Abstract
Lamellipodia of crawling cells represent both the motor for cell advance and the primary building site for the actin cytoskeleton. The organization of actin in the lamellipodium reflects actin dynamics and is of critical importance for the mechanism of cell motility. In previous structural studies, the lamellipodial actin network was analyzed primarily by electron microscopy (EM). An understanding of lamellipodial organization would benefit significantly if the EM data were complemented and put into a kinetic context by establishing correspondence with structural features observable at the light microscopic level in living cells. Here, we use an enhanced phase contrast microscopy technique to visualize an apparent long-range diagonal actin meshwork in the advancing lamellipodia of living cells. Visualization of this meshwork permitted a correlative light and electron microscopic approach that validated the underlying organization of lamellipodia. The linear features in the light microscopic meshwork corresponded to regions of greater actin filament density. Orientation of features was analyzed quantitatively and compared with the orientation of actin filaments at the EM level. We infer that the light microscopic meshwork reflects the orientational order of actin filaments which, in turn, is related to their branching angle.
INTRODUCTION
Protrusion at the leading edge of most crawling cells is believed to be driven by assembly of actin filaments (Mogilner and Oster, 1996; Borisy and Svitkina, 2000; Pollard et al., 2000; Pantaloni et al., 2001). One of the major protrusive specializations of the cell edge is the lamellipodium, a flat, sheet-like region of the cell filled with a dense network of actin. The dynamics of actin assembly in lamellipodia and the nature of the molecular events that are orchestrated to produce protrusion represent central problems in the cell motility field. Conclusions about actin dynamics have been inferred from structural studies under the assumption that the dynamics of the actin network are imprinted in its structure. Given that the dimensions of actin filaments themselves as well as the average interfilament distances in the cell are well below the resolution of light microscopy, electron microscopy (EM) has long been the primary tool for analyzing the structure of lamellipodia.
Important structural features established by EM are the uniform polarity of actin filaments, the preferred end for elongation (barbed end) pointing toward the leading edge of the cell (Small et al., 1978; Svitkina et al., 1997), and the dendritic organization of the actin network (Svitkina et al., 1997) with the Arp 2/3 complex present at the branching points of the filaments (Svitkina et al., 1999). These findings in combination with biochemical data and functional analysis (Machesky and Insall, 1998; Loisel et al., 1999; Mullins and Pollard, 1999) led to the formulation of a conceptual framework for actin dynamics featuring dendritic nucleation of filaments mediated by the Arp 2/3 complex and rapid turnover of the dendritic network in an array treadmilling manner (Borisy and Svitkina, 2000; Pollard et al., 2000; Pantaloni et al., 2001).
A feature of the lamellipodial network that had long remained unexplained is the apparent nonrandom angular orientation of actin filaments with respect to the leading edge. Earlier electron microscopic images suggested a preferential diagonal orientation of actin filaments at approximately a 45 degree angle to the leading edge (Small et al., 1994, 1995), although the images were not analyzed at a quantitative level. Diagonal orientation was shown theoretically to favor efficient protrusion (Mogilner and Oster, 1996), but it was not clear how filaments become oriented in this way. Recently, Maly and Borisy (2001) formulated a mathematical model according to which the orientation of actin filaments in the lamellipodium is related to the angle between two filaments forming a branch. In this model, the approach of population dynamics was applied to the growth of the actin filament network, and it was demonstrated that filament families with mother and daughter filaments oriented symmetrically with respect to the edge have the best chances to survive and proliferate. Given the ∼70 degree branching angle (Mullins et al., 1998; Svitkina and Borisy, 1999; Blanchoin et al., 2000), the most favored family was predicted to consist of filaments oriented at an angle of ±35 degrees to the normal to the leading edge. Quantitative analysis of lamellipodial electron micrographs confirmed that filaments oriented at ±35 degrees were indeed the most abundant (Maly and Borisy, 2001).
These structural observations established essential features of the actin network. However, because structural studies are necessarily performed on static, fixed specimens, they are subject to qualifications stemming from limitations intrinsic to individual EM approaches. The dendritic character of the lamellipodial network was revealed (Svitkina et al., 1997; Svitkina and Borisy, 1999) by a metal-replica technique that tends to emphasize the uppermost layer of filaments; long and straight filaments in an apparent diagonal arrangement were emphasized in negative-stained preparations (Small, 1981; Small et al., 1994, 1995); and cryo-EM, recently used (Resch et al., 2002) in an attempt to overcome limitations of drying and contrasting, is limited to small regions over holes in the support film. Thus, uncertainties have remained regarding aspects of lamellipodial organization. The EM approaches are also limited in that they do not readily demonstrate how the ultrastructure of the actin network fits into the “bigger picture,” i.e., the overall organization of the cell, its shape, and dynamic behavior. In particular, it was not clear whether the orientation of actin filaments in the lamellipodia had any implications for the organization of the cell at a larger scale. To remove these uncertainties and to relate ultrastructural features to the overall organization and dynamic behavior of the cell, EM needs to be complemented by other methods of analysis.
One important approach is to correlate the EM data with light microscopic observations made on intact and, preferably, living cells (Svitkina and Borisy, 1998). Comparison to light microscopic observations helps to establish a relationship between ultrastructural and cellular levels of organization, permits the EM data to be put into a kinetic context, and also allows evaluation of possible artifacts of fixation and contrasting. Correlative microscopy has previously been used to analyze distinct supramolecular features that were observable at both the light microscopic and EM level, including clusters of myosin II filaments (Verkhovsky et al., 1995), individual microtubules (Svitkina and Borisy, 1998), and filopodia (Svitkina et al., 2003). Exact correspondence between light and electron microscopic images provided strong evidence for faithful representation of the living state in EM images and helped to establish the ultrastructural identity of subcellular features observable at the light microscopic level.
The correlative approach has not previously been applied to lamellipodia because it has generally been considered homogenous at the light microscopic level. A few studies compared light and EM data at the level of overall shape and movement of the lamellipodia (Rinnerthaler et al., 1991; Lewis and Bridgman, 1992), but no light/EM correspondence was established at the level of the lamellipodial actin network itself. However, examination of the literature reveals several reports suggesting the presence of individual high density features (DePasquale and Izzard, 1991) and of a network-like actin pattern (Heath and Holifield, 1991, 1993; Svitkina et al., 1997; for individual images, see also Euteneuer and Schliwa, 1984, Figure 1b; Steinmetz et al., 1997, Figure 2b; Small et al., 2002, Figure 1 and supplementary video). Here, we present results obtained by using an enhanced phase contrast microscopy technique to visualize an apparent diagonal meshwork in the advancing lamellipodia of living cells. Visualization of these features permitted a correlative light and electron microscopic approach that validated the underlying organization of lamellipodia. We infer that the light microscopic meshwork results from subtle variations of actin filament density and reflects the underlying orientational organization of individual actin filaments.
Figure 1.
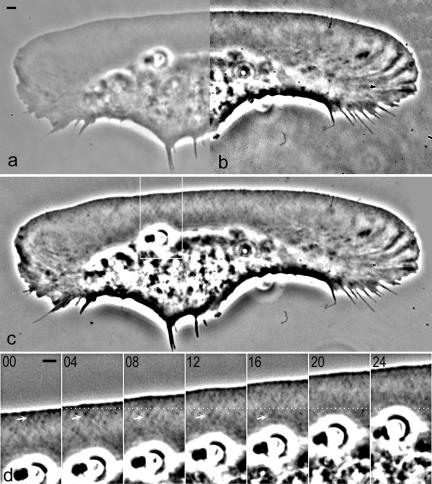
Diagonal meshwork in keratocyte lamellipodia revealed in a living cell by enhanced phase contrast microscopy. (a) Image displayed with a wide contrast range emulating appearance of a keratocyte when examined visually with a microscope. (b) Image displayed with a narrow contrast range to enhance subtle features of the lamellipodium; under these conditions, background mottle pattern due to the microscope optics also becomes apparent. (c) Background-subtracted image displayed with a narrow contrast range; background pattern is removed and lamellipodial features are clearly visible. (d) Time-lapse sequence of the area boxed in c. As the cell moves, meshwork pattern in the lamellipodium is stationary with respect to the substrate. Dotted line indicates fixed position with respect to the substratum; origin of one of the linear meshwork features is indicated by arrow. Bar, 2 μm (see supplementary material for a QuickTime movie).
Figure 2.
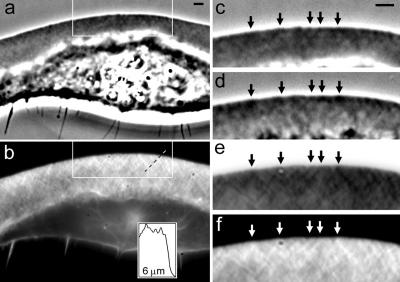
Correlative phase contrast (a, c, and d) and fluorescence (b, e, and f) microscopy of keratocyte lamellipodia. (a) General view of the same cell visualized in living state with phase contrast microscopy and with fluorescence microscopy after staining with rhodamine-phalloidin (b). Enlarged phase contrast image of the boxed region in living (c) and fixed (d) state, and the fluorescence image of the same region in inverted (e) and “normal” (f) contrast. Intersections of the meshwork linear features with the leading edge are marked with arrows. Dark phase contrast features coincide with dark features in inverted fluorescence (bright features in normal fluorescence), suggesting elevated density of actin filaments. Fluorescence intensity scan along the dashed line in b is shown in the inset to illustrate intensity variations associated with line features. Bars, 2 μm. To switch between phase contrast and fluorescence views in a single image, browse frame by frame the QuickTime movie provided in the supplementary material.
MATERIALS AND METHODS
Cell Culture
Black tetra (Gymnocorymbus ternetzi) keratocytes were cultured in DMEM supplemented with 10% fetal bovine serum and antibiotics, as described previously (Svitkina et al., 1997), with the following modification: after treatment of the migrated colonies with trypsin/EDTA solution, the cells were placed in 70% medium (30% purified water) for recovery and subsequent observation, because it was found that the cells migrating in diluted media exhibited more regular shape and behavior than the cells in undiluted media.
Optical Microscopy
Optical microscopy was performed using a Nikon Eclipse TE300 inverted microscope with CFI Plan 100× phase objective (numerical aperture 1.25) and 100-W halogen and 100-W HBO light sources for phase contrast and epifluorescence microscopy, respectively. Most of the images were captured with a Roper Scientific (Tucson, AZ) MicroMAX-1300PB cooled charge-coupled device (CCD) camera operated with MetaMorph software (Universal Imaging, West Chester, PA). For some experiments, MicroMAX 512FT and Photometrics Series 300 cooled CCD cameras were used (both from Roper Scientific). All the cameras used are characterized by large electron well capacity and low readout noise. These features were essential for the enhanced phase contrast microscopy, because to detect subtle contrast variations associated with fine lamellipodial features it was necessary to achieve an intensity readout exceeding the noise level by a factor of ∼5000 or more. For a reference, electron well capacity and readout noise of MicroMAX-1300PB camera are listed at 180,000 and six to nine electrons, respectively. Large well capacity is associated with the large pixel size (20 μm for MicroMAX 1300PB). To maintain spatial resolution despite the large pixel size of the camera, a 2.5× intervening adapter was used, so that one pixel corresponded to 80 nm in the object space. For enhanced phase contrast microscopy, single frame exposures of 500 - 800 ms were used to achieve intensity readout of ∼50% saturating level of the camera. Background images of the cell-free areas of the same dish were subtracted from the cell images (a constant was added to the result to avoid negative intensity values), and the resulting images were scaled to a narrow contrast range for optimal visualization of the lamellipodial substructure.
Correlative Phase Contrast, Fluorescence, and Electron Microscopy
Correlative microscopy was generally performed as described previously (Svitkina and Borisy, 1998), but the extraction and fixation procedures were modified to minimize the time between live observation and immobilization of rapidly moving fish keratocytes. In most cases, a single cell was observed by two of three microscopy techniques: phase microscopy and fluorescence microscopy, or fluorescence and electron microscopy. Thus, a correlation between phase contrast and electron microscopic images was established through an intermediate step of fluorescent microscopy. This strategy facilitated the processing and analysis of a large number of cells. A few cells, however, were analyzed by all three techniques: in the living state by phase contrast microscopy and by fluorescence and electron microscopy after extraction and fixation. For correlative phase contrast and fluorescence microscopy, keratocytes were observed on a coverslip mounted over a hole in an open 35-mm Petri dish containing 1 ml of diluted medium. Rapid fixation was achieved by pouring 1 ml of fixation solution (2% glutaraldehyde in culture medium) into the dish while still on the microscope stage. The fixation was achieved in ∼2 s (estimate based on the distance traveled by the cell since the last frame recorded before fixation) and resulted in a minimal distortion of the cell shape. Fixed cells were subsequently washed in phosphate-buffered saline (PBS), permeabilized in 1% Triton X-100 in PBS, treated with 2 mg/ml sodium borohydride in PBS (two treatments 5 min each), and stained with rhodamine-phalloidin to reveal F-actin.
For correlation with electron microscopy, cells were cultured and observed on gold-coated finder coverslips (Svitkina and Borisy, 1998) in Petri dishes containing 0.5 ml of diluted culture medium and extracted by pouring into the dish 2 ml of the extraction medium (1% Triton X-100 and 1 μM rhodamine-phalloidin in PEM buffer [100 mM PIPES-KOH pH 6.9, 1 mM MgCl2, and 1 mM EGTA] with or without 4% polyethylene glycol 40,000 [PEG]). Extraction both with and without PEG resulted in a rapid arrest of the cell movement with good preservation of the shape of the lamellipodia; however, partial collapse of the cell body was observed upon omission of PEG from the extraction medium. Extracted cells were washed briefly with PEM buffer, fixed with 2% glutaraldehyde in 0.1 M Na-cacodylate pH 7.3, and observed by fluorescence microscopy. Positions of the cells were recorded and the specimens were processed for EM as described previously (Svitkina and Borisy, 1998).
Image Analysis
For the analysis of angular distribution of linear features in the lamellipodia, fluorescence images and digitized electron micrographs of the same cells were aligned to each other by using Photoshop. Quantification of angular distribution was performed initially in MATLAB software (Mathworks, Natick, MA) by using the procedure described in Maly and Borisy (2001) based on edge detection with the Canny (1986) algorithm and Radon transform (Leavers, 1992; Helgason, 1999). The important differences from the previous study of Maly and Borisy (2001) were that the method had to be adapted to light microscopic images and also that to correlate the EM and fluorescence images, significantly bigger regions of the electron micrographs had to be analyzed. However, in the course of this analysis, we found that the Canny filter as implemented in MATLAB has nonuniform sensitivity to edges of different orientations. It preferentially detects diagonal and, to a lesser extent, vertical edges over horizontal ones. This effect was not important for images with strong orientational order but became significant for less ordered images, resulting in apparent detection of diagonal features in completely disordered test images. Consequently, we explored different methods of edge detection. Two approaches were implemented: 1) Canny filter with rotation. In this approach, nonuniform detection of differently oriented edges was compensated by rotating the image between 0° and 90° with an increment of 9, resulting in 11 rotated images. Each rotated image was subjected to the Canny filter and then rotated back to its initial orientation. Finally, 11 filtered images were superimposed using an algorithm that retained white pixels of every image in the result (“lighter” blending scheme). 2) “Glowing edges” filter available in Photoshop (Adobe Systems, Mountain View, CA) with subsequent binarization of the image. The detailed algorithm for this filter is not provided with the software, but the filter produces results similar to the sum of the squares of derivatives of the image along two orthogonal directions. Both the modified Canny and the glowing edges procedure were tested using simulated images and were confirmed to detect features of all orientations with uniform sensitivity. Both methods produced similar results, but the glowing edges filter apparently generated less noise. The results shown are produced with the glowing edges method.
The procedure for quantification of feature orientation consisted of the following steps: the images were subjected to a high-pass filter (size of 5 pixels, or 15 and 400 nm in object space for EM and light microscopy, respectively) to suppress intensity variations not associated with linear features, contrast-enhanced (“equalize” filter of the Photoshop, light microscopic images only), zoomed (two times for EM and two to four times for light microscopy), blurred with a Gaussian blur filter (size 2 pixels) to suppress noise, and subjected either to glowing edges or modified Canny procedure. Resulting images were used to quantify feature orientation with a modified Radon transform method.
In the Radon transform, the intensity of the image is projected onto a rotating axis. Strong signal in the projection is indicative of linear features oriented in the direction of the projection. We modified the Radon transform procedure implemented in MATLAB by using a rotating square as a mask to isolate the region of the image to be projected, rather than using a fixed square region. The rotating mask ensured that the features located at the center and near the boundaries of the mask contributed equally to the intensity of the projection. The corresponding areas in EM and fluorescence images were subdivided into several square regions with side dimensions of ∼1.5 and 5 μm for EM and fluorescence, respectively. The boundaries of each square region were rotated with an angle increment of 3 degrees, and the intensity of the image within the square was projected on the side of the square for each rotation angle (zero projection angle was aligned with the normal to the leading edge). Linear features oriented in the direction of the projection are expected to produce sharp intensity peaks. However, the signal could be lost due to a possible overlap of the projections of several features, this event being particularly likely for big images. To avoid this effect, the whole image was scanned with a relatively narrow band (100 and 60 pixels wide for EM and light microscopy, respectively), and the intensity was projected from this band rather than from an entire image.
Previously, Maly and Borisy (2001) quantified the signal in the projection by integrating intensity values above a threshold equal to the mean plus two standard deviations. However, this thresholding method leaves the possibility that weak linear features remain under the threshold and go undetected. To improve detection of weak signals, we used integration of the square of the derivative of the projection. Sharp intensity peaks in the projection produced by linear features are expected to result in high positive and negative values of the derivative. Consequently, the integral of the square of the derivative would exhibit high positive value. Integration of the square of the derivative of the projection is expected to provide a better measure than the integration of the projection itself because sharp intensity peaks produced by linear features would contribute significantly to the square of the derivative, whereas the smooth intensity variations would not. To further suppress the noise due to variations of projection intensity not associated with linear features, the derivative of the projection was computed with a step size of three pixels, and the values of the square derivative below a threshold value were discarded. The threshold was defined manually by analyzing a small portion of the image containing identifiable linear features in the direction of the projection as well as features oriented at an angle to the projection and isotropic features. The filtered integrals of the square of the derivative were averaged between all square regions contained within the chosen area of the image and plotted as a function of the projection angle. Processing of several types of test images containing defined complex patterns of the oriented features demonstrated that the resulting plot of the projection signal closely reflects the distribution of the feature's incidence angle.
RESULTS
Lamellipodia of moving fish epidermal keratocytes usually seem uniform, low in contrast, and lacking recognizable features when examined visually by phase contrast microscopy (Figure 1a). However, when images of lamellipodia were taken at nearly saturating light levels with a 16-bit cooled CCD camera, and then digitally scaled to a narrow contrast range (Figure 1b) and background-subtracted (Figure 1c), a meshwork of darker and brighter lines crossing each other was revealed. The meshwork lines were apparently oriented at two specific angles, approximately diagonal to the direction of the protrusion. This meshwork pattern should not be confused with ruffling areas and radial ridges, which were described previously (Lee et al., 1993). Ruffles and ridges frequently coincided with an irregular shape and rough edge of the cell. In contrast, the meshwork pattern was most evident in regularly shaped cells with a smooth edge lacking ruffles and ridges. The dark lines in the meshwork originated as dots at the front boundary of the lamellipodium, which then elongated as the edge moved forward, while remaining stationary with respect to the substrate (Figure 1d). This behavior suggested that the line pattern was due to the actin content of the lamellipodia rather than to the surface features, because actin filaments are stationary with respect to the substrate (Theriot and Mitchison, 1991), whereas the membrane components are not (Kucik et al., 1989; Lee et al., 1993).
Rapid fixation or extraction of the cells and visualization of F-actin with rhodamine-phalloidin confirmed the actin nature of the phase contrast pattern. The diagonal line pattern was apparent after phalloidin staining, with brighter phalloidin-stained lines coinciding with dark phase contrast features, and vice versa (Figure 2). Because fluorescence intensity after phalloidin staining is proportional to actin filament density, phase contrast features were likely due to the nonuniform density of actin filaments in lamellipodia.
The individual diagonal lines in the meshwork pattern seemed to have a length of several micrometers and were spaced at distances of ∼500-700 nm from each other. The apparent width of the features was close to the resolution limit of light microscopy, making it impossible to determine their true width. The differences in fluorescence intensity between bright and dark lines were relatively small and usually amounted to 5-15% of the mean intensity (see the intensity scan in Figure 2b, inset). In phase contrast images, the intensity of the features was closely related but not directly proportional to the fluorescence intensity of the same features, and contrast between the line features in phase microscopy amounted to only ∼1-3% of the mean image intensity. This explains why the line pattern was clearly visible only in the phase contrast and fluorescence images acquired with sufficient amount of light and after adequate contrast enhancement. The line pattern was also discernible in fluorescence images of living keratocytes injected with rhodamine-actin (our unpublished data), but the visualization was inferior to the phase contrast and phalloidin-fluorescence images. A likely explanation is that the background of labeled G-actin in rhodamine-actin-injected cells interfered with the visualization of F-actin pattern.
The light microscopic line pattern could not represent individual actin filaments, because the filaments are submicroscopic and too closely packed to be resolved by light microscopy. We will subsequently refer to the light microscopic pattern as a “long-range meshwork” or “apparent long-range organization” to indicate the fact that the pattern is visible at a bigger than ultrastructural scale. To determine what kind of supramolecular features corresponded to the long-range pattern, we performed correlative light and electron microscopy of the same cells. One possibility was that the line features corresponded to small bundles of actin filaments, which could have had previously escaped detection in EM preparations. More than 20 cells displaying clear line pattern at the light microscopic level were examined at the electron microscopic level by using the platinum replica technique. None of these cells revealed actin filament bundles in the lamellipodia. On the contrary, the cells displayed a more or less continuous and smooth network of actin filaments.
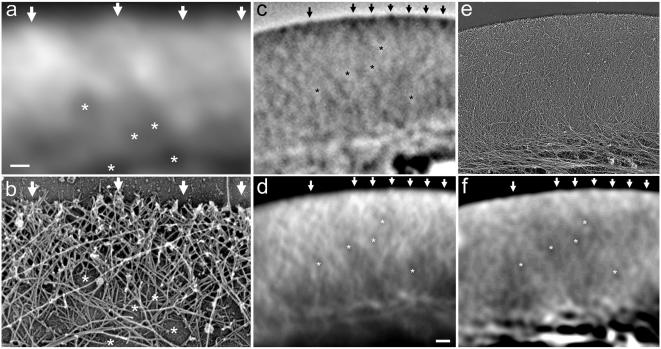
Closer examination of the images demonstrated that the light microscopic line pattern resulted from variation of filament density in the network. To account for the fluorescence intensity differences, the network density would only have to vary by ∼5-15%. These relatively small differences were easiest to detect in regions of lamellipodia where the actin network was very sparse, thus allowing most of the actin filaments to be exposed to the platinum coating and visualized in the images. In these regions, the correspondence between fluorescence intensity and actin network density was readily apparent (Figure 3, a and b).
Figure 3.
Correlative fluorescence (a and d), phase contrast (c), and electron microscopy (b, e, and f) of keratocyte lamellipodia. Region of lamellipodia displaying relatively sparse actin network revealed by fluorescence (a) and electron (b) microscopy; correspondence of the bright fluorescence features with the regions of high actin filament density is apparent. Large region of the lamellipodia with dense network of actin filaments in phase contrast after extraction of the cell (c), fluorescence (d), and electron (e) microscopy; the same electron microscopic image is blurred with Photoshop (blur radius ∼200 nm), helping to display the correspondence between the fluorescence and EM image (f). Selected features are highlighted: intersections of the high actin density features with the leading edge are marked with arrows, low actin density areas are marked with asterisks. Note that the significant difference between the fluorescence and blurred EM image (d and f) at the lamellipodium/cell body boundary (bottom of the images) is due to the presence of intermediate filaments, which contribute to the EM image but not to the fluorescence image. Intermediate filaments and microtubules are concentrated in the cell body but do not penetrate into the lamellipodia of moving keratocytes (Svitkina et al., 1997). Bars, 200 nm (a), 1 μm (d).
In the case of a denser actin network, the differences in filament density were difficult to appreciate from the EM image. EM images contain far more detail than the light microscopic images, and it is difficult to visually extract filament density information from such complex pictures. To make the light microscopy/EM comparison at the same level of resolution, we reduced the resolution of the EM image to that of the light microscopic level. This was achieved by application to the EM image of a Gaussian blur filter with a kernel size equivalent to the diffraction-limited spot of light microscopy, generating a lower resolution, “blurred EM” image (Figure 3f). The contrast of the blurred image could be expected to reflect the filament density in a way similar to the fluorescence image, namely, regions containing more filaments would seem brighter after blurring than regions with fewer filaments. The blur procedure produced a simulation of the light microscopic image, which was fairly similar to the experimental image both in general character and in fine details, and was useful to illustrate the EM/light microscopy correlation for relatively large regions of the actin network (Figure 3, c, d, and e). It should be noted, however, that the correspondence between the blurred EM images and fluorescence images was not exact, but exact correspondence is not to be expected because the replica reflects total mass in the cytoskeleton preparation, not only actin filaments. Replicas often displayed bright features not associated with actin filament density, such as heavily coated amorphous material. At the same time, dark regions in EM images accurately represented the regions of low filament density, contributing to the strong similarity between blurred EM and fluorescence images. Thus, the bright line features revealed by light microscopy corresponded to regions of actin filament network with elevated filament density, but which were not otherwise structurally distinct from the rest of the network.
The light microscopic images produced a visual impression of an ordered meshwork with a specific (approximately diagonal to the leading edge) angular orientation of the line features. To analyze objectively the orientation of the light microscopic features and to compare it to the orientation of actin filaments in the EM images, we used the Radon transform approach similar to that previously applied by Maly and Borisy (2001). The Radon transform (Helgason, 1999) detects linear objects in the image by computing projections of an image matrix along specified directions. Linear features oriented in the direction of the projection produce a stronger signal than features oriented at an angle to the projection, so that the integrated intensity of a signal in the projection becomes a function of the number of linear features in the direction of the projection. The projection signal plotted as a function of the projection angle reflects the distribution of the incidence angle of the linear features of the image.
Despite visually apparent orientational order, objective quantification of orientation in light microscopic images presented significant difficulties, because the linear features were relatively weak and the images displayed other prominent features, such as overall gradients of intensity due to variation of actin density from the front to the rear of the lamellipodium, amorphous features, and significant noise. To overcome these difficulties, we modified the edge enhancement procedure to extract linear features and the Radon transform procedure to derive orientational information (see MATERIALS AND METHODS).
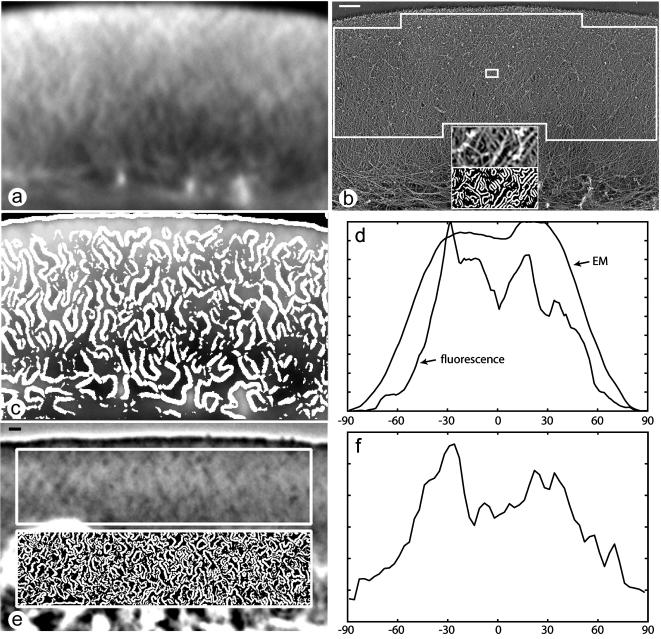
After edge detection (Figure 4, a and c), the modified Radon transform detected two symmetrically positioned peaks of the distribution of incidence angle in most of the fluorescence images of lamellipodia (Figure 4d). The positions of the two peaks varied in different cells approximately between ±25° and ±45°. These values were consistent with the results of visual estimation of the orientation of diagonal features in the criss-cross network. It should be noted that variability in position, shape, and amplitude of peaks of incidence angle distribution are expected, because only a small number of features contributed to the signal in individual images. The phase contrast images were generally noisier than fluorescence images, but, in favorable cases, also exhibited two symmetrically positioned peaks of incidence angle distribution (Figure 4, e and f). Thus, the Radon analysis confirmed the visual impression of an approximately diagonal orientational organization in the light microscopic actin network.
Figure 4.
Angular distribution of actin filaments and linear features revealed by fluorescence and phase contrast microscopy in keratocyte lamellipodia as evaluated by modified Radon transform. Fluorescence (a) and electron microscopic (b) image of the same keratocyte lamellipodia, the region analyzed with Radon transform is indicated with large box. The detection of filament edges with the Canny filter in the area marked with the small box is illustrated in the inset in b. Detection of edges in fluorescence image shown in c as overlay of detected edges over the original image. Bright detected edges flank fluorescence features. (e) Phase contrast image of a different cell; edge detection in boxed part of the image shown in the inset. Radon signal reflecting the density distribution of the incidence angle of the filaments and fluorescence line features (d) for the cell shown in a-c and of phase contrast features (f) for the cell shown in e. Zero angle is assigned to the features perpendicular to the leading edge; positive and negative angles indicate the orientation toward one or the other lateral side of the lamellipodia. Bars, 1 μm.
Whereas analysis of EM images by blurring consistently displayed diagonal features, the outcome of Radon analysis depended upon the density of actin filaments in the lamellipodial preparations. In EM images of lamellipodia with relatively sparse actin networks, the Radon transform showed two peaks of incidence angle distribution (Figure 4, b and d) with the peak positions similar to the ones determined for the corresponding fluorescence images (two of five cells analyzed by Radon procedure both at fluorescence and EM level). However, in cells with a very dense actin filament network, the Radon transform displayed a broad incidence angle distribution even though the same cells exhibited preferential feature orientations at two symmetrical angles at the light microscopic level. These cells were also characterized by an abundance of amorphous material on the surface of the platinum replicas, suggesting that the filament orientation throughout the network may not have been accurately detected. It is important to note, however, that when specific preferential feature orientations were detected both at the light and EM level in the same cell, the directions nearly coincided at the two microscopic levels (Figure 4d). These findings validate and reinforce the conclusion that the actin network has an ordered angular organization and further suggest that the organization at an individual filament scale is related to the larger scale features detectable in living, motile cells by light microscopy.
DISCUSSION
In this study, we visualized a meshwork of actin in the lamellipodium of living, moving keratocytes by an enhanced phase contrast microscopy procedure. The procedure is a noninvasive and simple technique that takes advantage of the high dynamic range and low noise of cooled CCD sensors to resolve the subtle differences in contrast within lamellipodia. Previously, images of an apparent light microscopic criss-cross network in lamellipodia occurred in several reports (see INTRODUCTION for references), but the significance of these observations was not clear and the nature of the network features was not investigated. The simplest interpretation of the light microscopic features in terms of supramolecular organization is that they result from groups of actin filaments rather than individual filaments. Although some of the previous EM studies visualized an apparent diagonal network of individual filaments (Small et al., 1994, 1995), no corresponding filament grouping was described. What then is the explanation for features at the light microscopic level but not at the EM level? Three possibilities are as follows: 1) the features were lost in preparing the specimens for EM, 2) the network pattern at the light microscopic level represented a rare event or an artifact, and 3) the features arise from variations in filament density as opposed to structural grouping.
Our observation of a meshwork pattern by enhanced phase contrast microscopy of living cells establishes that a diagonal meshwork is a consistent, characteristic feature of the lamellipodium. Moreover, we correlated the fine meshwork visualized by phase contrast microscopy to the actin density distribution revealed by fluorescence and electron microscopy and demonstrated that the light and electron microscopic images were consistent with each other. Thus, the features were not lost in preparing specimens for EM, nor were they a rare event or artifact of preparation. The network pattern did not result from recognizable actin bundles or other distinct features, but was due to subtle variations of actin filament density. When the light and electron microscopic images were displayed at a similar resolution, the close correspondence between the distributions of actin density revealed by the two techniques became apparent, reinforcing and validating the results of both techniques and suggesting that only negligible distortions are introduced during the processing of the specimen for electron microscopy.
Time-lapse observation demonstrated that the light microscopic diagonal features occurred concomitantly with protrusion and did not result from subsequent rearrangement of the lamellipodial network. This is consistent with mechanisms of self-organization operating in the process of network growth at the leading edge. Such mechanisms were proposed to be based on the biophysics of interaction of filament ends with the membrane (Mogilner and Oster, 1996), and, more recently, based on evolutionary selection of branching actin filament families oriented symmetrically with respect to the leading edge (Maly and Borisy, 2001). In the latter mechanism, the most favored filament orientation is predicted as half of the branching angle of actin filaments (±35°) with a distribution of approximately ± 10°. The orientation of the long-range meshwork features was consistent with this prediction, displaying a range of symmetric angle values from ±25° to ±45°.
The relative orientation of the light microscopic features at an angle close to the filament branching angle suggested that the orientational order at the light microscopic level reflected the underlying order of individual actin filaments.
However, although the light microscopic images mostly displayed features aligned along two symmetric directions, the replica images did not always show two distinct preferential orientations of actin filaments. It is not surprising that the light microscopic and replica images were not similar in all respects, because the phalloidin fluorescence images reflect the organization of actin filaments throughout the depth of the lamellipodium, whereas the replica images preferentially reveal the surface features accessible to platinum coating. Furthermore, the phalloidin signal is specific to actin, whereas the platinum replica will reflect all material in the cytoskeleton, not just actin filaments. One could expect that surface features would in some instances mask the overall orientation of filaments. We note that although the light microscopic and the replica images were not always consistent with respect to their orientational order, they were mostly consistent in the representation of overall actin density distribution. We take this remarkably close correspondence as an indication that the bulk of the mass in the cytoskeleton preparations of lamellipodia consists of actin filaments.
Orientation of the light microscopic network features along the suggested preferred directions of the growth of individual filaments (Mogilner and Oster, 1996; Maly and Borisy, 2001) suggests that the variations of actin density in the lamellipodia propagate along the direction of actin filament elongation. One can imagine several possible mechanisms of density propagation. One possibility is that filaments aligned in one direction are selectively protected from capping by a protein (e.g., Ena/VASP family member; Bear et al., 2002), which remains associated with the ends of protected filaments as they grow at the leading edge of the lamellipodia. If this is true, the network features could be related to the early filopodia precursors recently identified by Svitkina et al. (2003). However, unlike filopodia precursors, the high actin density lines in keratocyte lamellipodia did not merge to each other and did not develop into microspikes or filopodia, indicating that they were features of the lamellipodial network itself. Alternatively, a factor stimulating branching of actin filaments (e.g., an Arp2/3-activating protein) could be associated with growing filament ends resulting in a high-density feature. Another possibility is that the features at the light microscopic level may simply reflect that the cell has a mechanism for selecting the orientation of filaments but that their lateral position is essentially random. A spot of elevated actin density could arise due to a stochastic fluctuation in the nucleation and/or capping rate and propagate linearly as filaments grow. Fluctuations in filament density would then give the appearance of similarly oriented linear features at the lower resolution of the light microscope. Quantitative analysis of the light microscopic images in combination with the localization of potential regulatory proteins may help to distinguish these possibilities and provide further insights into actin dynamics at the leading edge.
Whatever the mechanism of the formation of the longrange meshwork pattern, it may have important implications for the control of protrusion. Protrusion of the lamellipodium is precisely orchestrated within the cell. The stability of the shape of the lamellipodium of the fish epidermal keratocyte is achieved through a graded distribution of extension along the length of the front edge of the cell (Lee et al., 1993). This implies that the characteristic spatial scale at which the extension control mechanism operates is comparable with the size of the cell. The organization of the lamellipodia is expected to reflect the long-range mechanisms, and the ordered mesh described in our study may be one of the manifestations of this process. Our observation of a large-scale meshwork in the living cell detectable by a simple enhanced phase microscopy technique provides the possibility of kinetic analysis with respect to cell shape and motility and could help to uncover the mechanism of the long-range control of extension.
Supplementary Material
Acknowledgments
We thank I.V. Maly for a critical reading of the manuscript. This study was supported by Swiss National Science Foundation grant 31-61589 (to A.B.V.) and National Institutes of Health grants GM-62431 and GM-64346 (to G.G.B.).
Online version of this article contains video material for some figures. Online version is available at www.molbiolcell.org.
References
- Bear, J.E., et al. (2002). Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109, 509-521. [DOI] [PubMed] [Google Scholar]
- Blanchoin, L., Amann, K.J., Higgs, H.N., Marchand, J.B., Kaiser, D.A., and Pollard, T.D. (2000). Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404, 1007-1011. [DOI] [PubMed] [Google Scholar]
- Borisy, G.G., and Svitkina, T.M. (2000). Actin machinery: pushing the envelope. Curr. Opin. Cell Biol. 12, 104-112. [DOI] [PubMed] [Google Scholar]
- Canny, J.F. (1986). A computational approach to edge detection. IEEE Trans. PAMI 8, 679-698. [PubMed] [Google Scholar]
- DePasquale, J.A., and Izzard, C.S. (1991). Accumulation of talin in nodes at the edge of the lamellipodium and separate incorporation into adhesion plaques at focal contacts in fibroblasts. J. Cell Biol. 113, 1351-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer, U., and Schliwa, M. (1984). Persistent, directional motility of cells and cytoplasmic fragments in the absence of microtubules. Nature 310, 58-61. [DOI] [PubMed] [Google Scholar]
- Heath, J., and Holifield, B. (1991). Cell locomotion. Actin alone in lamellipodia. Nature 352, 107-108. [DOI] [PubMed] [Google Scholar]
- Heath, J.P., and Holifield, B.F. (1993). On the mechanisms of cortical actin flow and its role in cytoskeletal organisation of fibroblasts. Symp. Soc. Exp. Biol. 47, 35-56. [PubMed] [Google Scholar]
- Helgason, S. (1999). The Radon Transform, Boston: Birkhauser.
- Kucik, D.F., Elson, E.L., and Sheetz, M.P. (1989). Forward transport of glycoproteins on leading lamellipodia in locomoting cells. Nature 340, 315-317. [DOI] [PubMed] [Google Scholar]
- Leavers, V.F. (1992). Shape Detection in Computer Vision Using the Hough Transform, New York: Springer.
- Lee, J., Ishihara, A., Theriot, J.A., and Jacobson, K. (1993). Principles of locomotion for simple-shaped cells. Nature 362, 167-171. [DOI] [PubMed] [Google Scholar]
- Lewis, A.K., and Bridgman, P.C. (1992). Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J. Cell Biol. 119, 1219-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel, T.P., Boujemaa, R., Pantaloni, D., and Carlier, M.F. (1999). Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401, 613-616. [DOI] [PubMed] [Google Scholar]
- Machesky, L.M., and Insall, R.H. (1998). Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8, 1347-1356. [DOI] [PubMed] [Google Scholar]
- Maly, I.V., and Borisy, G.G. (2001). Self-organization of a propulsive actin network as an evolutionary process. Proc. Natl. Acad. Sci. USA 98, 11324-11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner, A., and Oster, G. (1996). Cell motility driven by actin polymerization. Biophys. J. 71, 3030-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, R.D., Heuser, J.A., and Pollard, T.D. (1998). The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 95, 6181-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, R.D., and Pollard, T.D. (1999). Structure and function of the Arp2/3 complex. Curr. Opin. Struct. Biol. 9, 244-249. [DOI] [PubMed] [Google Scholar]
- Pantaloni, D., Le Clainche, C., and Carlier, M.F. (2001). Mechanism of actin-based motility. Science 292, 1502-1506. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., Blainchoin, L., and Mullins, R.D. (2000). Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29, 545-576. [DOI] [PubMed] [Google Scholar]
- Resch, G.P., Goldie, K.N., Krebs, A., Hoenger. A., and Small, J. V. (2002). Visualisation of the actin cytoskeleton by cryo-electron microscopy. J. Cell Sci. 115, 1877-1882. [DOI] [PubMed] [Google Scholar]
- Rinnerthaler, G., Herzog, M., Klappacher, M., Kunka, H., and Small, J.V. (1991). Leading edge movement and ultrastructure in mouse macrophages. J. Struct. Biol. 106, 1-16. [DOI] [PubMed] [Google Scholar]
- Small, J.V. (1981). Organization of actin in the leading edge of cultured cells: influence of osmium tetroxide and dehydration on the ultrastructure of actin meshworks. J. Cell Biol. 91, 695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, J.V., Herzog, M., Haner, M., and Abei, U. (1994). Visualization of actin filaments in keratocyte lamellipodia: negative staining compared with freeze-drying. J. Struct. Biol. 113, 135-141. [DOI] [PubMed] [Google Scholar]
- Small, J.V., Herzog, M., and Anderson, K. (1995). Actin filament organization in the fish keratocyte lamellipodium. J. Cell Biol. 129, 1275-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, J.V., Isenberg, G., and Celis, J.E. (1978). Polarity of actin at the leading edge of cultured cells. Nature 272, 638-639. [DOI] [PubMed] [Google Scholar]
- Small, J.V., Stradal, T., Vignal, E., and Rottner, K. (2002). The lamellipodium: where motility begins. Trends Cell Biol. 12, 112-120. [DOI] [PubMed] [Google Scholar]
- Steinmetz, M.O., Stoffler, D., Hoenger, A., Bremer, A., and Aebi, U. (1997). Actin: from cell biology to atomic detail. J. Struct. Biol. 119, 295-320. [DOI] [PubMed] [Google Scholar]
- Svitkina, T.M., and Borisy, G.G. (1999). Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament arrays in lamellipodia. J. Cell Biol. 145, 1009-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina, T.M., and Borisy, G.G. (1998). Correlative light and electron microscopy of the cytoskeleton of cultured cells. Methods Enzymol. 298, 570-592. [DOI] [PubMed] [Google Scholar]
- Svitkina, T.M., Bulanova, E.A., Chaga, O.Y., Vignjevic, D.M., Kojima, S., Vasiliev, J.M., and Borisy, G.G. (2003). Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160, 409-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina, T.M., Verkhovsky, A.B., McQuade, K.M., and Borisy, G.G. (1997). Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 139, 397-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot, J.A., and Mitchison, T.J. (1991). Actin microfilament dynamics in locomoting cells. Nature 352, 126-131. [DOI] [PubMed] [Google Scholar]
- Verkhovsky, A.B., Svitkina, T.M., and Borisy, G.G. (1995). Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J. Cell Biol. 131, 989-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.