibiotics against such pathogens as Plasmodium falciparum, Toxoplasma gondii, and HIV (7–10). An antisense PS-ODN for treatment of cytomegalovirus retinitis in AIDS patients became the first antisense ODN-based drug approved for human use in the U.S. (11).
In this report, we have utilized modified antisense ODNs, in which all internucleoside linkages are phosphorothioates (PS-ODNs), to study the expression and function of M. tuberculosis glutamine synthetase and the feasibility of using antisense PS-ODNs as antimicrobial agents against this pathogen. We find that glutamine synthetase-specific antisense PS-ODNs substantially inhibit the expression and activity of M. tuberculosis glutamine synthetase, the amount of the P-l-glx structure in the mycobacterial cell wall, and bacterial replication. These observations are consistent with entry of the antisense PS-ODNs into the bacterial cytoplasm.
Materials and Methods
PS-ODN Selection and Preparation.
Three target sites for the binding of the antisense PS-ODNs were chosen (Table 1). One site, located near the 5′ end of the glutamine synthetase mRNA, corresponds to codons 4–9 of the glutamine synthetase mRNA translation product, starting from the N-terminal methionine codon GTG. The other two sites, located in the vicinity of a catalytically important histidine residue (H276), correspond to codons 269–275 and 275–282, respectively. Antisense PS-ODNs were synthesized by using standard phosphoroamidite chemistry. Phosphorothioate bonds were introduced by oxidation with the Beaucage thiolating reagent (12), and assembled PS-ODNs were purified by HPLC and lyophilized. Amikacin derivatives were synthesized by a phosphorothioate-based methodology, linking the antibiotic via one of its amino groups to a phosphate residue at the end of a 3′-attached 18-atom spacer arm. PS-ODN stock solutions were prepared just before use and added to mycobacterial cultures after filter sterilization.
Table 1.
Target sites of M. tuberculosis glutamine synthetase-specific antisense PS-ODNs
| Antisense PS-ODN 4–9 | Antisense PS-ODN 269–275 | Antisense PS-ODN 275-282 | |
|---|---|---|---|
| glnA1 region 5′ > 3′ | rowAAGACGCCCGACGACGTC | cellGGCGACAACGGGTCCGGCATG | cellATGCACTGTCATCAGTCGCTGTGG |
| glnA1 mRNA 5′ > 3′ | rowAAGACGCCCGACGACGUC | rwoGGCGACAACGGGUCCGGCAUG | cellAUGCACUGUCAUCAGUCGCUGUGG |
| PS-ODN sequence 5′ > 3′ | rowGACGTCGTCGGGCGTCTT | cellCATGCCGGACCCGTTGTCGCC | cellCCACAGCGACTGATGACAGTGCAT |
| PS-ODN length | 18 nt | 21 nt | 24 nt |
| PS-ODN GC content | 66.7% | 71.4% | 54.2% |
| Propensity of PS-ODN to remain linear | Weak | Strong | Moderate |
glnA1, glutamine synthetase gene.
Bacterial Cultures.
M. tuberculosis strain Erdman (ATCC 35801) and Mycobacterium smegmatis 1–2c (13) were cultured in 7H9 medium (Difco) supplemented with 2% glucose at 37°C. M. tuberculosis was maintained in a 5% CO2/95% air atmosphere as unshaken cultures (because of safety considerations), and M. smegmatis was maintained at ambient conditions with vigorous shaking. For studies of the effect of PS-ODNs on the bacteria, M. tuberculosis and M. smegmatis were cultured in duplicate in 1 ml, 2 ml, or 5 ml of 7H9 broth in polystyrene tubes (Fisher) or tissue culture flasks (Costar) in the presence of medium alone, PS-ODNs at final concentrations of 0.1, 1, or 10 μM, or PS-ODNs plus subinhibitory concentrations of the antibiotics amikacin (0.058, 0.58, and 5.8 μg/ml), ethambutol (0.1, 0.25, and 0.5 μg/ml), or polymyxin B nonapeptide (0.1, 0.25, 0.5 μg/ml). All antibiotics were from Sigma. The minimal inhibitory concentrations (MIC) of the antibiotics for M. tuberculosis and M. smegmatis were established in bacterial cultures grown under the same conditions in the presence of antibiotics ranging in concentration from 0.01 to 32 μg/ml for amikacin and 0.1 to 25 μg/ml for ethambutol and polymyxin B nonapeptide.
Effect of PS-ODNs on Glutamine Synthetase Activity and Expression.
Aliquots were removed at weekly (M. tuberculosis) or daily (M. smegmatis) intervals from the cultures, and glutamine synthetase activity was assayed by transfer assay, calibrated with the purified M. tuberculosis enzyme representing 1 × 108 cell equivalents. An enzyme unit was defined as the amount of glutamine synthetase that produced 1 μmol γ-glutamylhydroxamate per min at 37°C (2). Proteins were analyzed by denaturing PAGE, and protein concentrations were determined by the bicinchoninic acid reagent (Pierce). The integrity of the bacteria was monitored by assaying the activity of the cytoplasmic marker protein lactate dehydrogenase during the 6-week/6-day growth period using a commercially available diagnostic kit (Sigma). Extracellular lactate dehydrogenase activity never exceeded 0.2% of intracellular activity.
Effect of PS-ODNs on the Amount of Cell Wall P-l-glx.
Bacterial cultures, inoculated at 1 or 5 × 105 cells/ml and grown for 6 weeks (M. tuberculosis) or 6 days (M. smegmatis), were analyzed for the amount of P-l-glx and d,l-alanine in the cell wall as described (14). Briefly, disrupted cells were treated with trypsin and chymotrypsin, delipidated by acetone and chloroform/methanol extractions, and acid-hydrolyzed. The redissolved hydrochloric acid precipitate contained the P-l-glx fraction, whereas the insoluble portion contained the peptidoglycan chain. Both fractions were analyzed for their amino acid content after complete acid hydrolysis.
Effect of PS-ODNs on the Growth of M. tuberculosis and M. smegmatis.
Growth of the cultures was monitored weekly (M. tuberculosis) or daily (M. smegmatis) by gently sonicating the cultures to break up clumped bacteria, removing a small aliquot, washing the bacteria, plating serial dilutions of the washed bacteria on 7H11 agar (Difco), and enumerating colony-forming units (cfu) after incubation for 2 weeks (M. tuberculosis) or 3 days (M. smegmatis). SDs are given in the relevant figure legends.
Results
Selection of PS-ODN Targets and Stoichiometric Considerations.
M. tuberculosis contains four genetic loci with domains that exhibit homologies to glutamine synthetase genes from other bacteria (2, 15). However, in the Erdman strain, and presumably other clinical strains, only one gene, designated glnA1 and located at map position Rv2220 of the M. tuberculosis H37Rv genome (15), expresses an active glutamine synthetase. This enzyme has an apparent molecular mass of ≈680,000 Da (12 subunits of ≈56,000 Da each) (2). It is not known whether the other loci (glnA2, glnA3, glnA4) are expressed. M. tuberculosis and other pathogenic mycobacteria express large amounts of the enzyme and export a large proportion of what is produced—one-third in the case of M. tuberculosis (2, 3). Nonpathogenic mycobacteria such as M. smegmatis and Mycobacterium phlei typically exhibit a lower glutamine synthetase expression level and export less than 1/100th of the total enzyme produced (2).
Because the rate of transcription under standard growth conditions is nearly constant (3) and the downstream effects of inhibition by antisense PS-ODNs, i.e., a decrease in detectable enzyme and its activity, can be tracked continuously and quantitatively, the glutamine synthetase transcript is a logical target with which to study regulation of gene expression by antisense PS-ODNs.
The glutamine synthetase coding region contains 1,434 bp, excluding the stop codon. The primary transcript under standard axenic growth conditions is 1,500–1,600 nt in length (3). Based on densitometric analyses of our Northern hybridizations, we calculated that ≈11 glutamine synthetase gene transcripts of ≈1,550 nt are present per cell.
We selected three target sites, one near the 5′ end of the glutamine synthetase transcript and two at a region encoding catalytically important residues. All three mRNA regions are devoid of any stable secondary structures, and, thus, they are theoretically readily accessible to complementary PS-ODNs. However, the possibility that the in vivo structure of the mRNA is much more complex because of the binding of proteins cannot be excluded. All PS-ODNs were at least 18 bases in length, making random hybridization to the M. tuberculosis genome [4.4 × 106 bp (15)] and even partial hybridization of short sequences very unlikely.
Effect of the Antisense PS-ODNs on the Detectable Activity and Amount of Glutamine Synthetase in M. tuberculosis Erdman.
We first cultured M. tuberculosis in 1 or 2 ml of broth medium for 2 weeks to provide a sufficient number of bacteria for reliable measurements of both intra- and extracellular glutamine synthetase activity, i.e., ≥5 × 106 cell equivalents per assay. We then inoculated the cultures with 0.1, 1, and 10 μM of the following PS-ODNs: (i) each antisense PS-ODN alone; (ii) combinations of 2 PS-ODNs; and (iii) the combination of all three PS-ODNs. We inoculated an identical set of cultures with antisense PS-ODNs containing amikacin tethered to the 3′ end and a third set of cultures with all the corresponding nonsense controls (all codons mismatched at the second nt position) to each of the three antisense PS-ODNs. We assayed the cultures for enzyme activity from week 2 to 6 (Fig. 1).
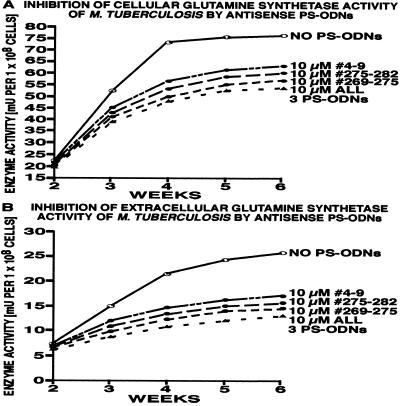
Figure 1.
Inhibition of M. tuberculosis Erdman glutamine synthetase activity by antisense PS-ODNs. Duplicate M. tuberculosis Erdman cultures were grown for 2 weeks in 1–2 ml of 7H9 medium alone and thereafter in the absence or presence of antisense PS-ODNs, individually or combined, at concentrations of 10 μM. At each time point indicated, cultures were harvested and analyzed for glutamine synthetase activity by the transfer assay as described (2). All data points had a SD of ≤10%, even when different PS-ODN preparations were used.
The results of these experiments can be summarized as follows. (i) At 10 μM antisense PS-ODN, the only effective concentration, the cellular glutamine synthetase activity decreased at 6 weeks from 76.5 milliunits per 108 cells (mU) in uninhibited cultures to 61.3 mU for PS-ODN 4–9, 57.1 mU for PS-ODN 275–282, and 54.8 mU for PS-ODN 269–275 (SD < 10% for all data). Combinations of 2 or 3 PS-ODNs at 10 μM each were slightly more inhibitory than single PS-ODNs; the combination of all 3 PS-ODNs was the most inhibitory, decreasing the activity by ≈33% to 51.4 mU. Interestingly, the inhibitory capacity of a PS-ODN was directly related to its propensity to remain in a linear conformation (Table 1), as assessed by the Sigma–Genosys oligo-5 secondary structure analysis program. Whereas the most effective PS-ODN (PS-ODN 269–275) has a strong potential to maintain a linear structure, the second most effective PS-ODN (PS-ODN 275–282) has a moderate potential to do so, and the least effective PS-ODN (PS-ODN 4–9) has a weak potential to do so. (ii) In the presence of antisense PS-ODNs, extracellular glutamine synthetase activity decreased from 25.8 mU under standard growth conditions to 17.1 mU in the presence of PS-ODN 4–9, 15.6 mU in the presence of PS-ODN 275–282, and 15.1 mU in the presence of PS-ODN 269–275. Combinations of 2 PS-ODNs were not significantly more inhibitory than 1 PS-ODN; however, the combination of all 3 PS-ODNs decreased enzyme activity to 13.3 mU, a reduction of 50% below the level of an uninhibited culture (SD < 10% for all data). Possibly, at these high concentrations of antisense PS-ODNs, the amount of intracellularly expressed enzyme is lowered to the point that the cell is no longer able to export the same proportion of enzyme molecules as it normally does. (iii) PS-ODNs tethered to an amikacin moiety, in theory to guide PS-ODNs to the ribosomes, had no greater inhibitory effect than PS-ODNs not tethered to amikacin in this as well as in all subsequent studies. Once tethered to a PS-ODN, the antibiotic itself was completely inactive, even though, at a PS-ODN concentration of 10 μM, it was present at a concentration (5.8 μg/ml) at which free amikacin is strongly bactericidal. Consistent with this, nonsense PS-ODNs tethered to amikacin had no growth-inhibitory effect whatsoever at 10 or 1 μM. (iv) At all concentrations and combinations tested, control PS-ODNs did not affect the detectable enzyme activity. (v) The observed reductions in detectable enzyme activity were mirrored by reductions in the amount of glutamine synthetase protein present on Coomassie blue-stained polyacrylamide gels after electrophoresis of bacterial cells and culture filtrates.
In addition to the experiments described above, we investigated the effect on glutamine synthetase expression of the antisense PS-ODN 269–275 with a single nt mismatch at codon 270 (third nt position) of the glutamine synthetase target site. This PS-ODN had the same effect on glutamine synthetase expression and M. tuberculosis growth as the perfectly matched antisense PS-ODN (data not shown).
Specifity of Antisense PS-ODNs for M. tuberculosis Glutamine Synthetase mRNA.
To confirm the specificity of the antisense PS-ODNs, we studied their effect on the transcript of glnA1 of M. smegmatis, a closely related gene [≈70% DNA sequence similarity (M. V. Tullius, G.H., and M.A.H., unpublished data)]. At the target sites, nucleotide differences between the two mycobacterial species are sufficient to anticipate that the antisense PS-ODNs, based on the DNA sequence of the M. tuberculosis glnAl gene, would bind only to the M. tuberculosis gene (Table 1). The aligned RNA sequences demonstrate that the target site for PS-ODN 4–9 is different in 3 nt positions (5′-AAG ACG CCC GAC GAC GUC-3′ for M. tuberculosis and 5′-AAG ACG UCG GAC GAC AUC-3′ for M. smegmatis), the target site for PS-ODN 269–275 is different in 2 nt positions (5′-GGC GAC AAC GGG UCC GGC AUG-3′ for M. tuberculosis and 5′-GGC GAC AAC GGU UCG GGC AUG-3′ for M. smegmatis), and the target site for PS-ODN 275–282 is different in 4 nt positions (5′-AUG CAC UGU CAU CAG UCG CUG UGG-3′ for M. tuberculosis and 5′-AUG CAC GCC CAC CAG UCG CUG UGG-3′ for M. smegmatis). To compare the effect of the antisense PS-ODNs on M. smegmatis and M. tuberculosis glutamine synthetase mRNA, we added the PS-ODNs to cultures of the M. smegmatis 1–2c wild-type strain and its recombinant isotype expressing the M. tuberculosis Erdman glutamine synthetase from a mycobacterial shuttle vector (3). We added the PS-ODNs to M. smegmatis cultures after 2 days of growth and monitored the cultures for 4 days, by which time they had reached stationary phase. We differentiated the recombinant and endogenous glutamine synthetases by analyzing the first amino acid residue of each of the 12 identical enzyme subunits, a threonine in M. tuberculosis and an alanine in M. smegmatis (3). Moreover, virtually all extracellular enzyme activity in recombinant M. smegmatis cultures is exported recombinant M. tuberculosis glutamine synthetase (3).
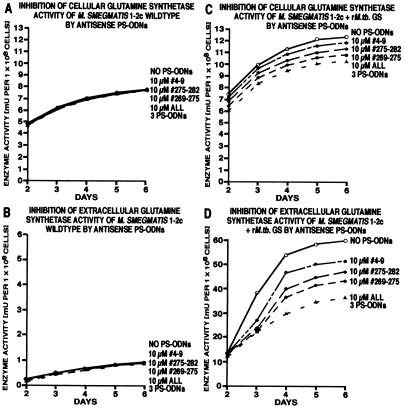
The results of these experiments can be summarized as follows (Fig. 2). (i) At 10 μM antisense PS-ODNs (perfectly matched with the recombinant M. tuberculosis transcript but mismatched at 2–4 nt positions with the endogenous M. smegmatis transcript), the cellular glutamine synthetase activity in the parent strain remained essentially unchanged at 7.2 mU per 1 × 108 cells when compared with uninhibited cultures (Fig. 2A). Neither the various combinations of 2 PS-ODNs nor the addition of all 3 PS-ODNs decreased the detectable cellular enzyme activity. Similarly, extracellular glutamine synthetase activity remained unchanged in the parent strain at barely detectable levels amounting to ≤0.1 mU of enzyme activity (Fig. 2B). (ii) In contrast, the cellular glutamine synthetase activity of the recombinant strain, 60% of which is endogenous M. smegmatis enzyme activity and 40% is recombinant M. tuberculosis enzyme activity, decreased from 12.4 mU in uninhibited cultures to 11.7 mU for cultures growing in the presence of PS-ODN 4–9, 11.4 mU in the presence of PS-ODN 275–282, 11.0 mU in the presence of PS-ODN 269–275, and 10.7 mU in the presence of all 3 PS-ODNs (Fig. 2C). Assuming the entire 1.7-mU decrease in cellular enzyme activity in the presence of 3 PS-ODNs only involved recombinant M. tuberculosis glutamine synthetase (≈5 mU in a total of 12.4 mU), this fraction of activity was decreased 34%, similar to the decrease observed in M. tuberculosis cultures. The extracellular enzyme activity of the recombinant M. smegmatis strain, of which ≥99% is due to the export of recombinant M. tuberculosis glutamine synthetase, decreased from 61.9 mU in uninhibited cultures to 55.2 mU in the presence of PS-ODN 4–9, 47.3 mU in the presence of PS-ODN 275–282, 44.6 mU in the presence of PS-ODN 269–275, and 36.7 mU in the presence of all 3 PS-ODNs (SD < 15% for all data). The 41% reduction in the presence of all 3 PS-ODNs was similar to the decrease observed in M. tuberculosis cultures (Fig. 2D). Combinations of 2 or 3 PS-ODNs at 10 μM each were somewhat better than single PS-ODNs. (iii) At all concentrations and combinations tested, the control PS-ODNs did not affect the detectable enzyme activity. (iv) The observed reduction in detectable enzyme activity in recombinant M. smegmatis (Fig. 2D) was mirrored by a reduction in the detectable amount of glutamine synthetase protein molecules (data not shown). It is not known whether regulation of glutamine synthetase expression in M. smegmatis is patterned according to the mechanisms governing glutamine synthetase expression in M. tuberculosis (3).
Figure 2.
Inhibition of glutamine synthetase activity in M. smegmatis 1–2c wild type and its recombinant isotype expressing the M. tuberculosis glutamine synthetase. Duplicate M. smegmatis cultures were grown for 2 days in 1–2 ml of 7H9 medium alone and thereafter in the absence or presence of antisense PS-ODNs, individually or combined, at concentrations of 10 μM. At each time point indicated, cultures were harvested and analyzed for glutamine synthetase activity by the transfer assay as described (2). Note that the cellular enzyme activity of the recombinant strain is a mixture of two enzyme activities—the endogenous glutamine synthetase (60%) and recombinant M. tuberculosis glutamine synthetase (40%). In contrast, the extracellular enzyme activity of the recombinant strain is almost exclusively M. tuberculosis glutamine synthetase (≥99%). All data points had a SD of ≤15%.
Our finding that expression of recombinant cellular and exported M. tuberculosis glutamine synthetase is decreased in M. smegmatis to approximately the same degree as in M. tuberculosis suggests strongly that the PS-ODNs are taken up into M. smegmatis and bind to their cognate target.
Determination of the Amount of P-l-glx in the Cell Wall of M. tuberculosis.
Pathogenic, but not nonpathogenic mycobacteria, release abundant amounts of glutamine synthetase and contain a large amount of P-l-glx in the bacterial cell wall. The high correlation between glutamine synthetase export and the presence of P-l-glx in the cell wall suggests the possibility that extracellular glutamine synthetase is involved in the synthesis of this cell wall structure. If so, then inhibition of extracellular glutamine synthetase might reduce the amount of this structure. We investigated this hypothesis by determining the amount of this cell wall structure in M. tuberculosis grown in the presence of various antisense and control PS-ODNs at their effective concentration of 10 μM. The heteropolymer was isolated from the mycobacterial cell wall to a purity of 90–95% and, along with the copurified mycobacterial peptidoglycan moiety, subjected to total amino acid hydrolysis (Table 2).
Table 2.
Effect of antisense PS-ODNs on the M. tuberculosis cell wall component P-l-glx and the peptidoglycan chain
| P-l-glx, μg/1 × 1010 bacteria | d,l-alanine, μg/1 × 1010 bacteria | |
|---|---|---|
| Wild type | 50.3 ± 0.6† | 20.6 ± 0.4 |
| PS-ODN 4–9* | 46.7 ± 0.6 | 19.0 ± 0.6 |
| PS-ODN 269–275 | 43.5 ± 1.6 | 18.5 ± 0.4 |
| PS-ODNs 4–9 + 269–275 | 38.3 ± 1.1 | 17.5 ± 0.4 |
| Control PS-ODNs 4–9 + 269–275 + 275–282 | 49.5 ± 0.9 | 20.4 ± 0.5 |
*All PS-ODNs used at 10 μM.
†Mean ± SD for two independent analyses.
PS-ODN 4–9 or 269–275, separately or in combination, reduced the amount of P-l-glx from 50.3 μg per 1 × 1010 cells in untreated control cultures to 46.7 μg in the presence of PS-ODN 4–9, 43.5 μg in the presence of PS-ODN 269–275, and 38.3 μg in the presence of both PS-ODNs. The combination of all control PS-ODNs together did not significantly decrease the amount of this structure (Table 2). Quantitatively, the decrease in P-l-glx (23.8 ± 3.1%) compares favorably with the decrease in detectable extracellular glutamine synthetase activity described above. Because the isolation procedure resulted in the hydrolysis of glutamine, we could not determine the exact ratio of glutamate/glutamine residues in the heteropolymer.
By comparison, PS-ODNs decreased the amount of d,l-alanine (present in equimolar amounts) in the peptidoglycan fraction (≥90% homogeneous) from 20.5 μg per 1 × 1010 cells in control cultures to 19.0 μg in the presence of PS-ODN 4–9, 18.5 μg in the presence of PS-ODN 269–275, and 17.4 μg in the presence of both PS-ODNs, a reduction of 15.1 ± 0.3%. The combination of all control PS-ODNs did not decrease the amount of this structure. These results indicated that the inhibitory effect of the PS-ODNs was somewhat greater on P-l-glx than on d,l-alanine.
Effect of Antisense PS-ODNs on M. tuberculosis and M. smegmatis Proliferation.
In the course of evaluating the cellular and extracellular glutamine synthetase activity of the two mycobacterial species cultured in the presence of various combinations of antisense PS-ODNs at concentrations of 10 μM, we noticed a reduction in cell density in M. tuberculosis but not M. smegmatis cultures after their standard growth period. We therefore explored the capacity of antisense PS-ODNs to inhibit bacterial growth. We added PS-ODNs to cultures of M. tuberculosis Erdman and cultures of both the parent and recombinant strain of M. smegmatis and quantified cfu weekly for 6 weeks (M. tuberculosis) or daily for 6 days (M. smegmatis) (Fig. 3).
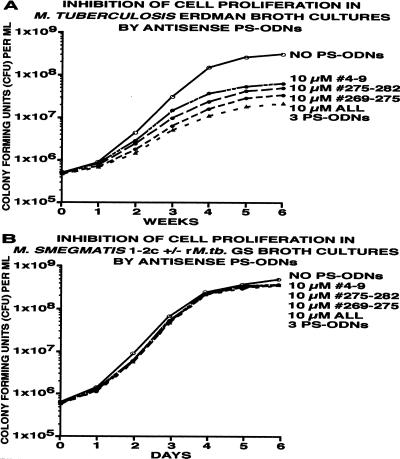
Figure 3.
Inhibition of cell proliferation of M. tuberculosis Erdman and M. smegmatis 1–2c wild type and recombinant strain by antisense PS-ODNs. Duplicate bacterial cultures were grown for 6 weeks (M. tuberculosis) or 6 days (M. smegmatis) in 1–2 ml of 7H9 medium in the presence of the antisense PS-ODNs, individually or combined, at concentrations of 10 μM. At each time point, cultures were harvested, washed, serially diluted, and plated on 7H11 agar medium. Viable bacteria were enumerated after an incubation period of 2 weeks (M. tuberculosis) or 3 days (M. smegmatis). SDs varied between 15 and 20%.
All three antisense PS-ODNs exerted a substantial inhibitory effect on M. tuberculosis growth; inhibited cultures lagged 0.7–0.9 log units (±0.1–0.2 log units; SD) behind uninhibited cultures at the end of the growth period (Fig. 3A). The magnitude of an individual PS-ODN′s capacity to inhibit growth paralleled its capacity to inhibit M. tuberculosis glutamine synthetase activity: PS-ODN 269–275 > 275–282 > 4–9. Combinations of 2 PS-ODNs had a slightly greater inhibitory effect than single PS-ODNs, decreasing growth by up to 1.0 log units, and the combination of all three PS-ODNs decreased growth by 1.25 log units (±0.2 log units; SD). Interestingly, in contrast to the results with glutamine synthetase activity, at a concentration of 1 μM, there was a small but measurable growth-inhibitory effect of the PS-ODNs, 0.1–0.25 log units. There was no evidence that any of the M. tuberculosis cultures were able to develop resistance to the antisense PS-ODNs. At all concentrations and combinations tested, the control PS-ODNs did not affect M. tuberculosis multiplication (data not shown).
None of the antisense or control PS-ODNs (mismatched at 2–4 nt positions with the endogenous M. smegmatis enzyme), in the range of 0.1–10 μM, in any possible combination, either in the free form or tethered to an amikacin moiety, had any growth-inhibitory effect on either strain of M. smegmatis (Fig. 3B).
Effect of Combinations of Antituberculous Drugs and Antisense ODNs on M. tuberculosis and M. smegmatis Proliferation.
Several antituberculous drugs, such as isoniazid or ethambutol, affect the membrane metabolism of M. tuberculosis (16, 17). At high drug concentrations, the bacteria lyse. We reasoned that subinhibitory concentrations of such drugs might “soften” the cell wall and promote the influx of PS-ODNs, thereby enhancing their effectiveness. Two drugs, ethambutol and polymyxin B nonapeptide, were studied.
We first determined that the MIC of ethambutol was ≈5 μg/ml for M. tuberculosis and ≈10 μg/ml for M. smegmatis and that the MIC of polymyxin B nonapeptide was ≈5 μg/ml for both species. The MICs for the parent and recombinant strains of M. smegmatis were identical.
We then incubated the three strains of bacteria with subinhibitory concentrations of 0, 0.1, 0.25, and 0.5 μg/ml ethambutol, polymyxin B nonapeptide, or both antibiotics and with or without each PS-ODN alone or all 3 PS-ODNs in combination.
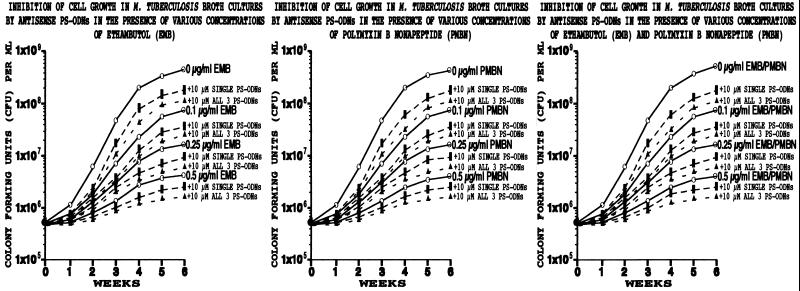
The addition of antisense PS-ODNs to M. tuberculosis cultures containing subinhibitory concentrations of one or both antibiotics resulted in additional growth inhibition beyond that caused by the antibiotic alone (Fig. 4). However, the incremental increase in growth inhibition was not greater than the amount of growth inhibition observed in cultures treated with PS-ODNs in the absence of antibiotics. Indeed, with increasing concentrations of antibiotic, the magnitude of the incremental increase in growth inhibition caused by the PS-ODNs diminished.
Figure 4.
Inhibition of cell proliferation of M. tuberculosis by antisense PS-ODNs in the presence of various concentrations of ethambutol or polymyxin B nonapeptide. Duplicate bacterial cultures were grown for 6 weeks in 1–2 ml of 7H9 medium in the presence of the antisense PS-ODNs, individually or combined, at concentrations of 10 μM and 0, 0.1, 0.25, or 0.5 μg/ml ethambutol (EMB) and/or polymyxin B nonapeptide (PMBN) to “soften” the cell wall. At each time point, cultures were harvested, washed, serially diluted, plated on 7H11 agar medium, and cfu enumerated after 2 weeks. SDs varied between 15 and 20%. Values for single PS-ODNs were combined to yield one dashed line; the range of values for single PS-ODNs is indicated by the broad, solid, vertical bar.
None of the antisense or control PS-ODNs, in the range of 0.1–10 μM, in any possible combination, either in their free form or tethered to an amikacin moiety, enhanced the growth-inhibitory effects of ethambutol and/or polymyxin B nonapeptide on either strain of M. smegmatis (data not shown).
Discussion
This study demonstrates the feasibility of using antisense PS-ODN technology to combat M. tuberculosis, an organism whose cell wall presents a formidable barrier to the entry of most biomolecules. The antisense PS-ODNs substantially altered M tuberculosis metabolism and growth, undoubtedly by entering the bacterial cytoplasm and binding to their cognate targets. The effect of the antisense PS-ODNs on M. tuberculosis was highly specific. Whereas the antisense PS-ODNs inhibited the growth of M. tuberculosis and the expression of its glutamine synthetase, they had no effect on M. smegmatis growth or the expression of its closely related glutamine synthetase. Moreover, control PS-ODNs had no effect on M. tuberculosis growth or glutamine synthetase expression.
The antisense PS-ODNs exhibit three simultaneous effects on M. tuberculosis: reduction in intracellular and extracellular glutamine synthetase activity, reduction in the formation of the P-l-glx cell wall structure, and inhibition of bacterial growth. Except that it acts almost exclusively on extracellular glutamine synthetase, the glutamine synthetase inhibitor l-methioninine-S-sulfoximine exerts parallel effects on M. tuberculosis (4), and we have postulated that the reduction in the P-l-glx heteropolymer is a consequence of reduced extracellular glutamine synthetase activity. It seems possible that the PS-ODNs reduce the amount of the heteropolymer by the same mechanism. The reduction in the heteropolymer, in turn, likely affects mycobacterial growth by disturbing the integrity of the cell wall. As an aside, on a more speculative note, the recent observations that poly-l-glutamine degradation in mammalian cells results in intracellular protein aggregation raises the possibility of a novel mechanism by which M. tuberculosis might subvert host cell function—release of portions of its cell wall P-l-glx (18–20).
Although the effects of the antisense PS-ODNs on M. tuberculosis were highly consistent, they were relatively weak compared, for example, with l-methionine-S-sulfoximine (4). At 2 μM, the latter compound inhibited bacterial growth by 1.5–2 log units, and at 20 μM, it reduced bacterial growth by ≥2.5 log units, extracellular glutamine synthetase activity by 80%, and the amount of the P-l-glx cell wall structure by 93%. In contrast, the combination of all 3 PS-ODNs at a concentration of 10 μM each, the most inhibitory preparation, reduced bacterial growth by only 1.25 logs, extracellular glutamine synthetase activity by only 48%, and the amount of the heteropolymer by only 24%. Even if inhibition is incomplete, the use of concentrations of PS-ODNs higher than 10 μM generally is not desirable because of the rise in non-sequence-specific interactions with host proteins and nucleic acids (8, 21, 22).
Two potential explanations for the weak effect of the antisense PS-ODNs in our study are inefficient uptake across the mycobacterial cell wall and intracellular instability. With respect to uptake, both strategies to improve PS-ODN uptake—tethering the PS-ODN to an amikacin moiety and “softening” the mycobacterial cell wall with antibiotics—were unsuccessful. In future studies, we hope to improve transport of PS-ODNs across the mycobacterial cell wall by using alternative transporters such as a covalently linked ethambutol residue (23).
Although the experiments were not designed to investigate rigorously the effect of mismatches in the PS-ODNs, the data suggested that the antisense PS-ODNs had to be highly specific for their target site. PS-ODNs perfectly matched with the M. tuberculosis glutamine synthetase mRNA transcript but mismatched at 2–4 nt positions with the M. smegmatis glutamine synthetase transcript inhibited expression of the recombinant M. tuberculosis enzyme but not the endogenous M. smegmatis enzyme in M. smegmatis. However, one mismatch was tolerated because a PS-ODN mismatched at 1 nt with the M. tuberculosis mRNA transcript inhibited both expression of the enzyme in M. tuberculosis and bacterial growth to the same extent as its perfectly matched PS-ODN counterpart.
Finally, our study demonstrates that antisense PS-ODNs can add to the inhibitory effect of conventional antibiotics such as ethambutol and/or polymyxin B nonapeptide. Tuberculosis typically is treated with a mixture of antibiotics to prevent the emergence of resistant organisms. Although in this study, because of safety considerations, we demonstrated an effect of antisense PS-ODNs against a drug-sensitive M. tuberculosis strain, it is likely that the PS-ODNs also would be effective against drug-resistant strains because resistance mechanisms generally are antibiotic-specific and PS-ODNs differ substantially from conventional antibiotics.
Pharmacokinetic studies by Agrawal and colleagues indicate that therapeutic concentrations of ODNs are achievable in blood and tissues at nontoxic doses (22, 24); tissue concentrations remain relatively high for several days. With respect to tuberculosis, it is noteworthy that when FITC-labeled ODNs are administered i.v. to mice, the ODNs are preferentially taken up by mononuclear phagocytes (25), the host cells for M. tuberculosis. Thus, our study and the pharmacologic work of Agrawal and colleagues (22, 24, 25) suggest the feasibility of employing modified nuclease-resistant ODNs as part of a mixture of antituberculous drugs.
Acknowledgments
We thank Barbara Jane Dillon for expert technical assistance with the PS-ODN inhibition assays and Audree Fowler of the University of California at Los Angeles Protein Microsequencing Facility for performing amino acid analyses on M. tuberculosis and M. smegmatis glutamine synthetases and cell wall P-l-glx. The facility is supported by Grant 1 S10RR05554 from the National Institutes of Health. This work was supported by Grants AI 42925 and AI 31338 from the National Institutes of Health. P.Z. is a member of the Board of Directors of and a consultant to Hybridon, Inc.
Abbreviations
- cfu
colony-forming unit
- nt
nucleotide
- ODN
oligodeoxyribonucleotide
- P-l-glx
poly-l-glutamate/glutamine
- PS-ODN
phosphorothioate-modified ODN
- MIC
minimal inhibitory concentration
- mU
milliunits
References
- 1.Pablo-Mendez A, Raviglione M C, Laszlo A, Binkin N, Rieder H L, Bustreo F, Cohn D L, Lambregts-van Weezenbeek C S B, Kim S J, Chaulet P, et al. New Engl J Med. 1998;338:1641–1649. doi: 10.1056/NEJM199806043382301. [DOI] [PubMed] [Google Scholar]
- 2.Harth G, Clemens D L, Horwitz M A. Proc Natl Acad Sci USA. 1994;91:9342–9346. doi: 10.1073/pnas.91.20.9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harth G, Horwitz M A. J Biol Chem. 1997;272:22728–22735. doi: 10.1074/jbc.272.36.22728. [DOI] [PubMed] [Google Scholar]
- 4.Harth G, Horwitz M A. J Exp Med. 1999;189:1425–1435. doi: 10.1084/jem.189.9.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zamecnik P C, Stephenson M L. Proc Natl Acad Sci USA. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephenson M L, Zamecnik P C. Proc Natl Acad Sci USA. 1978;75:285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zamecnik P C. Antisense Nucleic Acid Drug Dev. 1997;7:199–202. doi: 10.1089/oli.1.1997.7.199. [DOI] [PubMed] [Google Scholar]
- 8.Barker R H, Jr, Metelev V, Rapaport E, Zamecnik P. Proc Natl Acad Sci USA. 1996;93:514–518. doi: 10.1073/pnas.93.1.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakaar V, Samuel B U, Ngo E O, Joiner K A. J Biol Chem. 1999;274:5083–5087. doi: 10.1074/jbc.274.8.5083. [DOI] [PubMed] [Google Scholar]
- 10.Lisziewicz J, Sun D, Klotman M, Agrawal S, Zamecnik P, Gallo R C. Proc Natl Acad Sci USA. 1992;89:11209–11213. doi: 10.1073/pnas.89.23.11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson K P, Fox M C, Brown-Driver V, Martin M J, Azad R F. Antimicrobiol Agents Chemother. 1996;40:2004–2011. doi: 10.1128/aac.40.9.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padmapriya A A, Tang J, Agrawal S. Antisense Res Dev. 1994;4:185–199. doi: 10.1089/ard.1994.4.185. [DOI] [PubMed] [Google Scholar]
- 13.Garbe T R, Barathi J, Barnini S, Zhang Y, Abou-Zeid C, Tang D, Mukherjee R, Young D B. Microbiology. 1994;140:133–138. doi: 10.1099/13500872-140-1-133. [DOI] [PubMed] [Google Scholar]
- 14.Wietzerbin J, Lederer F, Petit J-F. Biochem Biophys Res Commun. 1975;62:246–252. doi: 10.1016/s0006-291x(75)80130-6. [DOI] [PubMed] [Google Scholar]
- 15.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, III, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 16.Mdluli K, Slayden R A, Zhu Y, Ramaswamy S, Pan X, Mead D, Crane D D, Musser J M, Barry C E., III Science. 1998;280:1607–1610. doi: 10.1126/science.280.5369.1607. [DOI] [PubMed] [Google Scholar]
- 17.Telenti A, Philipp W J, Sreevatsan S, Bernasconi C, Stockbauer K E, Wieles B, Musser J M, Jacobs W R., Jr Nat Med. 1997;3:567–570. doi: 10.1038/nm0597-567. [DOI] [PubMed] [Google Scholar]
- 18.Green H. Cell. 1993;74:955–956. doi: 10.1016/0092-8674(93)90718-6. [DOI] [PubMed] [Google Scholar]
- 19.Karpuj M V, Garren H, Slunt H, Price D L, Gusella J, Becher M W, Steinman L. Proc Natl Acad Sci USA. 1999;96:7388–7393. doi: 10.1073/pnas.96.13.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Proc Natl Acad Sci USA. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein C A, Chen Y-C. Science. 1993;261:1004–1012. doi: 10.1126/science.8351515. [DOI] [PubMed] [Google Scholar]
- 22.Agrawal S, Zhao Q. Curr Opin Chemical Biol. 1998;2:519–528. doi: 10.1016/s1367-5931(98)80129-4. [DOI] [PubMed] [Google Scholar]
- 23.Rapaport E, Levina A, Metelev V, Zamecnik P C. Proc Natl Acad Sci USA. 1996;93:709–713. doi: 10.1073/pnas.93.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agrawal S, Zhang R. CIBA Found Symp. 1997;209:60–78. doi: 10.1002/9780470515396.ch6. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Q, Zhou R, Temsamani J, Zhang Z, Roskey A, Agrawal S. Antisense Nucleic Acid Drug Dev. 1998;8:451–458. doi: 10.1089/oli.1.1998.8.451. [DOI] [PubMed] [Google Scholar]