Abstract
Purpose
BRCAPRO, a BRCA mutation carrier prediction model, was developed on the basis of studies in individuals of Ashkenazi Jewish and European ancestry. We evaluated the performance of the BRCAPRO model among clinic-based minority families. We also assessed the clinical utility of mutation status of probands (the first individual tested in a family) in the recommendation of BRCA mutation testing for other at-risk family members.
Patients and Methods
A total of 292 minority families with at least one member who was tested for BRCA mutations were identified through the Breast Cancer Family Registry and the University of Chicago. Using the BRCAPRO model, the predicted likelihood of carrying BRCA mutations was generated. Area under the receiver operating characteristic curves (AUCs) were calculated.
Results
There were 104 African American, 130 Hispanic, 37 Asian-American, and 21 other minority families. The AUC was 0.748 (95% CI, 0.672 to 0.823) for all minorities combined. There was a statistically nonsignificant trend for BRCAPRO to perform better in Hispanic families than in other minority families. After taking into account the mutation status of probands, BRCAPRO performance in additional tested family members was improved: the AUC increased from 0.760 to 0.902.
Conclusion
The findings support the use of BRCAPRO in pretest BRCA mutation prediction among minority families in clinical settings, but there is room for improvement in ethnic groups other than Hispanics. Knowledge of the mutation status of the proband provides additional predictive value, which may guide genetic counselors in recommending BRCA testing of additional relatives when a proband has tested negative.
INTRODUCTION
Germline mutations in the BRCA1 and BRCA2 genes significantly increase the risks of breast and ovarian cancer. A recent meta-analysis of 10 studies estimated that the risk of developing breast cancer by age 70 years was 57% and 49% for BRCA1 and BRCA2 mutation carriers, respectively.1 The corresponding risks for ovarian cancer were 40% and 18%, respectively. As risk-reduction strategies, including chemoprevention and prophylactic surgeries, have been demonstrated to be effective in high-risk individuals,2 genetic testing for BRCA gene mutations has been increasingly utilized in clinical settings. Because genetic testing is expensive, genetic counselors and third-party payers often use the pretest probability that a woman carries a deleterious mutation in BRCA1 or BRCA2 as a guide in the recommendation of genetic testing. It is, therefore, important to ensure that the prediction models are accurate and appropriate in the populations to which they are applied.
Several carrier prediction models have been developed by using either a Mendelian approach3,4 or empirical data.5–8 Of these, the BRCAPRO model has been widely used and extensively evaluated, mostly in non-Hispanic white populations.9–16 This model, like other carrier prediction tools, was developed on the basis of mutation rates and penetrances observed mainly in women of Ashkenazi Jewish and European ancestry. To our knowledge, no large study has evaluated the utility of the BRCAPRO model in African American, Hispanic and Asian American populations.17,18 This is not surprising, given that minority populations remain underrepresented in genetic counseling and testing programs. A recent study identified large racial disparities between African Americans and whites in the use of BRCA genetic testing.19,20 Data on BRCA testing from Myriad Genetic Laboratories showed that less than 10% of individuals tested were from minority populations,7 though minorities constitute 31% of the US population.21
In clinical counseling, it is not always clear whether to offer genetic testing to family members for BRCA mutations after the proband has tested negative, because the proband's breast cancer could be nonattributable to BRCA mutations. Recalculation of the carrier probability for other family members by incorporating the proband's test result could be helpful in decision making, but the magnitude of improvement in risk prediction is unknown, because no empirical data are available. Another issue that confronts genetic counselors is that increasing numbers of individuals from high-risk families have undergone prophylactic interventions, such as oophorectomy or mastectomy. Prophylactic interventions alter mutation penetrance, and relatives who believe themselves to be at higher risk of developing cancer or of carrying a BRCA1 or BRCA2 mutation may be more likely to opt for prophylactic intervention. Therefore, ignoring medical interventions could diminish the accuracy of carrier prediction models.22 Although BRCAPRO now accounts for medical intervention and genetic testing results of family members,22,23 no study, to our knowledge, has evaluated these options by using empirical data.
This study was conducted to validate the performance of the BRCAPRO model among ethnic minority families seen in clinical settings and to examine whether the incorporation of prophylactic oophorectomy and/or genetic test results of probands improves the accuracy of carrier status prediction.
PATIENTS AND METHODS
Study Sample
The study sample consists of individuals from 292 families identified through two sources. The first group of families was identified through the Breast Cancer Family Registry (BCFR),24 a consortium established by the National Cancer Institute in 1995. Three (New York City, Philadelphia, and Salt Lake City) of the six sites identified families through clinic-based and/or community-based recruitment between 1996 and 2005 and were included in this study. All BCFR participants were administered a questionnaire to obtain information on demographics, ethnicity, personal history of cancer, and family history of cancer in at least first- and second-degree relatives. Pathology report verification was sought to confirm reported breast and ovarian cancer history. The second group of families was identified through the Cancer Risk Clinic at the University of Chicago between 1993 and 2006. Family history was assessed by experienced genetic counselors, and a pedigree of at least three generations was constructed. All participants were informed that their DNA samples would be used for mutation screening under protocols approved by the institutional review board at each institution.
This study included self-reported African American, Hispanic, Asian-American, and Native American families. Families with Ashkenazi Jewish ancestry were excluded from the analysis. A family was eligible if at least one member had been tested for BRCA1 and BRCA2 mutations. For families with two or more members tested, the first family member enrolled at the respective institutions was designated as the proband. Appendix Table A1 (online only) lists characteristics of families by study centers.
Mutation Detection
Genomic DNA samples of participants from the University of Chicago and from the the New York City and Salt Lake City sites of the BCFR were tested at Myriad Genetic Laboratories (Salt Lake City, UT). Full sequencing analysis was done for probands, and single-site testing for the family-specific mutation was done for relatives of mutation-positive probands. For participants from the BCFR Philadelphia site, polymerase chain reaction fragments that covered the BRCA1 and BRCA2 genes were analyzed by using the enzyme mutation detection assay or heteroduplex analysis. Candidate mutations were confirmed by using direct sequencing.
Test results were considered positive if the mutation was protein truncating (ie, nonsense, frame-shifting insertions or deletions, splice site mutations) or a known deleterious missense mutation according to the Breast Cancer Information Core (http://research.nhgri.nih.gov/projects/bic). In some instances, mutation classification was determined in consultation with Myriad Genetic Laboratories. Variants of unknown significance were considered negative.
Estimation of Mutation Probability
Probability that a proband or relative carried a BRCA1 and BRCA2 mutation was calculated by using the BRCAPRO model on the basis of her personal and first- and second-degree family history of breast and ovarian cancers.3 We used the version implemented in the BayesMendel 1.3-2 package,23 a library of the R statistical software.25 The default penetrances1 and allele frequencies4 for non–Ashkenazi Jewish populations were used in this analysis. The allele frequencies of BRCA1 and BRCA2 mutations were 0.058% and 0.068%, respectively, which corresponded to a mutation carrier frequency of 0.25%.4 For each proband, two sets of carrier probabilities were calculated by ignoring or incorporating family history of prophylactic oophorectomy. We used the full set of individuals and incorporated information on prophylactic oophorectomy when available. Postintervention penetrances were calculated with hazard ratios, as detailed by Katki.22 For each tested relative, two sets of carrier probabilities were calculated by ignoring or considering the proband's mutation test result.
Statistical Analysis
Performance of the BRCAPRO model in ethnic minorities was evaluated in terms of its discriminatory ability and calibration.26,27 Regarding discriminatory ability, receiver operating characteristic (ROC) curves were constructed by plotting the sensitivities against 1 minus specificities by using each value of BRCAPRO prediction probabilities as a cutoff point. The area under the ROC curve (AUC)—that is, the concordance index—is a measure of the overall ability of discriminating BRCA mutation carriers from noncarriers. The AUC is also the likelihood that a randomly selected positively tested individual will have a higher predicted probability of BRCA mutation than a randomly selected negatively tested individual. An AUC of 0.5 indicated no discriminating ability, and an AUC of 1 meant perfect discrimination. AUCs were compared between ethnic groups and between predictions with or without incorporation of prophylactic oophorectomy by using the method described by DeLong et al.28 CIs for tested relatives' mutation probabilities with or without incorporation of test results of probands were evaluated by using the bootstrap method.29 For comparison with previous studies, the sensitivity and specificity of the model that used 10% mutation carrier probability as a cutoff were also presented.
Regarding calibration, the observed and predicted proportions of mutation carriers were compared in each ethnic group. Because the sensitivity of BRCA mutation detection techniques ranged from 63% to 85%,11,30 with the full sequencing method being the most sensitive method, whereas the specificity was believed to be 100%, the carrier probability calculated by the BRCAPRO model was converted for fair evaluation. Because a majority of the probands was screened by the full sequencing method, we assumed a sensitivity of 85% and calculated the carrier probabilities as the probabilities from the BRCAPRO model multiplied by 0.85. The assumed 15% false negative rate was due to undetected large deletions or rearrangements in BRCA1/2, deleterious missense variants in BRCA1/2, and breast and ovarian cancer susceptibility genes other than BRCA1/2 (D. Berry, personal communication, September 2008). We also grouped probands into four categories according to the converted probability: less than 0.05, 0.05 to 0.09, 0.10 to 0.24, and ≥ 0.25. Within each category, we calculated the average probability of positive tests (predicted proportion) and compared it with the observed proportion of mutations and the corresponding 95% CI (calculated by using the exact binomial distribution). The mean square error of prediction (ie, Brier score) was calculated to quantify the overall accuracy.31 The smaller the Brier score, the closer the predicted probability to the observed mutation status. Statistical analysis was performed by using SAS 9.1 (SAS Institute, Cary, NC) and Stata 10.0 (Stata Corp, College Station, TX).
RESULTS
Family Characteristics
In total, 292 minority families were identified through the BCFR and the University of Chicago, which included 104 African Americans, 130 Hispanics, 37 Asian-Americans, 16 Native Americans, and five families of multiple ethnicity. Because of their small numbers, Asian-American, Native American, and multiple-ethnicity families were combined and were referred to as other ethnicity. Table 1 lists the characteristics of families from the three ethnic groups. Fifty-eight (19.9%) of 292 probands tested positive for deleterious mutations: 32 in BRCA1 and 26 in BRCA2. Cancer history data were collected on 5,265 family members, and the average number was 18 individuals per family.
Table 1.
Characteristics of Probands and Families by Ethnicity
| Characteristic | Ethnicity |
|||||||
|---|---|---|---|---|---|---|---|---|
| African American |
Hispanic |
Other |
Total |
|||||
| No. | % | No. | % | No. | % | No. | % | |
| Total No. of families/probands | 104 | 130 | 58 | 292 | ||||
| Female probands | 103 | 99.0 | 127 | 97.7 | 56 | 96.6 | 286 | 98.0 |
| Probands with unilateral breast cancer | 80 | 76.9 | 93 | 71.5 | 41 | 70.7 | 214 | 73.3 |
| Probands with bilateral breast cancer | 14 | 13.5 | 16 | 12.3 | 9 | 15.5 | 39 | 13.4 |
| Probands with ovarian cancer | 6 | 5.8 | 13 | 10.0 | 3 | 5.2 | 22 | 7.5 |
| Probands with BRCA1 deleterious mutation | 17 | 16.3 | 8 | 6.2 | 7 | 12.1 | 32 | 11.0 |
| Probands with BRCA2 deleterious mutation | 15 | 14.4 | 8 | 6.2 | 3 | 5.2 | 26 | 8.9 |
| Mutation-tested individuals | 148 | 173 | 72 | 393 | ||||
| Families with ≥ 2 individuals tested | 18 | 17.3 | 18 | 13.8 | 7 | 12.1 | 43 | 14.7 |
| Families with oophorectomy | 19 | 18.3 | 26 | 20.0 | 10 | 17.2 | 55 | 18.8 |
| Individuals per family | ||||||||
| Mean | 18.6 | 17.8 | 17.6 | 18.0 | ||||
| SD | 8.4 | 10.0 | 8.7 | 9.2 | ||||
| Individuals with breast cancer per family | ||||||||
| Mean | 2.23 | 1.55 | 1.66 | 1.81 | ||||
| SD | 1.26 | 1.14 | 1.00 | 1.20 | ||||
| Individuals with bilateral breast cancer per family | ||||||||
| Mean | 0.27 | 0.17 | 0.19 | 0.21 | ||||
| SD | 0.51 | 0.40 | 0.44 | 0.45 | ||||
| Individuals with ovarian cancer per family | ||||||||
| Mean | 0.25 | 0.32 | 0.33 | 0.30 | ||||
| SD | 0.59 | 0.66 | 0.76 | 0.66 | ||||
| Age at breast cancer diagnosis | ||||||||
| Mean | 46.5 | 43.7 | 44.8 | 45.1 | ||||
| SD | 12.8 | 12.1 | 12.6 | 12.6 | ||||
| Age at ovarian cancer diagnosis | ||||||||
| Mean | 55.7 | 47.6 | 49.3 | 50.4 | ||||
| SD | 13.7 | 14.4 | 13.3 | 14.3 | ||||
Abbreviation: SD, standard deviation.
Performance by Ethnicity
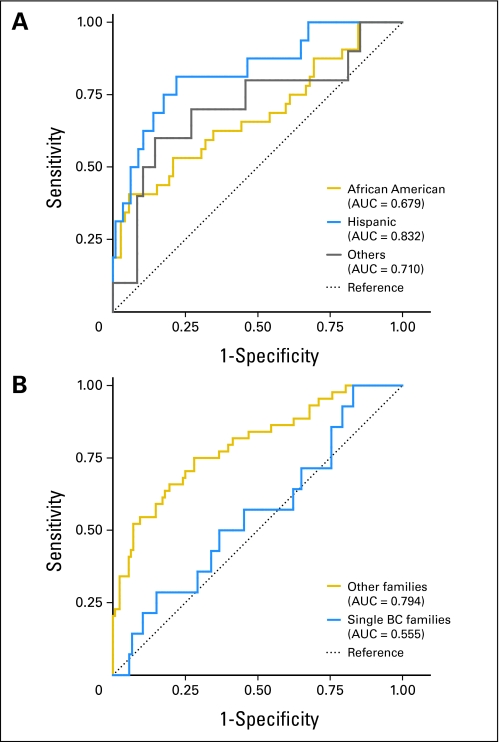
The mean carrier probability per BRCAPRO was 22.4% overall: 47.6% in probands who tested positive and 16.2% in those who tested negative (Wilcoxon rank sum test; P < 10−8). The AUC was 0.748 (95% CI, 0.672 to 0.823) for all minority groups combined. The AUCs were 0.832 (95% CI, 0.716 to 0.947), 0.679 (95% CI, 0.560 to 0.799), and 0.710 (95% CI, 0.505 to 0.916) for Hispanics, African Americans, and other minority groups, respectively (Table 2; Fig 1A).
Table 2.
Discriminatory Ability of BRCAPRO Stratified by Ethnicity
| Ethnicity by Mutation | Discriminatory Ability Analysis Data |
||||
|---|---|---|---|---|---|
| Sensitivity* | Specificity* | AUC | 95% CI | P† | |
| BRCA1/2 | |||||
| All | 0.707 | 0.645 | 0.748 | 0.672 to 0.823 | |
| African American | 0.656 | 0.528 | 0.679 | 0.560 to 0.799 | |
| Hispanic | 0.813 | 0.684 | 0.832 | 0.716 to 0.947 | |
| Other | 0.700 | 0.729 | 0.710 | 0.505 to 0.916 | .18 |
| BRCA1 | |||||
| All | 0.750 | 0.750 | 0.796 | 0.708 to 0.884 | |
| African American | 0.765 | 0.736 | 0.780 | 0.635 to 0.924 | |
| Hispanic | 0.750 | 0.762 | 0.836 | 0.716 to 0.956 | |
| Other | 0.714 | 0.745 | 0.759 | 0.554 to 0.964 | .75 |
| BRCA2 | |||||
| All | 0.423 | 0.771 | 0.634 | 0.518 to 0.749 | |
| African American | 0.267 | 0.684 | 0.509 | 0.348 to 0.670 | |
| Hispanic | 0.750 | 0.811 | 0.762 | 0.543 to 0.982 | |
| Other | 0.333 | 0.818 | 0.618 | 0.270 to 0.966 | .19 |
NOTE. BRCAPRO is a BRCA carrier prediction model.
Abbreviation: AUC, area under the receiver operating characteristic curve.
Sensitivity and specificity at 10% cutoff point of BRCA mutation probability.
P value for testing differences in area under the receiver operating characteristic curve among three ethnic groups.
Fig 1.
Receiver operating characteristic curves that compare the performance of BRCAPRO, a BRCA carrier prediction model, (A) among three ethnic minority groups and (B) between probands with breast cancer (BC) diagnosis but no family history and probands with a family history of BC. AUC, area under the receiver operating characteristic curve.
Table 3 shows the observed and predicted proportion of mutations, stratified by risk categories, ethnicity, and family structure. Overall, BRCAPRO predicted 56 detectable mutation carriers (19.1%), when that the sensitivity of genetic testing techniques was assumed to be 0.85. This was close to the observed number of 58 mutations (19.9%). However, the expected proportion was higher than the observed proportion in high-risk probands (≥ 0.25 prior probability), and the opposite was seen in low-risk probands (< 0.05 prior probability). As indicated by Brier scores, the prediction accuracy (or calibration) was the highest in Hispanics, followed by other minority groups, and was the lowest in African Americans.
Table 3.
BRCAPRO-Predicted Proportion of Deleterious BRCA1 or BRCA2 Mutations and Observed Proportion
| Variable | No. of Probands | Proportion Data |
Brier Score | ||
|---|---|---|---|---|---|
| Expected (%)* | Observed† |
||||
| % | 95% CI | ||||
| Ethnicity | |||||
| African American | 104 | 23.3 | 30.8 | 22.1–40.6 | 0.208‡ |
| Hispanic | 130 | 15.9 | 12.3 | 7.2–19.2 | 0.089 |
| Other | 58 | 18.6 | 17.2 | 8.6-29.4 | 0.139‡ |
| All probands | 292 | 19.1 | 19.9 | 15.4–24.9 | 0.141‡ |
| < 0.05 | 149 | 1.9 | 10.7 | 6.2–16.9 | |
| 0.05 to < 0.10 | 25 | 7.2 | 8.0 | 1.0–26.0 | |
| 0.10 to < 0.25 | 42 | 15.0 | 16.7 | 7.0–31.4 | |
| ≥ 0.25 | 76 | 65.5 | 43.4 | 32.1–55.3 | |
| Single breast cancer families | 120 | 7.1 | 11.7 | 6.5–18.8 | 0.124‡ |
| < 0.05 | 92 | 2.0 | 10.9 | 5.3–19.1 | |
| 0.05 to < 0.10 | 10 | 7.4 | 10.0 | 0.3–44.5 | |
| 0.10 to < 0.25 | 9 | 15.0 | 11.1 | 0.3–48.2 | |
| ≥ 0.25 | 9 | 51.0 | 22.2 | 2.8–60.0 | |
| Other families | 172 | 27.4 | 25.6 | 19.2–32.8 | 0.153‡ |
| < 0.05 | 57 | 1.8 | 10.5 | 4.0–21.5 | |
| 0.05 to < 0.10 | 15 | 7.0 | 6.7 | 0.2–31.9 | |
| 0.10 to < 0.25 | 33 | 15.0 | 18.2 | 7.0–35.5 | |
| ≥ 0.25 | 57 | 59.9 | 46.3 | 34.0–58.9 | |
NOTE. BRCAPRO is a BRCA carrier prediction model.
Calculated as BRCAPRO-predicted probability multiplied by 0.85.
95% confidence interval is for the true proportion.
P < .01
There were 120 probands with breast cancer but with no family history of breast cancer. BRCAPRO had extremely low discriminatory value in these families (AUC, 0.555; Fig 2B). Predicted and observed mutation proportions for these individuals were quite different (Table 3). For the remaining 172 probands, BRCAPRO had good discriminatory value (AUC, 0.794).
Fig 2.
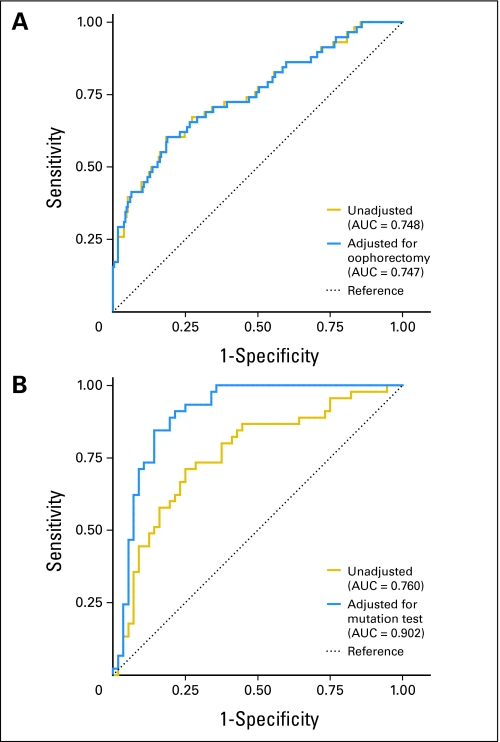
Receiver operating characteristic curves that compare the performance of BRCAPRO, a BRCA carrier prediction model, in (A) probands who ignore and incorporate prophylactic oophorectomy and in (B) relatives who ignore and incorporate genetic test results of probands. AUC, area under the receiver operating characteristic curve.
Incorporation of Prophylactic Oophorectomy
Prophylactic oophorectomy was performed in 62 individuals from 55 families (19%). After a history of prophylactic oophorectomy was accounted for, the AUC, sensitivity, and specificity at a 10% threshold changed only slightly (Appendix Table A2, online only; Fig 2A). For all 55 probands, the predicted carrier probability was increased, but the absolute change was small (mean, 0.015) and the increase in probability of greater than 0.05 was noted in only five probands.
Incorporation of Mutation Results of Probands
In total, 393 individuals from the 292 families were tested for BRCA1 and BRCA2 mutations, and 43 families (15%) had more than one person tested. The mutation test results of probands were incorporated in the mutation prediction of the 101 additional family members tested by using a specific option in BRCAPRO, which resulted in a significant increase in discriminatory ability. Figure 2B and Appendix Table A2 show that the AUC increased from 0.760 to 0.902 for prediction of BRCA1 or BRCA2 (P = .02). In the 17 families in whom probands tested negative, 22 additional family members were tested for BRCA mutations. Their mean predicted probability decreased from 0.146 to 0.086 after incorporation of probands' test results.
DISCUSSION
In this study, we evaluated the performance of the BRCAPRO model in nearly 300 ethnic minority families in the United States and demonstrated that the model had good overall discrimination between BRCA mutation carriers and noncarriers in minority families. The AUC was 0.75 for all minority groups combined, which is within the range of those reported in non-Hispanic white populations (0.71 to 0.83).9,10,13,14,16,32–34
Of the minority groups examined, BRCAPRO performed the best in Hispanics, as indicated by the highest AUC (0.83) and the smallest Brier score (0.089), in which the AUC was higher than in a previous observation in Hispanics (0.77).18 The model had the worst performance in African Americans (AUC, 0.68; Brier score, 0.208), in which the AUC was lower than our previous observation in African Americans (0.77).17 The performance of BRCARPO was intermediate for the other minority groups, mainly Asian-Americans and Native Americans (AUC, 0.71; Brier score, 0.139), and was similar to that observed in Han Chinese (0.70).35 However, the difference between ethnic groups was not statistically significant. Thus, the data suggest that BRCAPRO performs reasonably well and is applicable to all minorities. However, the sample size of this study may not be sufficient for subgroup comparisons. Real differences possibly exist in BRCAPRO performance across ethnic minority groups, because the same penetrances and non-Ashkenazi white allele frequencies were used in the calculation. These genetic parameters likely vary across ethnicity. Better BRCAPRO performance in Hispanics might occur because Hispanics are genetically closer to other white populations. Thus, mutation prediction could improve if population-specific allele frequencies and penetrance data were available. Our findings underscore the need for population-based studies in these minority populations.36,37
A single instance of breast cancer in families that have limited family structure presents a challenge for mutation prediction with the BRCAPRO model and probably with other models. Weitzel et al15 reported an AUC of 0.67 for the BRCAPRO model in women with breast cancer who did not have any first- or second-degree relatives with breast or ovarian cancers. We observed a similar finding in the current cohort. In terms of accuracy of the model, we showed that the numbers of mutations predicted and observed were virtually the same when the mutation detection method is assumed to have 85% sensitivity. Consistent with previous studies in predominantly white populations,9–11 we found that BRCAPRO overestimated mutation probability for high-risk persons and underestimated mutation probability for low-risk persons. Interestingly, this pattern was also observed for the Breast and Ovarian Analysis of Disease Incidence and Carrier Estimation Algorithm (BOADICEA) model, which allows for polygenic locus effects,10 and for the Log Odds of the Probability of Carrying an Ancestral Mutation in BRCA1 or BRCA2 for a Defined Personal and Family Cancer History in an Ashkenazi Jewish Woman (LAMBDA) model, which was developed empirically.34 Reasons for this pattern are unclear and may be related to families that have a single breast cancer diagnosis and limited family structure, familial clustering caused by environmental factors, heterogeneous penetrance caused by genetic/environmental modifiers and mutation spectrum, other low penetrance genes, and imperfect sensitivity of molecular methodology. We showed that lack of information in families that have a single breast cancer diagnosis is an important cause of this pattern. Whatever the reasons, genetic counselors should treat the calculated mutation probability from pretest prediction models with caution and should consider the amount of available information when interpreting mutation probability. Given the performance in both discrimination and accuracy, these results indicate that BRCAPRO is a useful risk assessment tool in clinical settings for minority families, but clinicians should exercise judgment in using the numbers to make recommendations.
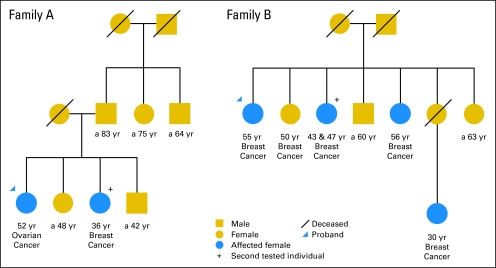
Incorporation of the proband's test result significantly improved the ability to distinguish likely mutation carriers from noncarriers (ie, AUC increased from 0.76 to 0.90). This is the first study, to our knowledge, to estimate the magnitude of improvement in clinical settings. Thus, it is helpful to recalculate the mutation probability for other family members after the proband has been tested, especially if the proband tests negative. In the two examples shown in Figure 3, the probands tested negative. For the 36-year-old breast cancer patient in family A, her mutation probability was 0.254 before the proband was tested, and the recalculated probability from BRCAPRO incorporation of the proband's test result was 0.075. Thus, a recommendation not to test for BRCA mutations could be entertained. (Indeed, her BRCA test result was negative.) In contrast, the family member with bilateral breast cancer in family B had a mutation probability of 0.882 before the proband's test result was considered, and her recalculated probability was 0.740 after incorporation of the proband's test result. Thus, a strong recommendation to test for BRCA mutation could be made. (In fact, she had a BRCA1 mutation).
Fig 3.
Two family trees with breast and ovarian cancer history.
We did not find that the inclusion of information on prophylactic oophorectomy, a new feature of BRCAPRO, improved mutation prediction. Improvement may be relatively small in the current cohort, because less than 20% of families had at least one member with a history of oophorectomy. Of the 62 individuals who underwent prophylactic oophorectomy, 22 later developed breast cancer after 11 years, on average. As illustrated by Katki,22 distortion caused by ignorance of information on prophylactic oophorectomy is large if the family member has lived many years after the surgery. Therefore, we may not be able to see an influence yet, as prophylactic interventions for familial breast cancer are still relatively recent. Another explanation is that the proband may not know whether her relatives had a prophylactic oophorectomy.
In conclusion, this study demonstrates that the BRCAPRO model performs well in clinic settings and supports its routine use in pretest prediction of BRCA mutations in minority families, especially Hispanic families. Mutation test results of probands, especially if negative, provide additional discriminatory ability for counselees, which may help counselors decide whether to offer other family members testing when one member has already tested negative. The study also highlights the need for population-based studies to estimate penetrance and allele frequencies of BRCA genes in minority populations. Lastly, the inaccuracy in carrier prediction using BRCAPRO for families with a single breast cancer diagnosis is a challenge worthy of additional investigation. Genetic counselors should recognize this limitation when using BRCAPRO or other models to recommend genetic testing in single-diagnosis families.
Acknowledgment
We thank Jeniffer Iriondo-Perez and Chaehyung Ahn at RTI International for database management; Alice Whittemore, PhD, Hormuzd Katki, PhD, Giovanni Parmigiani, PhD, Edwin Iverson, PhD, Esther M. John, PhD, Rita Nanda, MD, James Fackenthal, PhD, and Betsy Bove, PhD, for advice; Masha Kocherginsky, PhD, for R programming help; and Michelle Porcellino for critical reading of the manuscript.
Appendix
Table A1.
Characteristics of Probands and Families by Study Center
| Characteristic | Study Center Location |
|||||||
|---|---|---|---|---|---|---|---|---|
| Philadelphia |
New York |
Salt Lake City |
Chicago |
|||||
| No. | % | No. | % | No. | % | No. | % | |
| Total No. of families/probands | 41 | 166 | 3 | 82 | ||||
| Female probands | 39 | 95.1 | 162 | 97.6 | 3 | 100 | 82 | 100 |
| Probands with unilateral breast cancer | 35 | 85.4 | 123 | 74.1 | 1 | 33.3 | 55 | 67.1 |
| Probands with bilateral breast cancer | 4 | 9.8 | 20 | 12.0 | 2 | 66.7 | 13 | 15.9 |
| Probands with ovarian cancer | 1 | 2.4 | 13 | 7.8 | 0 | 0 | 8 | 9.8 |
| Probands with BRCA1 deleterious mutation | 4 | 9.8 | 12 | 7.2 | 3 | 100 | 13 | 15.9 |
| Probands with BRCA2 deleterious mutation | 5 | 12.2 | 9 | 5.4 | 0 | 0 | 12 | 14.6 |
| Mutation-tested individuals | 41 | 218 | 9 | 125 | ||||
| Families with ≥ 2 individuals tested | 0 | 24 | 14.5 | 3 | 100 | 16 | 19.5 | |
| Families with oophorectomy | 9 | 22.0 | 38 | 23.9 | 3 | 100 | 5 | 6.1 |
| No. of individuals per family | ||||||||
| Mean | 19.8 | 15.7 | 37.3 | 21.1 | ||||
| SD | 7.2 | 9.3 | 7.1 | 8.0 | ||||
| No. of individuals with breast cancer per family | ||||||||
| Mean | 1.95 | 1.50 | 3.00 | 2.33 | ||||
| SD | 0.92 | 1.01 | 1.00 | 1.46 | ||||
| No. of individuals with bilateral breast cancer per family | ||||||||
| Mean | 0.20 | 0.15 | 1.00 | 0.30 | ||||
| SD | 0.46 | 0.36 | 1.00 | 0.54 | ||||
| No. of individuals with ovarian cancer per family | ||||||||
| Mean | 0.10 | 0.31 | 1.00 | 0.35 | ||||
| SD | 0.30 | 0.67 | 1.00 | 0.71 | ||||
Abbreviation: SD, standard deviation.
Table A2.
Performance of Prediction of Deleterious Mutations by BRCAPRO After Adjustment for Mutation Results of Probands or History of Oophorectomy
| Analysis Type by Group | Prediction Analysis Data |
|||||
|---|---|---|---|---|---|---|
| No. | Sensitivity* | Specificity* | AUC | 95% CI | P† | |
| Probands | ||||||
| Unadjusted | 292 | 0.707 | 0.645 | 0.748 | 0.672 to 0.823 | |
| Incorporation of oophorectomy | 292 | 0.707 | 0.641 | 0.747 | 0.672 to 0.823 | .84 |
| Relatives | ||||||
| Unadjusted | 101 | 0.800 | 0.607 | 0.760 | 0.612 to 0.884 | |
| Incorporation of mutation results of probands | 101 | 1.000 | 0.607 | 0.902 | 0.830 to 0.962 | .02 |
NOTE. BRCAPRO is a BRCA carrier prediction model.
Abbreviation: AUC, area under the receiver operating characteristic curve.
Sensitivity and specificity at 10% cutoff point of BRCA mutation probability.
P value for testing differences from unadjusted area under the receiver operating characteristic curve.
Footnotes
Supported by National Cancer Institute Grant No. CA-RO1 89085-01A, by the Falk Medical Research Trust, and by the Entertainment Industry National Women's Cancer Research Alliance. O.I.O. is a McArthur Fellow.
Presented in part at the 30th Annual San Antonio Breast Cancer Symposium, December 13-16, 2007, San Antonio, TX; and at the 43rd Annual Meeting of the American Society of Clinical Oncology, June 1-5, 2007, Chicago, IL.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Dezheng Huo, Olufunmilayo I. Olopade
Financial support: Olufunmilayo I. Olopade
Administrative support: Shelly Cummings, Kisha Hope, Olufunmilayo I. Olopade
Provision of study materials or patients: Ruby T. Senie, Mary Daly, Saundra S. Buys, Shelly Cummings, Kisha Hope, Olufunmilayo I. Olopade
Collection and assembly of data: Ruby T. Senie, Mary Daly, Shelly Cummings, Jacqueline Ogutha, Kisha Hope, Olufunmilayo I. Olopade
Data analysis and interpretation: Dezheng Huo, Ruby T. Senie
Manuscript writing: Dezheng Huo, Olufunmilayo I. Olopade
Final approval of manuscript: Dezheng Huo, Ruby T. Senie, Mary Daly, Saundra S. Buys, Shelly Cummings, Jacqueline Ogutha, Kisha Hope, Olufunmilayo I. Olopade
REFERENCES
- 1.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25:1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robson M, Offit K. Management of an inherited predisposition to breast cancer. N Engl J Med. 2007;357:154–162. doi: 10.1056/NEJMcp071286. [DOI] [PubMed] [Google Scholar]
- 3.Parmigiani G, Berry D, Aguilar O. Determining carrier probabilities for breast cancer-susceptibility genes BRCA1 and BRCA2. Am J Hum Genet. 1998;62:145–158. doi: 10.1086/301670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoniou AC, Pharoah PD, McMullan G, et al. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer. 2002;86:76–83. doi: 10.1038/sj.bjc.6600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couch FJ, DeShano ML, Blackwood MA, et al. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N Engl J Med. 1997;336:1409–1415. doi: 10.1056/NEJM199705153362002. [DOI] [PubMed] [Google Scholar]
- 6.Evans DG, Eccles DM, Rahman N, et al. A new scoring system for the chances of identifying a BRCA1/2 mutation outperforms existing models including BRCAPRO. J Med Genet. 2004;41:474–480. doi: 10.1136/jmg.2003.017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank TS, Deffenbaugh AM, Reid JE, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: Analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 8.Apicella C, Andrews L, Hodgson SV, et al. Log odds of carrying an Ancestral Mutation in BRCA1 or BRCA2 for a defined personal and family history in an Ashkenazi Jewish woman (LAMBDA). Breast Cancer Res. 2003;5:R206–R216. doi: 10.1186/bcr644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marroni F, Aretini P, D'Andrea E, et al. Evaluation of widely used models for predicting BRCA1 and BRCA2 mutations. J Med Genet. 2004;41:278–285. doi: 10.1136/jmg.2003.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barcenas CH, Hosain GM, Arun B, et al. Assessing BRCA carrier probabilities in extended families. J Clin Oncol. 2006;24:354–360. doi: 10.1200/JCO.2005.02.2368. [DOI] [PubMed] [Google Scholar]
- 11.Berry DA, Iversen ES, Jr, Gudbjartsson DF, et al. BRCAPRO validation, sensitivity of genetic testing of BRCA1/BRCA2, and prevalence of other breast cancer susceptibility genes. J Clin Oncol. 2002;20:2701–2712. doi: 10.1200/JCO.2002.05.121. [DOI] [PubMed] [Google Scholar]
- 12.Euhus DM, Smith KC, Robinson L, et al. Pretest prediction of BRCA1 or BRCA2 mutation by risk counselors and the computer model BRCAPRO. J Natl Cancer Inst. 2002;94:844–851. doi: 10.1093/jnci/94.11.844. [DOI] [PubMed] [Google Scholar]
- 13.James PA, Doherty R, Harris M, et al. Optimal selection of individuals for BRCA mutation testing: A comparison of available methods. J Clin Oncol. 2006;24:707–715. doi: 10.1200/JCO.2005.01.9737. [DOI] [PubMed] [Google Scholar]
- 14.Kang HH, Williams R, Leary J, et al. Evaluation of models to predict BRCA germline mutations. Br J Cancer. 2006;95:914–920. doi: 10.1038/sj.bjc.6603358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weitzel JN, Lagos VI, Cullinane CA, et al. Limited family structure and BRCA gene mutation status in single cases of breast cancer. JAMA. 2007;297:2587–2595. doi: 10.1001/jama.297.23.2587. [DOI] [PubMed] [Google Scholar]
- 16.Parmigiani G, Chen S, Iversen ES, Jr, et al. Validity of models for predicting BRCA1 and BRCA2 mutations. Ann Intern Med. 2007;147:441–450. doi: 10.7326/0003-4819-147-7-200710020-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nanda R, Schumm LP, Cummings S, et al. Genetic testing in an ethnically diverse cohort of high-risk women: A comparative analysis of BRCA1 and BRCA2 mutations in American families of European and African ancestry. JAMA. 2005;294:1925–1933. doi: 10.1001/jama.294.15.1925. [DOI] [PubMed] [Google Scholar]
- 18.Vogel KJ, Atchley DP, Erlichman J, et al. BRCA1 and BRCA2 genetic testing in Hispanic patients: Mutation prevalence and evaluation of the BRCAPRO risk assessment model. J Clin Oncol. 2007;25:4635–4641. doi: 10.1200/JCO.2006.10.4703. [DOI] [PubMed] [Google Scholar]
- 19.Armstrong K, Micco E, Carney A, et al. Racial differences in the use of BRCA1/2 testing among women with a family history of breast or ovarian cancer. JAMA. 2005;293:1729–1736. doi: 10.1001/jama.293.14.1729. [DOI] [PubMed] [Google Scholar]
- 20.Hall M, Olopade OI. Confronting genetic testing disparities: Knowledge is power. JAMA. 2005;293:1783–1785. doi: 10.1001/jama.293.14.1783. [DOI] [PubMed] [Google Scholar]
- 21.US Census Bureau. Census 2000 data for the United States. http://www.census.gov/census2000/states/us.html.
- 22.Katki HA. Incorporating medical interventions into carrier probability estimation for genetic counseling. BMC Med Genet. 2007;8:13. doi: 10.1186/1471-2350-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S, Wang W, Broman KW, et al. BayesMendel: An R environment for Mendelian risk prediction. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1063. Article21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.John EM, Hopper JL, Beck JC, et al. The Breast Cancer Family Registry: An infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:R375–R389. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R Development Core Team: R. Vienna, Austria: R Foundation for Statistical Computing; 2006. A Language and Environment for Statistical Computing. [Google Scholar]
- 26.DeGroot M, Fienberg S. The comparison and evaluation of forecasters. Statistician. 1983;32:12–22. [Google Scholar]
- 27.Spiegelhalter DJ. Probabilistic prediction in patient management and clinical trials. Stat Med. 1986;5:421–433. doi: 10.1002/sim.4780050506. [DOI] [PubMed] [Google Scholar]
- 28.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 29.Pepe MS. New York City, NY: Oxford University Press; 2003. The Statistical Evaluation of Medical Tests for Classification and Prediction. [Google Scholar]
- 30.Ford D, Easton DF, Stratton M, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families: The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brier G. Verification of forecasts expresssed in terms of probability. Monthly Weather Rev. 1950;78:1–3. [Google Scholar]
- 32.Antoniou AC, Durocher F, Smith P, et al. BRCA1 and BRCA2 mutation predictions using the BOADICEA and BRCAPRO models and penetrance estimation in high-risk French-Canadian families. Breast Cancer Res. 2006;8:R3. doi: 10.1186/bcr1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oros KK, Ghadirian P, Maugard CM, et al. Application of BRCA1 and BRCA2 mutation carrier prediction models in breast and/or ovarian cancer families of French Canadian descent. Clin Genet. 2006;70:320–329. doi: 10.1111/j.1399-0004.2006.00673.x. [DOI] [PubMed] [Google Scholar]
- 34.Apicella C, Dowty JG, Dite GS, et al. Validation study of the LAMBDA model for predicting the BRCA1 or BRCA2 mutation carrier status of North American Ashkenazi Jewish women. Clin Genet. 2007;72:87–97. doi: 10.1111/j.1399-0004.2007.00841.x. [DOI] [PubMed] [Google Scholar]
- 35.Rao NY, Hu Z, Li WF, et al. Models for predicting BRCA1 and BRCA2 mutations in Han Chinese familial breast and/or ovarian cancer patients. Breast Cancer Res Treat. 2009;113:467–477. doi: 10.1007/s10549-008-9965-9. [DOI] [PubMed] [Google Scholar]
- 36.Huo D, Olopade OI. Genetic testing in diverse populations: Are researchers doing enough to get out the correct message? JAMA. 2007;298:2910–2911. doi: 10.1001/jama.298.24.2910. [DOI] [PubMed] [Google Scholar]
- 37.John EM, Miron A, Gong G, et al. Prevalence of pathogenic BRCA1 mutation carriers in 5 US racial/ethnic groups. JAMA. 2007;298:2869–2876. doi: 10.1001/jama.298.24.2869. [DOI] [PubMed] [Google Scholar]