Abstract
We report the characterization of rdh54+, the second fission yeast Schizosaccharomyces pombe Rad54 homolog. rdh54+ shares sequence and functional homology to budding yeast RDH54/TID1. Rdh54p is present during meiosis with appropriate timing for a meiotic recombination factor. It interacts with Rhp51 and the meiotic Rhp51 homolog Dmc1 in yeast two-hybrid assays. Deletion of rdh54+ has no effect on DNA damage repair during the haploid vegetative cell cycle. In meiosis, however, rdh54Δ shows decreased spore viability and homologous recombination with a concomitant increase in sister chromatid exchange. The rdh54Δ single mutant repairs meiotic breaks with similar timing to wild type, suggesting redundancy of meiotic recombination factors. Consistent with this, the rdh54Δ rhp54Δ double mutant fails to repair meiotic double strand breaks. Live cell analysis shows that rdh54Δ rhp54Δ asci do not arrest, but undergo both meiotic divisions with near normal timing, suggesting that failure to repair double strand breaks in S. pombe meiosis does not result in checkpoint arrest.
INTRODUCTION
DNA double-strand breaks (DSBs), whether caused by exogenous damaging agents or introduced to initiate meiotic recombination, must be repaired for a cell to survive. Homologous recombination (HR) serves as one means to repair DSBs in cycling cells and seems to be the major repair mechanism during meiosis. In budding yeast and other systems, a large group of proteins, known as the Rad52 epistasis group, is involved in HR (for reviews, see Paques and Haber, 1999; Symington, 2002). HR involves resection of the break to generate a 3′ single-stranded DNA end, invasion of the resected end into a homologous double-strand of DNA, repair DNA synthesis, and resolution of the synapsed molecule to generate intact double-stranded DNA.
A key step in HR is the formation of the displacement loop (D-loop). In this process, Rad51, a homolog of the Escherichia coli recA protein, forms filaments on the resected, single-stranded DNA end (Sung and Robberson, 1995). This nucleoprotein filament invades into homologous double-stranded DNA creating the D-loop. In vitro experiments have shown that Rad51 alone can catalyze D-loop formation on linear naked DNA but that the reaction requires Rad54 for strand invasion into a more complex double-stranded (ds)DNA template (Petukhova et al., 1998). Rad54 is a member of the Swi2/Snf2 family of proteins, which contain a DNA-stimulated ATPase domain. Many Swi2/Snf2 proteins have been implicated in chromatin remodeling (Fyodorov and Kadonaga, 2001). Although bacterial RecA lacks the ability to catalyze D-loop formation on chromatin, Rad51/Rad54 together perform strand invasion into DNA packaged into nucleosomes at least as readily as with naked DNA (Alexiadis and Kadonaga, 2002; Jaskelioff et al., 2003), leading to a model suggesting that the role of Rad54 is to alter chromatin structure to allow access of the Rad51-coated single-stranded (ss)DNA into the homologous template. Additionally, budding yeast Rad54 can alter the position of nucleosomes on DNA, and this activity is enhanced in the presence of Rad51 (Alexeev et al., 2003). Biochemical assays have shown that Rad54 can induce transient DNA strand separation, and this separation likely allows access of the Rad51 DNA filament (Van Komen et al., 2000). The critical role of budding yeast Rad51 and Rad54 in DSB repair is demonstrated by the severe sensitivity of the respective mutants to X-rays (Paques and Haber, 1999).
During meiosis in budding yeast, RAD51 and RAD54 together are not sufficient for DSB repair. A homolog of RAD51, DMC1, is present during meiosis and both RAD51 and DMC1 are involved in meiotic DSB repair (Bishop et al., 1992). RDH54/TID1, another Rad54 homolog in budding yeast, has been shown to play a role in meiotic recombination and is a contributor to DSB repair during the vegetative cell cycle, especially in diploids (Dresser et al., 1997; Klein, 1997; Shinohara et al., 1997). During meiosis, RAD54 and RDH54 are partially redundant. Both single mutants have somewhat reduced spore viability, but when the two mutations are combined the double mutant fails to repair DSBs and arrests at meiotic prophase (Shinohara et al., 1997). As with RAD54, in vitro assays have identified a role for RDH54 in promoting D-loop formation by a RAD51 nucleoprotein filament (Petukhova et al., 2000).
In the fission yeast Schizosaccharomyces pombe, Rhp51 and Rhp54 have been shown to have damage sensitivity during vegetative growth reminiscent of their budding yeast counterparts Rad51 and Rad54, respectively, although the mutations cause greater UV sensitivity in fission than in budding yeast (Muris et al., 1993, 1996). Rhp51p is critical for meiosis in fission yeast, because its absence yields spores with <2% viability (Muris et al., 1997). In contrast, a disruption of S. pombe dmc1+, homologous to Saccharomyces cerevisiae DMC1, modestly reduces mitotic recombination but has little effect on overall meiotic progression or spore viability (Fukushima et al., 2000) and does not seem to affect repair of meiotic DSBs (Shimada et al., 2002). This contrasts with the severe meiotic phenotype of Δdmc1 in budding yeast, which results in meiotic delay or arrest, depending on strain background (Bishop et al., 1992; Rockmill and Roeder, 1994), suggesting SpDmc1 has a reduced role compared with ScDMC1 in meiosis.
Fission yeast Δrhp54 shows little or no reduction in meiotic recombination (Muris et al., 1997; van den Bosch et al., 2002) and produces some viable spores (27%) (Muris et al., 1997). This suggests that a second Rad54 homolog in S. pombe may work together with Rhp54p to repair DSBs during meiosis.
We report here the cloning and characterization of the fission yeast gene rdh54+, homologous to budding yeast RDH54/TID1. We examined the level of Rdh54p during vegetative and meiotic growth and see that the protein is present with appropriate timing for a meiosis specific recombination factor. Two-hybrid analysis shows that Rdh54 interacts with Dmc1 and with Rhp51. Characterization of a strain lacking rdh54+ shows that the single mutant has no vegetative defects. In meiosis, there is a modest effect on meiotic recombination and spore viability, but no overall defect in DSB repair. However, an rdh54Δ rhp54Δ double mutant is unable to repair meiotic DSBs. Live cell analysis reveals that the double mutant undergoes both meiotic divisions, despite the absence of DSB repair. These divisions are highly disordered and viable spores are not produced but they occur with near normal timing. Thus, severe defects in meiotic homologous recombination are not sufficient to arrest meiotic progression in S. pombe.
MATERIALS AND METHODS
Media and Strains
Strains (Table 1) were constructed using standard techniques (Moreno et al., 1991). The rhp54Δ allele was described in Muris et al. (1996 and was a kind gift of V. Bashkirov (Russian Academy of Sciences, Moscow, Russia). Fission yeast strains were grown in yeast extract plus supplements (YES) or in Edinburgh minimal medium (EMM) with appropriate supplements as described previously (Moreno et al., 1991). Synthetic sporulation agar (SPAS) (Gutz et al., 1974) was used for matings. Transformations were carried out by electroporation (Kelly et al., 1993). For yeast two-hybrid assays, budding yeast was cultured on YPD or SD lacking appropriate amino acids (Guthrie and Fink, 1991). UV sensitivity assays were performed at 32°C as described previously (Snaith et al., 2000).
Table 1.
Strains used in this study
| Name | Genotype | Source |
|---|---|---|
| FY252 | h— can1-1 leu1-32 ade6-M210 | Our laboratory stock |
| FY254 | h— can1-1 leu1-32 ura4-D18 ade6-M216 | Our laboratory stock |
| FY255 | H+ can1-1 leu1-32 ura4-D18 ade6-M210 | Our laboratory stock |
| FY259 | h+ can1-1 leu1-32 ade6-M216 | Our laboratory stock |
| FY572 | h— pat1-114 ura4-D18 leu1-32 ade6-M210 | Our laboratory stock |
| FY766 | mat2-102 ura4-D18 leu1-32 ade6-M210 | Our laboratory stock |
| FY1251 | h+ ade6-M26 his4-239 | G.R. Smith |
| FY1252 | h— ade6-52 lys4-95 | G.R. Smith |
| FY1454 | h— rdh54□::ura4+ ura4-D18 leu1-32 ade6-M210 | This study |
| FY1455 | h+ rdh54□::ura4+ ura4-D18 leu1-32 ade6-M216 | This study |
| FY1456 | h— pat1-114 rdh54□::ura4+ ura4-D18 leu1-32 ade6-M210 | This study |
| FY1596 | h— pat1-114 rdh54-SV5::leu1+ ura4 leu1 ade6-M210 | This study |
| FY1664 | h+ rhp54Δ::ura4+ ade6-52 lys4-95 ura4-D18 | This study |
| FY1665 | h— smt-0 rhp54Δ::ura4+ ade6-M26 his4-239 ura4-D18 | This study |
| FY1744 | h+ pat1-114 rhp54-HA::leu1+ ura4 leu1 ade6-M210 | This study |
| FY1773 | h+ pat1-114 rdh54-SV5::leu1+ cdc22-M45 ura4-D18 leu1-32 ade6-M210 | This study |
| FY1774 | h— pat1-114 rdh54-SV5::leu1+ cdc10-V50 ura4-D18 leu1-32 ade6-M210 | This study |
| FY1775 | h+ pat1-114 rdh54-SV5::leu1+ mei4□::ura4+ ura4-D18 leu1-32 ade6-M210 | This study |
| FY1776 | h+ pat1-114 rdh54-SV5::leu1+ rep1::ura4+ ura4-D18 leu1-32 ade6-M210 | This study |
| FY1777 | h— rdh54-V5::leu1+ leu1-32 ade6-M216 ura4-D18 | This study |
| FY1778 | h+ rhp54-HA::leu1+ leu1-32 ade6-M210 ura4-D18 | This study |
| FY1801 | h+ pat1-114 rec12Δ::LEU2 rdh54-V5::leu1+ ura4-18 ade6-M210 | This study |
| FY1898 | h— rdh54Δ::ura4+ ade6-L469/pUC8/his3+/ade6-M375 ura4-D18 leu1-32 his3-D1 | This study |
| FY1866 | h— smt-0 rhp54Δ::ura4+ ura4-D18 leu1-32 ade6-M216 | This study |
| FY1901 | h— smt-0 rhp54Δ::ura4+ ade6-L469/pUC8/his3+/ade6-M375 ura4-D18 leu1-32 his3-D1 | This study |
| FY1921 | h— rdh54Δ::ura4+ ade6-M26 his4-239 ura4-D18 | This study |
| FY1941 | h+ rdh54Δ::ura4+ ade6-52 lys4-95 ura4-D18 | This study |
| FY1942 | h+ rdh54Δ::ura4+ ade6-M375-M210 leu1-32 ura4-D18 his3-D1 | This study |
| FY1944 | h+ rhp54Δ::ura4+ ade6-M375-M210 leu1-32 ura4-D18 his3-D1 | This study |
| FY1946 | h— smt-0 rhp54Δ::ura4+ rdh54Δ::ura4+ ade6-M26 his4-239 ura4-D18 | This study |
| FY1978 | h+ rhp54Δ::ura4+ rdh54Δ::ura4+ ade6-52 lys4-95 ura4-D18 | This study |
| FY2122 | h+ rhp54Δ::ura4+ ura4-D18 leu1-32 ade6-M210 | This study |
| FY2126 | h+ rhp54Δ::ura4+ rdh54Δ::ura4+ leu1-32 ade6-M216 ura4-D18 | This study |
| FY2127 | h— smt-0 rhp54Δ::ura4+ rdh54Δ::ura4+ ura4-D18 leu1-32 ade6-M210 | This study |
| FY2132 | h+ ade6-L469/pUC8/his3+/ade6-M375 leu1-32 his3-D1 | This study |
| FY2133 | h— ade6-M375-M210 leu1-32 his3-D1 | This study |
| FY2142 | h— smt-0 pat1-114 rhp54Δ::ura4+ rdh54Δ::ura4+ ura4-D18 leu1-32 ade6-M210 | This study |
| FY2299 | h— smt-0 rhp54Δ::ura4+ rdh54-V5::leu1+ ura4-D18 ade6-M216 leu1-32 | This study |
| FY2300 | h+ rhp54Δ::ura4+ rdh54-V5::leu1+ ura4-D18 ade6-M210 leu1-32 | This study |
| FY2344 | h— rhp54-HA::leu1+ leu1-32 ade6-M216 ura4-D18 | This study |
| FY2353 | h— smt-0 rhp54Δ::ura4+ pat1-114 ura4 leu1 ade6-M210 | This study |
| FY2400 | h+ rdh54-V5::leu1+ leu1-32 ade6-M210 ura4-D18 | This study |
Deletion of rdh54+
The 1-kb region 5′ of the START codon was polymerase chain reaction (PCR) amplified to add a 5′ SalI site and 3′ BglII site and the 0.8-kb region 3′ of the stop codon was amplified to add a 3′ SalI site (the endogenous BglII site at the stop codon was used as the 5′ site) by using primers 5′-CGTCGACCTCAGCACAGTATGACTATCC-3′ and 5′-CAGATCTGGTTTCAATTCTCTTTGGTAAC-3′ for promoter region and primers 5′-GTTCGAATAGATCTATTTCGGACT-3′ and 5′-CGTCGACCCGAAGCATGAAGATGGAGAAG-3′ for 3′ untranslated region. PCR products were cut with SalI and BglII and both were simultaneously cloned into pBluescript II KS+ (Stratagene, La Jolla, CA) cut with SalI. The resulting construct was cut with BglII and blunted and a HindIII cut and blunted fragment containing ura4+ from pSLF173 (Forsburg and Sherman, 1997) was ligated in to generate pMGC58. The SalI fragment from pMGC58 was transformed into a wild-type diploid, and a stable transformant was sporulated to generate FY1454. Proper integration was confirmed by Southern blot (our unpublished data). This disruption removes the region from 27 nucleotides 5′ of the start codon to the stop codon of rdh54+ and replaces it with ura4+.
Rhp54p and Rdh54p Epitope Tags
The 3′ end of each coding region was PCR amplified to add a 5′ SalI and KpnI site and in-frame 3′ NotI site (primers 5′-GTCGACGGTACCCCTGATCTATTGAATTTAACTG-3′ and 5′-GCGGCCGCAATGAGATTTGTATTGGAAAACGG-3′ for Rhp54 and primers 5′-GTCGACGGTACCCCGCCAAGAAGTATGCAAATA-3′ and 5′-GCGGCCGCTTTCGAACTTTTTATGCATTAG-3′ for Rdh54). The PCR product was cut with SalI and NotI and ligated into XhoI and NotI cut pSLF172 (Forsburg and Sherman, 1997) for Rhp54-HA or pSLF972 (pSLF172 with the hemagglutinin [HA] tag replaced with V5) for Rdh54-V5. The resulting constructs were cut with KpnI and SalI and dropped into pJK148 (Keeney and Boeke, 1994) to generate pMGC59 (Rhp54-HA) and pMGC62 (Rdh54-V5). Each construct was then cut with BbsI and transformed into a pat1-114 haploid and stable leu+ transformants selected to generate FY1744 and FY1596.
Time-Course Analysis
pat1-114 strains were grown and induced for meiosis, and progression through meiotic S phase and meiotic divisions was followed as described previously (Forsburg and Hodson, 2000), except that 10 mM hydroxyurea was used instead of 15 mM. Diploids were selected for using ade6-M210/ade6-M216 intragenic complementation. Diploid liquid cultures were grown in EMM lacking adenine and induced for meiosis by washing into EMM lacking adenine and nitrogen.
Pulsed Field Gels
Cells were first embedded in InCert agarose (BMA, Rocklin, ME) and then processed as described previously (Young et al., 2002). In brief, the NotI-digested plugs were molded into 1% certified megabase agarose (Bio-Rad, Hercules, CA) and electrophoresed in a CHEF apparatus (CBS Scientific, Del Mar, CA) at 14°C in 0.5× Tris borate-EDTA for 24 h by using 6 V/cm with an initial switch time of 8 s and an increment of 0.03 s, 120° included angle. After electrophoresis, gels were UV irradiated (60 mJ) and transferred to Gene-Screen Plus membrane (PerkinElmer Life Sciences, Boston, MA) in 0.5 M NaOH, 1.5 M NaCl for 48 h. The membrane was hybridized with a 32P-labeled PCR-amplified ura1+ fragment (primers 5′-AGGTACCGGCTGTTGCGAAAATGTTG-3′ and 5′-GAGCTCAGCAATGAAGTCAGTTGGTG-3′), which generates a probe essentially the same as described previously (Young et al., 2002). Images were collected and analyzed using a PhosphorImager and ImageQuant software (Amersham Biosciences, Piscataway, NJ).
Recombination Assays
To determine recombination frequency between his4-239 and lys4-95 and between ade6-M26 and ade6-52, strains were mated on SPAS plates for 4 d and then 1000 spores (wild type and rdh54Δ) or 2500 spores (rhp54Δ, to compensate for low viability) were plated to each of five YES plates and grown at 32°C until colonies grew, and then replica plated to EMM lacking histidine and lysine and to EMM lacking adenine to select for recombinants. To determine recombination frequency at ade6-L469/pUC8/his3+/ade6-M375, a fresh his+ isolate containing ade6-L469/pUC8/his3+/ade6-M375 was mated to an ade6-M375-M210 strain for 4 d on SPAS and then 1000 spores (wild type and rdh54Δ) or 2500 spores (rhp54Δ) were plated to a YES plate (titer plate), and 10,000 spores (wild type and rdh54Δ) or 25,000 spores (rhp54Δ) were plated to EMM lacking adenine and grown at 32°C for 7 d. Adenine prototrophs were then counted and total viable spores plated was determined from titer plates. For recombination assays, P values were determined using an unpaired t test. P values ≤ 0.05 were considered significant. ade6-M210 is useable to block interhomolog repair of ade6-L469 because the ade6-M210 mutation (G to A at nucleotide 1470 from Start; Greg Freyer, personal communication) is only three nucleotides downstream from the ade6-L469 mutation (C to T at nucleotide 1467 from ATG, Szankasi et al., 1988). The ade6-/his3+/ade6- alelle was a gift from Fekret Osman (Oxford University, Oxford, United Kingdom). The ade6-M375-M210 allele was a gift from Gerald Smith (Fred Hutchinson Cancer Research Center, Seattle, WA).
Spore Viability
Strains were mated on SPAS for 1.5 d at 25°C and then asci were dissected onto rich media (YES) and grown at 32°C. Spore viability was calculated as (number of colonies formed)/(number of asci dissected × 4).
Protein Extractions and Westerns
Protein extractions were made by trichloroacetic acid (TCA) extraction as described previously (Foiani et al., 1994). Total protein concentrations were determined by BCA protein assay (Pierce Chemical, Rockford, IL). Protein (15 μg) was loaded per lane and was separated using 12% SDS-PAGE (Protogel; National Diagnostics, Atlanta, GA) and transferred to Immobilon-P (Millipore, Bedford, MA). Rdh54-V5 was detected using mouse anti-PK tag antibody (Serotec, Raleigh, NC) at a 1:2500 dilution, Rhp54-HA was detected with antibody 12CA5 (a kind gift of Jill Meisenhelder and Tony Hunter, Salk Institute, La Jolla, CA) at 1:5000 dilution, and proliferating cell nuclear antigen (PCNA) was detected with PCNA (PC10) (Santa Cruz Biotechnology., Santa Cruz, CA) antibody at 1:1000 dilution. Secondary antibody was horseradish peroxidase-conjugated anti-mouse antibody (Jackson Immunoresearch Laboratories, West Grove, PA) at 1:2000 detected with enhanced chemiluminescence (Amersham Biosciences).
To generate the Rhp51 polyclonal antibody, the region coding for the first 123 amino acids of Rhp51 was PCR amplified, and a 5′ BamHI site and 3′ EcoRI site were added using primers 5′-CGCGGATCCATGGCAGATACAGAGGTG-3′ and 5′-CGGAATTCCGAATCGTAATTAATTCA-3′. The PCR product was cut with BamHI and EcoRI and then cloned into pRSET-A (Invitrogen, San Diego, CA) or pGEX-KG (Amersham Biosciences) cut with same to generate pJMB124 (N-terminal His-tagged Rhp51) or pJMB125 (N-terminal GST-tagged Rhp51). Purified GST-Rhp51 was then used to generate antibodies in rabbits (Covance Research Products, Denver, PA). The polyclonal antibody was affinity purified using His-Rhp51 and used at 1:2500 dilution. Secondary antibody was HRP-conjugated anti-mouse antibody (Jackson Immunoresearch Laboratories). This antibody was specific to Rhp51 by Western (our unpublished data).
Yeast Two-Hybrid Analysis
Full-length coding regions for Rdh54, Rhp54, Rhp51, Dmc1, Rad22, and Rti1 were PCR amplified from a meiotic cDNA library with primers that added a 5′ SalI site and 3′ NotI site and then cloned into pDBLEU (Invitrogen) for GAL4-DNA binding domain bait constructs or pEXP-AD502 (Invitrogen) for GAL4 activation domain prey constructs. Primers used were 5′-GTCGACCATGAAACGAAGAGCTACTTTTC-3′ and 5′-GCGGCCGCTATTCGAACTTTTTATGCATTAG-3′ for Rdh54, 5′-GTCGACCATGATTCAGCAACCAACAACTGC-3′ and 5′-GCGGCCGCTTAATGAGATTTGTATTGGAAAACG-3′ for Rhp54, 5′-GTCGACCATGGCGATACAGAGGTGGAAATGC-3′ and 5′-GCGGCCGCTTAGACAGGTGCGATAATTTCC-3′ for Rhp51, 5′-GTCGACCATGGAAGAATTCGCAGAGGGG-3′ and 5′-GCGGCCGCTTAGGAAACGTCTGCTATGCC-3′ for Dmc1, 5′-GTCGACCATGTCTTTTGAGCAAAAACAG-3′ and 5′-GCGGCCGCTTATCCTTTTTTGGCTTTCTTATC-3′ for Rad22, and 5′-GTCGACCATGGGCTCGCTACCTGATCAATCG-3′ and 5′-GCGGCCGCTTATTTCGTTGAGAACGTGTTTGC-3′ for Rti1. Bait and prey plasmids were transformed into S. cerevisiae strain AH109 (BD Biosciences Clontech, Palo Alto, CA) by standard lithium acetate transformation, and transformants were tested for interaction by culturing in a 96-well microtiter tray followed by spotting onto SD minimal media lacking appropriate amino acids. β-Galactosidase activity was determined as described previously (Duttweiler, 1996).
Live Observation of Meiosis
h+ and h- strains were transformed with pSGP502-SV40 (SV40 NLS-GFP-lacZ) (Pasion and Forsburg, 1999) and grown on EMM without leucine plus 15 μM thiamine. Strains were restreaked to EMM without leucine or thiamine, 32°C 2 d before mating, and were then crossed on SPAS at 25°C for 8 h and resuspended in SPAS liquid. The mating mixture was placed on a coverslip onto which 30 μl of 1 mg/ml concanavalin A (Sigma-Aldrich, St. Louis, MO) had been air dried. The coverslip was then placed over a 50-μl depression microscope slide (VWR, West Chester, PA) filled with SPAS liquid and sealed with VALAP (1:1:1 petroleum jelly, lanolin, and paraffin). Asci that were in horse-tail movements (based on green fluorescent protein [GFP] signal) were imaged every 5 min until both meiotic divisions were complete. Images are flat projections of 10 optical sections, 0.6 μM apart of GFP and differential interference contrast (DIC)/Nomarski collected using an IX70 microscope (Olympus, Melville, NY) and DeltaVision SoftWoRx software (Applied Precision, Issaquah, WA).
4,6-Diamidino-2-Phenylindole (DAPI)-stained Acsi
Strains were mated for 24 h on SPAS plates, 25°C and then suspended in 70% ethanol. Cells were washed in water, air dried onto a slide, and stained with DAPI. Images were collected using a Leica DMR microscope with a Hamamatsu digital camera and Openlab software (Improvision, Lexington, MA).
RESULTS
Identification of S. pombe rdh54+
The S. pombe genome contains an uncharacterized open reading frame with a high degree of homology to budding yeast RDH54/TID1 (Wood et al., 2002). The sequencing project predicted a coding region interrupted by 14 introns. Using gene-specific primers, we sequenced the rdh54+ transcript from a meiotic cDNA library and confirmed the presence of 14 introns, although the 3′ end of the third intron was spliced differently than originally predicted. The 5′ splice site was correct as predicted, but the 3′ splice site is at nucleotide 33,922 of SPAC22F3, as opposed to the original prediction of 34,052. Thus, the fourth exon codes from nucleotide 33,923-34,137. The corrected cDNA sequence has been submitted to GenBank as accession number AY293737. Although this gene was originally predicted to encode 768 amino acids, our sequence data suggests a protein of 811 amino acids, with residues 119-162 from our sequence not in the original predicted protein. These additional residues do not add significantly to the conservation of sequence identity in the domains discussed below.
Conceptual translation of rdh54+ predicts a protein of 811 amino acids, with substantial sequence conservation in the C-terminal two-thirds of the protein to the ATPase containing domain of other Rad54 homologs (Figure 1A). The percentage of sequence identity of the full-length Rdh54 to budding yeast RDH54/TID1 (ScRDH54) or its putative human counterpart Rad54B (HsRad54B) is not substantially different than to the fission yeast Rad54 homolog (SpRhp54), budding yeast RAD54 (ScRAD54), or human Rad54 (HsRad54) (Figure 1A). However, SpRdh54p contains a region near the N terminus that shows sequence conservation with budding yeast RDH54/TID1 and its putative human counterpart Rad54B (Figure 1, A and B). This region of homology has previously been noted (Tanaka et al., 2002). This region is not conserved in the SpRhp54, ScRAD54, or HsRad54 proteins. SpRhp54 and ScRad54 instead show sequence conservation to one another over this N-terminal region, and this homology is very weakly conserved in HsRad54 (Figure 1B). Thus sequence comparisons led us to suspect that rdh54+ might be a functional homolog of budding yeast RDH54/TID1.
Figure 1.
Comparison of Rad54 and Rdh54 homologs from fission yeast, budding yeast, and humans. (A) Cartoons of Rad54 homologs show the relative size of the proteins and the C-terminal ATPase domain (hatched boxes) (amino acids 193-702 in SpRdh54) with percentage of identity to SpRdh54 within this domain shown within boxes. The N-terminal region of similarity within Rdh54 homologs (SpRdh54, ScRdh54, and HsRad54B) is represented by the solid gray boxes, whereas the N-terminal region of similarity within Rad54 homologs (SpRhp54, ScRad54, and HsRad54B) is shown by cross-hatched boxes. Percentage of identity to full-length SpRdh54 or SpRhp54 is shown to right. (B) Sequence alignment of N-terminal region of similarity as shown in A. Identical amino acids are boxed in black and similar amino acids are boxed in gray. DE, FY, ILMV, KR, NQ, or ST are considered similar. The residue position at the beginning and end of each line is shown. Accession numbers for protein sequences used are SpRdh54 (AY293737), ScRdh54 (NP_009629), HsRad54B (AAD34331.1), SpRhp54 (S41886), ScRad54 (NP_011352), and HsRad54 (CAA66379.1).
Expression of SpRdh54p
To begin our characterization of rdh54+, we tagged endogenous rdh54+, rhp54+, and rhp51+ with HA or V5 epitopes (see MATERIALS AND METHODS). Strains bearing Rdh54-V5 and Rhp54-HA showed wild-type spore viability (Table 2), suggesting both are functional proteins in meiosis. UV treatment of rhp54+-HA gave the same survival rate as wild type (see Figure 4), further confirming that Rhp54-HA is a functional protein. However, Rhp51-HA was phenotypically identical to rhp51Δ for damage sensitivity (our unpublished data), indicating the tagged protein is nonfunctional. Therefore, we raised a polyclonal antibody to the N-terminal region of Rhp51p (see MATERIALS AND METHODS). We examined the levels of Rdh54-V5, Rhp54-HA, and Rhp51p during a synchronized haploid meiosis using pat1-114, a temperature-sensitive allele of the kinase that negatively regulates entry into meiosis (Iino and Yamamoto, 1985). We monitored DNA content by fluorescence-activated cell sorting and monitored progression of meiotic divisions by nuclear morphology (Figure 2A). Samples were harvested every 2 h over a 10-h period and total proteins extracted by TCA precipitation. We saw that while Rhp54-HAp and Rhp51p were present in asynchronously growing haploids, Rdh54-V5p was seen only in meiotic cells (Figure 2, B and C). All three proteins were absent from nitrogen starved cells (arrested in G1, t = 0), but protein levels increased before meiotic S phase and then declined as the bulk of cells began the meiotic divisions (Figure 2, B and C). The same protein species was present whether meiosis was induced by pat1 or in normal diploids (our unpublished data).
Table 2.
Spore viability by tetrad dissection
| Genotype | Strain | Viability (%) | Asci dissected |
|---|---|---|---|
| Wild type | FY252×FY259 | 92.5 | 50 |
| Rdh54-V5 | FY1777×FY2400 | 92.5 | 30 |
| Rhp54-HA | FY1778×FY2344 | 95.83 | 30 |
| rdh54Δ | FY1454×FY1455 | 54.0 | 50 |
| rhp54Δ | FY1866×FY2122 | 21.5 | 50 |
| rdh54Δ rhp54Δ | FY2126×FY2127 | 0.0 | 50 |
| rhp54Δ Rdh54-V5 | FY2299×FY2300 | 22.5 | 30 |
Figure 4.
Deletion of fission yeast rdh54+ does not alter UV sensitivity of vegetative haploids. Cells were exposed to level of UV radiation shown and viability calculated based on number of colonies formed at given dose relative to 0 J/m2 control. Data from a representative experiment are shown.
Figure 2.
Protein levels of Rdh54-V5p, Rhp54-HA, and Rhp51 during synchronized meiosis and vegetative growth. (A) Fluorescence-activated cell sorting data and nuclei counts for pat1-114 rdh54-V5 (FY1596) show that S phase occurred between 2 and 4 h and meiotic divisions began by 6 h. These data are representative of typical timing in pat1-114-induced haploid meiosis under conditions used. (B) Western blot of TCA extracted lysates from meiotic time course with and without 10 mM HU for pat1-114 rdh54-V5 (FY1596) blotted for Rhp51 and of pat1-114 rhp54-HA (FY1744) blotted for HA. PCNA serves as a loading control. (C) Western blot of TCA extracted lysates from time course of pat1-114 rdh54-V5 (FY1596) with and without HU, or pat1-114 rdh54-V5 with the addition of cdc22-M45 (FY1773), cdc10-V50 (FY1774), mei4Δ (FY1775), rep1Δ (FY1776), or rec12Δ (FY1801). (D) Protein (15 μg) from TCA-extracted lysates were blotted for V5 to detect Rdh54 and for PCNA as loading control. Lane 1, wild-type vegetative haploid (FY254). Lane 2, rdh54-V5 vegetative haploid (FY1777). Lane 3, h-/h+ rdh54-V5/rdh54+ vegetative diploid (FY1777/FY255). Lane 4, h-/mat2-102 rdh54-V5/rdh54+ vegetative diploid (FY1777/FY766). Lane 5, pat1 rdh54-V5 haploid induced for meiosis. Two exposures of the V5 blot are shown to reveal the small but detectable amount of V5 signal in vegetative cells.
To determine whether S-phase progression is required for meiotic protein expression, we used 10 mM hydroxyurea (HU) to arrest cells in early meiotic S phase. Rdh54-V5p and Rhp54-HAp occurred with normal timing and then remained present throughout the period of arrest, whereas Rhp51p levels seemed to increase in HU (Figure 2, B and C). The level of Rdh54-V5p was further examined by crossing additional temperature-sensitive mutations into the pat1-114 rdh54-V5 background. We saw that Rdh54-V5p levels remained high in cells blocked before meiotic S phase by cdc10 (a transcription factor controlling S-phase genes; Aves et al., 1985) or in early meiotic S phase by cdc22 (ribonucleotide reductase; Fernandez Sarabia et al., 1993). A deletion of mei4 (a meiotic transcription factor required for the meiotic divisions (Horie et al., 1998), results in the temporary decrease in Rdh54p followed by the reappearance of the protein. Rdh54p occurs and disappears normally in strains deleted for rep1+ (a meiosis specific partner of the cdc10+ transcription factor; Sugiyama et al., 1994) or rec12+ (the fission yeast homolog of SPO11, the endonuclease that initiates meiotic DSBs; Lin and Smith, 1994) (Figure 2C).
Budding yeast RDH54 has been implicated in diploid-specific mitotic recombination (Klein, 1997), suggesting it may be sensitive to ploidy. Therefore, we examined Rdh54-V5p in fission yeast diploids and haploids grown vegetatively. There was no difference due to ploidy: compared with the levels observed in meiotic cells, Rdh54-V5p was essentially undetectable in vegetative cells unless we carried out prolonged exposure of the film (Figure 2D). Additionally, there was no change in Rdh54-V5 protein in either haploids or after treatment with DNA-damaging agents (our unpublished data), suggesting Rdh54p is not involved in mitotic damage response. This agrees with phenotypic analysis of the Δrhp54 mutant (see below).
SpRdh54p Interactions
We attempted to identify interactions between these proteins by using coimmunoprecipitations from standard, non-denaturing cell lysates. However, we found that nearly all of the Rhp51, Rdh54-V5, and Rhp54-HA proteins were insoluble, even under conditions where the chromatin was solubilized (our unpublished data). We thus turned to the yeast two-hybrid system. Full-length Rdh54, Rhp54, Rhp51, Dmc1, Rti1, and Rad22 were cloned into Gal4-DNA binding (bait) and Gal4 activation domain (prey) fusion vectors. Rad22 and Rti1 are homologues of budding yeast Rad52 (Ostermann et al., 1993; Suto et al., 1999). Pairwise combinations of bait and prey were tested for interactions. On plates lacking histidine, and thus requiring activation of the least stringent reporter for cell growth, we saw that Rdh54 interacted with both Dmc1 and Rhp51. On selection for ade+ or for β-galactasidase activity, however, Rdh54 only interacted with Dmc1 (Figure 3). These data suggest that Rdh54 can interact with both Dmc1 and Rhp51 but that the interaction is stronger between Rdh54 and Dmc1. In contrast, Rhp54 interacted only with Rhp51 and not Dmc1 (Figure 3). Rhp51 also gave a positive signal with itself, with Rti1, and with Rad22 (Figure 3). The Rhp51 interactions with Rhp54, Rad22, and Rti1 have been observed previously, thus confirming our constructs behaved as expected (Tsutsui et al., 2001; Kim et al., 2002). We only observed interactions for Rdh54 or Rhp54 when these proteins were fused to the DNA-binding domain; the reciprocal assay using either Rad54 homolog fused to the activation domain failed to show an interaction (Figure 3) as was observed previously with Rhp54 (Tsutsui et al., 2001). Our two-hybrid data confirm that Rdh54 can interact with the RecA homologs of S. pombe, as expected of a true Rad54 homolog.
Figure 3.
Two-hybrid interactions between Rdh54, Rhp54, Rhp51, Dmc1, Rti1, and Rad22. Pairwise tests between full-length Gal4 DNA-binding domain fusion proteins (in columns) and Gal4 activation domain fusions (in rows). Strains were initially grown on plates lacking only leucine and tryptophan and then spotted to plates lacking leucine and tryptophan as well as histidine (left), adenine (middle), or assayed for β-galactosidase activity (right).
Characterization of rdh54Δ
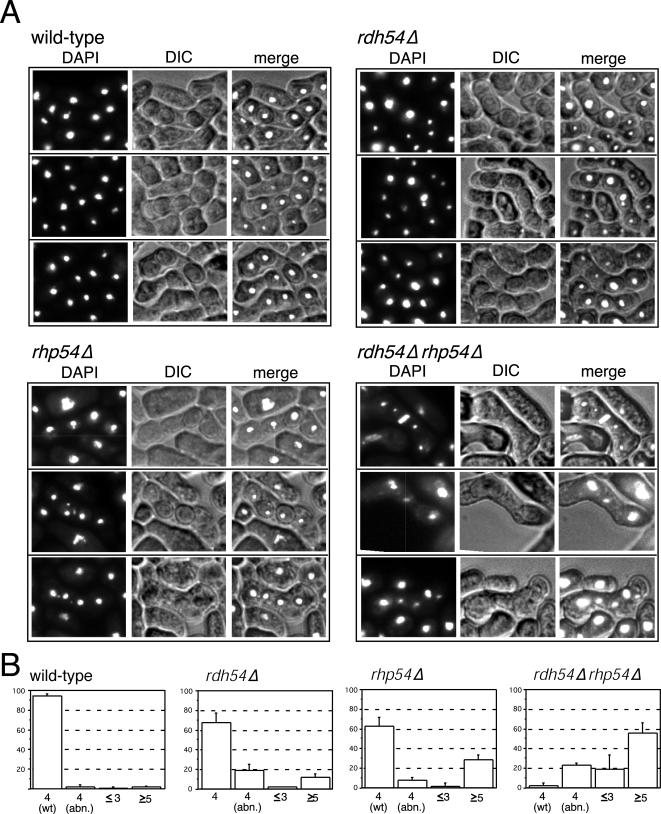
We constructed a strain deleted for the entire open reading frame of rdh54+ and found that a haploid rdh54Δ strain was indistinguishable from a wild-type strain for vegetative growth and for resistance to UV (Figure 4) and to gamma irradiation and MMS (our unpublished data). We found that an rdh54Δ rhp54Δ haploid was equally resistant to UV (Figure 4) and to gamma or MMS (our unpublished data) as was the rhp54Δ single mutant, consistent with Rdh54p functioning only in meiotic cells. We assayed spore viability with tetrad dissections and found that rdh54Δ gave about half as many viable spores as wild type, whereas only about one-quarter of the rhp54Δ spores were viable (Table 2). Strikingly, we found that when rdh54Δ rhp54Δ double mutants were crossed, no viable spores were produced (Table 2) Additionally, random spore analysis of >25,000 spores also failed to produce any viable progeny (our unpublished data). In fact, asci from rdh54Δ rhp54Δ rarely produced discrete spores, but instead contained what seemed to be cellular debris (punctuated by DAPI staining; Figure 5A). This suggests that the two fission yeast Rad54 homologs may be partially redundant for formation of viable spores.
Figure 5.
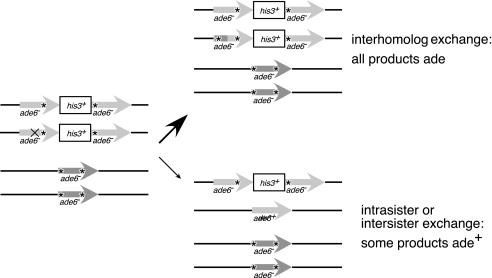
Cartoon of assay for meiotic intrasister or intersister repair. A haploid strain containing the nontandem heteroallelic ade6-L469/pUC8/his3+/ade6-M375 construct, shown as light gray arrows separated by boxed his3+, is mated to a haploid strain with ade6-M375-M210, shown as a dark gray arrow. Position of point mutations within ade6 are represented by *. Pairs of sister chromatids following meiotic S phase are shown on left. If a DSB occurs in one of the ade6/his+/ade6 sisters (“X” in light gray arrow on left), repair from the homologous chromosome will generate ade- progeny (top right), but repair using the homologous sister chromatid or within the chromatid containing the break can generate ade+ progeny (bottom right).
Work from budding yeast suggests that a role of Rdh54 in meiotic recombination is to promote the use of the homologous chromosome as the template for repair of meiotic DSBs as opposed to the sister chromatid (Arbel et al., 1999). To examine whether SpRdh54p affects meiotic recombination frequency or partner choice, we constructed strains to assay recombination between two sets of interhomolog alleles as well as within the nontandem heteroallelic ade6-L469/pUC8/his3+/ade6-M375 construct (Osman et al., 2000). Strains with this heteroallelic marker were mated to a strain with two corresponding mutations within its single ade6 locus (see MATERIALS AND METHODS). Because of this double mutant allele, the only way ade+ progeny can arise is via intrasister or intersister exchange (Figure 5). Details of repair events leading to ade+ products are discussed in Osman et al. (2000). Whereas rhp54Δ did not affect the recombination rate between either pair of interhomolog markers, rdh54Δ decreased the rate of meiotic recombination approximately threefold for each of these marker sets (Table 3). Both rdh54Δ and rhp54Δ increased the recovery of ade+ from the ade-/his+/ade- heteroallele, with an approximately threefold or twofold increase in intra- or intersister recombination, respectively (Table 3). These data are consistent with the idea that Rdh54p promotes interhomolog repair during meiosis, because interhomolog recombination is decreased and intra- and intersister recombination is increased in the rdh54Δ strain. Because the rdh54Δ rhp54Δ double mutant does not produce viable spores, we could not test it using these assays.
Table 3.
Meiotic recombination1
| Interhomolog recombination | ||||||
|---|---|---|---|---|---|---|
| Genotypea | his+lys+ % | P value | Fold Δ | ade+ rate (× 10-3) | P value | Fold Δ |
| Wild type | 7.683 ± 2.291 | 1 | 2.500 ± 0.661 | 1 | ||
| rdh54Δ | 2.740 ± 0.135 | 0.0203 | 2.8 ↓ | 0.837 ± 0.429 | 0.0217 | 3.0 ↓ |
| rhp54Δ | 7.463 ± 0.970 | 0.8857 | 1 | 2.860 ± 0.387 | 0.4614 | 1 |
| Sister chromatid exchangeb | ||||||
|---|---|---|---|---|---|---|
| Genotypec | ade+ rate (× 10-3) | P value | Fold Δ | |||
| Wild type | 2.260 ± 0.994 | 1 | ||||
| rdh54 | 7.763 ± 3.605 | .0258 | 3.4 ↑ | |||
| rhp54 | 5.393 ± 1.472 | .0124 | 2.4 ↑ | |||
Means of at least three experiments ± standard deviation shown. P values based on unpaired t tests versus wild type.
Strains crossed were wild type (FY1251×FY1252), rdh54Δ (FY1921×FY1941), and rhp54Δ (FY1664×FY1665)
Rate of ade+ from ade6-L469/pUC8/his3+/ade6-M375 crossed to ade6-M375-M210
Strains crossed were wild type (FY2132×FY2133), rdh54Δ (FY1898×FY1942), and rhp54Δ (FY1901×1944)
Double-Strand Breaks in rdh54Δ
To examine the formation and repair of DSBs in rdh54Δ strains directly, we used pulsed field gel electrophoresis (PFGE) to detect meiotic breaks. This assay has been used previously to show that meiotic DSBs in fission yeast occur in specific chromosomal regions as opposed to being equally distributed throughout the genome in pat1-114 meiosis (Young et al., 2002). We used a ura1+ probe to examine NotI digested chromosomes from numerous time points in a pat1-114-induced synchronous meiosis. pat1-114 alone serves as the “wild-type” control. Samples from each time course were taken to follow meiotic S phase and nuclear divisions. All four strains tested completed S phase within 3 to 4 h of meiotic induction (our unpublished data). On pulsed field gel electrophoresis, the intact NotI J fragment containing ura1+ migrates at ∼500 kb, whereas chromosomes that contain meiotic DSBs within the NotI fragment migrate faster (Young et al., 2002; Figure 6B). Distinct bands at ∼250, 350, and 450 kb occur below the larger 500-kb intact NotI fragment in pat1-114 wild-type cells 4 h after meiotic induction, and these lower migrating bands were very faint by 6 h and had completely disappeared by 8 h (Figure 6B). This indicates that meiotic DSBs have been formed before the 4-h time point and are repaired by 8 h. We next examined DSB formation and repair in pat1-114 fission yeast disrupted for the Rad54 homologs. We saw that timing of break formation and repair in rdh54Δ mirrored the timing in wild type (Figure 6B). However, a pat1-114 strain containing rhp54Δ showed the maximum amount of DSBs at 5 h as opposed to 4 h in the wild-type control, and then showed only limited repair of these breaks at later time-points. The pat1-114 rdh54Δ rhp54Δ double mutant formed DSBs normally but seemed to be completely defective for double-strand break repair, as the amount of broken NotI fragment continued to increase from 4 through 8 h and then showed only a minor decrease at 10 h (Figure 6B).
Figure 6.
Pulsed field gel analysis of formation and repair of meiotic double-strand breaks in rdh54Δ and rhp54Δ strains. Samples were collected from pat1-114-induced meiotic time courses at indicated times for pat1-114 control strain (FY572), pat1-114 rdh54Δ (FY1456), pat1-114 rhp54Δ (FY2353), and pat1-114 rdh54Δ rhp54Δ (FY2142). Time points are in hours. (A) Progression through the meiotic divisions was quantitated by counting of DAPI-stained nuclei for each time course. Percentage of cells with one nucleus (circles), two nuclei (squares), or three or more nuclei (triangles) are shown for each time point. (B) NotI digested chromosome plugs from cells collected at indicated time points separated by pulsed field gel electrophoresis and then probed for ura1+ as described in MATERIALS AND METHODS. (C) Data in B were quantitated by dividing the signal for the lowest break fragment (at ∼250 kb) by the signal in the uncut NotI J fragment (at ∼500 kb) to give “% cut.” Each signal was first corrected by subtracting out background based on signal below lowest break fragment.
We quantitated the signal in the ∼250-kb broken fragment relative to the signal in the intact NotI J fragment (each signal was first corrected for background) to give a relative measure of the percentage of breakage at each time point. This analysis confirmed that rdh54Δ rhp54Δ continued to accumulate DSBs, whereas breaks were repaired in wild type and rdh54Δ (Figure 6C). Additionally, it is clear that the maximum level of breaks is lower in rhp54Δ than in the double mutant and that some repair does occur in the rhp54Δ strain (Figure 6C). However, these differing levels of DSB repair did not affect progression through the meiotic divisions, because all four strains began divisions by 6 h, and the majority of cells had completed the meiotic divisions by 10 h (Figure 6A). These data suggest that although rdh54Δ rhp54Δ is profoundly defective in meiotic DSB repair, the cells nevertheless go on to divide.
Meiotic Divisions in Cells Lacking either Rad54 Homolog
We next sought to determine whether the meiotic divisions also occur in wild-type (pat1+) strains deficient for the Rad54 homologs. Initially, we planned to examine synchronous meiosis in nitrogen-starved diploids in liquid culture. We found that rhp54Δ/rhp54Δ diploids (with or without rdh54Δ) were difficult to isolate and maintain and could not be propagated to induce meiosis in bulk liquid culture (see DISCUSSION). However, we were able to examine meiotic progression in asci resulting from mating an h+ to an h- haploid on a sporulation plate. Meiosis in fission yeast is normally coupled to conjugation, so this condition is the most “normal” meiosis for this organism. After crosses, we examined zygotes by DAPI staining and DIC/Nomarski microscopy. Nearly all asci resulting from the crossing two wild-type strains showed four equal-sized DAPI bodies (Figure 7, A and B), indicating that both meiotic divisions had occurred and that each spore received a complete haploid genome. When two rdh54Δ strains or two rhp54Δ strains were crossed, approximately two-thirds of asci contained four equally sized DAPI bodies, and the remaining asci contained either less than three, greater than four, or four unequal DAPI signals (Figure 7, A and B). In contrast, few if any normal asci were produced in rdh54Δ rhp54Δ x rdh54Δ rhp54Δ crosses, and more than half contained greater than four DAPI bodies (Figure 7, A and B). In many of the asci, DAPI material was seen at the midplane of the cell, suggesting that DNA may have been left behind at the division plane as the first meiotic division occurred (most notable in the upper double mutant ascus shown in Figure 7A). Thus, rdh54Δ rhp54Δ does not arrest meiotic progression. Instead, homozygous double mutants continue into the meiotic divisions despite the presence of unrepaired DSBs.
Figure 7.
Nuclear morphology of asci from crosses of rdh54Δ, rhp54Δ, and rdh54Δ rhp54Δ strains. Haploid strains were crossed on SPAS mating plates for 24 h, fixed in ethanol, and stained with DAPI to allow visualization of DNA. (A) Several representative asci for each cross, shown in horizontal rows. The image obtained for DAPI (left), DIC/Nomarski (middle), and the merged image (right) is shown. (B) Quantitation of DAPI morphologies. One hundred asci from each cross were scored as having four evenly sized DAPI bodies [4 (wt)], four unequal DAPI bodies [4 (abn.)], three or fewer DAPI bodies (≤3), or five or more DAPI bodies (≥5). Averages with error bars = ± 1 SD of three independent crosses are shown. Two strains sets were used; they gave similar results so data are shown combined. Strains used were wild-type (FY252 × FY259 or FY1251 × FY1252), rdh54Δ (FY1454 × FY1455 or FY1921 × FY1941), rhp54Δ (FY1866 × FY2122 or FY1664 × FY1665), and rdh54Δ rhp54Δ (FY2126 × FY2127 or FY1946 × FY1978).
Live Observation of Meiosis in Cells Lacking the Rad54 Homologs
To better understand what steps lead to the terminal DAPI morphology phenotypes seen in Figure 7, we observed meiosis in live fission yeast cells by using a methodology similar to those previously developed (Hiraoka et al., 2000; Molnar et al., 2001). Both an h+ and an h- haploid were transformed with a construct expressing GFP with a nuclear localization signal, which labels nuclei in live cells. Haploid cells of the desired genotype were allowed to conjugate on a mating plate for 8 h and then examined for GFP signal. Shortly after nuclear karyogamy in fission yeast, the nucleus begins telomere lead movements back and forth throughout the ascus, know as the horse-tail phase. During this phase, both meiotic S phase and meiotic recombination occur (for review, see Yamamoto and Hiraoka, 2001). Zygotes in the horse-tail phase of meiosis, indicated by an elongated nuclear GFP signal, were located and followed until the completion of meiosis. We used serial Z-sections of both GFP and DIC/Nomarski acquired at 5-min intervals to observe progression through meiosis.
When wild-type strains were mated, we saw that the nucleus exits horse-tail phase, pauses, and then two distinct meiotic divisions occur, producing four evenly segregated GFP signals (Figure 8A; Video 1). When either rdh54Δ haploids or rhp54Δ haploids were crossed, nuclear movements and production of four evenly divided GFP bodies were generally similar to wild-type (our unpublished data). However, we did occasionally observe abnormal division for either single mutant (Figure 8B; our unpublished data; Videos 2 and 3). The example of the rdh54Δ ascus on the left of Figure 8B (Video 2) shows a case in which the final meiotic products seem quite similar to those produced in wild type, but live observation revealed that the first meiotic division was unequal, with the bulk of nuclear material moving to the left at MI. We also observed meiotic divisions that showed nuclear material left behind at the division plane during MI (Figure 8B, right ascus; Video 3). Although normal horse-tail movements and two distinct meiotic divisions were observed in rdh54Δ rhp54Δ double mutants, the divisions as well as size and number of final nuclear GFP signals were aberrant (Figure 8C; Videos 4 and 5). In the left-hand ascus of Figure 8C (Video 4), nearly all of the nuclear material segregates to the left at MI, with only a small GFP signal detected moving to the opposite end of the ascus. The second meiotic division of this ascus resulted in the production of five nuclear localization signal-GFP signals. We also observed rdh54Δ rhp54Δ asci that showed a large fraction of the nuclear material left behind at meiosis I, followed by an unequal meiosis II (right ascus, Figure 8C; Video 5). We did not observe any rdh54Δ rhp54Δ asci with nuclear divisions that resembled wild type. Thus, live observation of meiosis in fission yeast confirms that cells lacking both Rad54 homologs do not arrest but instead complete MI and MII to produce abnormal meiotic products.
Figure 8.
Live observation of meiosis in wild-type, rdh54Δ, and rdh54Δ rhp54Δ crosses. Haploid strains of wild-type (FY252 and FY259), rdh54Δ (FY1454 and FY1455), and rdh54Δ rhp54Δ (FY2126 and FY2127) were transformed with a construct expressing GFP with a nuclear localization sequence and then crossed. A serial Z-section, shown as a flat projection, of GFP and DIC/Nomarski were captured every 5 min from asci in horse-tail phase until the completion of the meiotic divisions. Representative frames from wild-type (A), rdh54Δ (B), or rdh54Δ rhp54Δ (C) asci are shown. Frames shown horse-tail movement, posthorse-tail nucleus, early MI, mid-MI, post-MI, early MII, and final meiotic products form top to bottom. For each ascus, the first image is a merge of GFP and DIC/Nomarki, and the remaining images are GFP alone with the outline of the ascus based on the DIC/Nomarski image shown as a dashed line. Times are in minutes and were defined by arbitrarily setting the first time point with a posthorse-tail nucleus as t = 0. The second ascus for rdh54Δ rhp54Δ begins with the early MI time point.
DISCUSSION
Our results demonstrate that the fission yeast gene rdh54+ is involved in the repair of meiotic double-strand breaks. The conserved domain seen at the N terminus of the respective amino acid sequences of Sp rdh54+, Sc RDH54/TID1, and Hs Rad54B (Figure 1; Tanaka et al., 2002) suggests that these proteins are homologous. Our functional characterization of fission yeast rdh54+ implicates this gene specifically in meiotic recombination. Likewise, budding yeast RDH54/TID1 is most important during meiosis (Klein, 1997; Shinohara et al., 1997). Although the meiotic role of Hs Rad54B is not yet known, its strong up-regulation in the testes (Hiramoto et al., 1999) implies it may play a role in human meiosis. Human tissue cultures cells lacking Rad54B do not show increased sensitivity to DNA-damaging agents (Miyagawa et al., 2002), consistent with our analysis of fission yeast rdh54Δ haploids. However, human cells without Rad54B do show a defect in gene targeting (Miyagawa et al., 2002), showing that Rad54B does play at least a limited role in outside of meiosis in humans.
Western blot analysis of Rdh54-V5p, Rhp54-HAp, and Rhp51p shows these proteins are present during meiosis before the initiation of S phase and then disappear as meiotic divisions begin. Treatment with HU or inclusion of additional temperature-sensitive alleles in the pat1-114 rdh54-V5 background shows that Rdh54p occurs before meiotic S phase and is not degraded until its completion, with the slightly puzzling exception of rep1Δ. Initiation of meiotic recombination is not necessary for degradation of Rdh54p, because disruption of rec12+ blocks formation of DSBs but does not affect Rdh54p protein levels. However, Rdh54-V5p, unlike Rhp54-HAp or Rhp51p, is not present in vegetative haploids. Additionally, rdh54Δ affects neither the damage sensitivity of wild-type haploids or rhp54Δ haploids, suggesting that rdh54+ is indeed meiosis specific. The budding yeast homolog RDH54 has been implicated in DSB repair in diploids (Klein, 1997). Because of the difficulty of isolating a diploid homozygous for rhp54Δ (discussed below), we have yet to address whether rdh54Δ would increase sensitivity to DSBs in a diploid rhp54Δ background. However, we have observed that a diploid rdh54Δ srs2Δ/rdh54Δ srs2Δ is viable (our unpublished data), whereas the equivalent strain in budding yeast shows diploid specific synthetic lethality (Klein, 1997). This leads us to suggest that fission yeast rdh54+ is truly meiosis specific.
Rdh54p is important for meiotic recombination. We see that rdh54Δ reduces the recombination between homologous chromosomes about threefold and results in a threefold increase in intrahomolog recombination at the ade6-L469/pUC8/his3+/ade6-M375 heteroallele. This suggests that the Rdh54 protein may promote DSB repair by using the homologous chromosome, rather than the sister chromatid. Our pulsed field gel electrophoresis data (described below) show that Rdh54p is dispensable for sister chromatid repair, as has been observed in budding yeast (Arbel et al., 1999). Additionally, we see that Rdh54 and Dmc1 interact by two-hybrid assay. This further implicates Rdh54p in interhomolog repair, because Dmc1 has been shown to be involved in promoting the use of the homologous chromosome as the repair template in budding yeast (Schwacha and Kleckner, 1997). Together, these data could lead to a model in which two separate pathways of repair function in meiosis. The Rhp51/Rhp54 (RAD51/RAD54) pathway would result in repair via the sister, whereas the Dmc1/Rdh54 pathway would repair strictly via the homolog.
However, our data do not support the rigid separation of these proteins into two distinct pathways. We see that rhp54Δ does not show any change in the rate of recombination between homologous chromosomes, in agreement with a previous study (van den Bosch et al., 2002). Others have reported that rhp54Δ results in a 1.7-fold decrease for intergenic recombination (Muris et al., 1997) by using different loci than those analyzed here. In any case, it is clear that rhp54Δ has at best a minor effect on the production of meiotic recombinants. However, the above-mentioned model would predict that the rate of interhomolog recombination should be increased in rhp54Δ because all breaks would be repaired by Rdh54p and thus via the homolog. Our data show that this is not the case. Likewise, some interhomolog recombinants are produced in rdh54Δ, so Rhp54p must be capable of promoting some repair via the homolog. A role for budding yeast Rad54 in interhomolog recombination has also been suggested (Schmuckli-Maurer and Heyer, 2000). We thus conclude that partial functional redundancy exists between Rhp54 and Rdh54 during meiosis; some overlap exists between the intersister and interhomolog repair pathways.
The pulsed field gel data presented here directly demonstrate a role for rhp54+ and rdh54+ in DSB repair. We show that an rdh54Δ strain is completely proficient for meiotic DSB repair, whereas in rhp54Δ there is a residual level of DSBs that persist though many breaks are resolved. In contrast, the rdh54Δ rhp54Δ strain is completely defective in meiotic DSB repair, showing no evidence of break resolution. Importantly, these assays were performed in haploid strains. Therefore, all repair must occur via the sister chromatid, in contrast to the case in a true wild-type meiosis. Residual DSBs in rhp54Δ, as observed, are expected if most intrachromatid repair is via Rhp54p. Similar assays in haploid budding yeast that tested DSB repair in RAD54 and RDH54 haploids gave results quite similar to those presented here (Arbel et al., 1999).
We attempted to assay break repair in meiotic diploids but were unable to induce meiosis in the rhp54Δ/rhp54Δ background. In addition, we consistently see that homozygous rhp54Δ diploids are difficult to isolate and maintain. If isolated, these diploids fail to enter meiosis when shifted to nitrogen-free liquid media; the cells simply stop growing and seem to arrest permanently (our unpublished data). Similar difficulties have been observed obtaining and propagating rhp51Δ diploids, as well as inducing meiosis in this background (Grishchuk and Kohli, personal communication). Recent reports also indicate severe genetic instability in budding yeast rad54Δ/rad54Δ diploids, but not haploids (Schmuckli-Maurer et al., 2003). Thus, it seems that the recombination machinery may be important for a stable diploid state, perhaps by monitoring the presence of homologous chromosomes. Although the mechanism is unclear, unfortunately this prevented performing our assays on rhp54Δ diploid strains.
Although we have been unable to address the question directly, we speculate that rhp54Δ must be reasonably proficient for DSB repair in a diploid meiosis, because a cross of rhp54Δ produces a reasonable proportion of viable spores (21.5% from Table 2, similar to data in Muris et al., 1997). Because the recombination rate in fission yeast has been calculated to be 12.5 crossovers per bivalent (Cox, 1995) and because these crossovers must arise via the production and repair of DSBs, it follows that nearly all individual DSBs produced in rhp54Δ must be repaired successfully to yield such a high percentage of viable spores. Shinohara et al. (1997) showed a defect in break resolution in rdh54Δ budding yeast diploids by pulsed field gel analysis. The lack of a similar repair defect in our hands in fission yeast rdh54Δ may be due to the use of haploids, or it could represent a true difference between the yeasts. We believe that the high spore viability of rdh54Δ and the near normal nuclear movements during meiotic divisions in asci of this genotype suggests that Rhp54p activity can compensate for loss of Rdh54p in fission yeast in diploids as well as haploids.
Our results suggest that DSBs are not repaired during meiosis in rdh54Δ rhp54Δ double mutants. However, DAPI staining of fixed meiotic asci and live observation of nuclear movements during meiosis show that this failure of break repair does not result in meiotic arrest. This was a surprising result because the equivalent strain in budding yeast arrests in meiotic prophase (Shinohara et al., 1997). However, increasing evidence indicates that the two yeasts respond very differently to the presence of unresolved recombination intermediates. Recent work has shown that a fission yeast strain deleted for meu13+ goes on to complete meiosis (Nabeshima et al., 2001), whereas the deletion of the budding yeast homolog HOP2 results in pachytene arrest (Leu et al., 1998). These gene products have been shown to be involved in meiotic homologous chromosome pairing (Leu et al., 1998; Nabeshima et al., 2001), and hop2Δ cells do not repair meiotic DSBs (Leu et al., 1998), whereas meu13Δ repairs breaks more slowly than wild type (Shimada et al., 2002). Furthermore, it has been shown that meu13Δ results in a short delay in entry into the first meiotic division and that this delay can be abrogated by deletion of mek1+ (Shimada et al., 2002; Perez-Hidalgo et al., 2003), the fission yeast homolog of budding yeast MEK1, a kinase implicated in the meiotic recombination checkpoint (Xu et al., 1997). Despite the unresolved DSBs in rdh54Δ rhp54Δ, we saw no evidence for a meiotic delay in pat1-114 haploids. For technical reasons, we were unable to observe sufficient numbers of complete meioses (karyogamy to completion of divisions) by using live cell observation in wild-type and rdh54Δ rhp54Δ asci to directly compare the timing of meiotic events. However, from these live cell assays as well as the DAPI staining analysis presented in Figure 7, it is clear that timing of meiotic events is similar in rdh54Δ rhp54Δ and wild type and that any delay, if it exists, is transitory. From our results and the results from meu13Δ discussed above, we propose that failure to resolve meiotic double-strand break intermediates does not cause meiotic arrest in S. pombe. How the cell coordinates the two RAD54 homologs in meiosis remains to be determined.
Supplementary Material
Acknowledgments
We thank Gerald Smith and Randy Hyppa for advice on pulsed field gel conditions, John Marlett for help with Western blots, and John Marlett and Vanessa Angeles for strain construction. Jen Gilbert aided in two-hybrid screening. We are indebted to Julie Bailis for valuable advice and discussions throughout this research project, as well as for work on the Rhp51 antibody. Strains used in this study were derived from kind gifts from Vladimir Bashkirov, Gerald Smith, Fekret Osman, and Chikashi Shimoda. Special thanks to Greg Freyer, Alexandra Grishchuk, and Jurg Kohli for sharing data before publication. Thanks to Julie Bailis, Will Dolan, and Dan Pankratz for careful reading of the manuscript before submission. M.G.C. was supported by National Institutes of Health training grant T32 CA64041. This work was supported by National Institutes of Health grants R29-GM54797 and R01-GM59321 to S.L.F.
Online version of this article contains video material for some figures. Online version is available at www.molbiolcell.org.
References
- Alexeev, A., Mazin, A., and Kowalczykowski, S.C. (2003). Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 10, 182-186. [DOI] [PubMed] [Google Scholar]
- Alexiadis, V., and Kadonaga, J.T. (2002). Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 16, 2767-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbel, A., Zenvirth, D., and Simchen, G. (1999). Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 18, 2648-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aves, S., Durkacz, B., Carr, T., and Nurse, P. (1985). Cloning, sequencing and transcriptional control of the Schizosaccharomyces pombe cdc10 “start” gene. EMBO J. 4, 457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, D.K., Park, D., Xu, L., and Kleckner, N. (1992). DMC1: a meiosis-specific yeast homolog of E. coli RecA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69, 439-456. [DOI] [PubMed] [Google Scholar]
- Cox, B.S. (1995). Genetic Analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. In: The Yeasts, ed. A.H. Rose and J.S. Harrison, London: Academic Press, 7-67.
- Dresser, M.E., Ewing, D.J., Conrad, M.N., Dominguez, A.M., Barstead, R., Jiang, H., and Kodadek, T. (1997). DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147, 533-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttweiler, H.M. (1996). A highly sensitive and non-lethal beta-galactosidase plate assay for yeast. Trends Genet. 12, 340-341. [DOI] [PubMed] [Google Scholar]
- Fernandez Sarabia, M-J., McInerny, C., Harris, P., Gordon, C., and Fantes, P. (1993). The cell cycle genes cdc22+ and suc22+ of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol. Gen. Genet. 238, 241-251. [DOI] [PubMed] [Google Scholar]
- Foiani, M., Marini, F., Gamba, D., Lucchini, G., and Plevani, P. (1994). The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14, 923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg, S.L., and Hodson, J.A. (2000). Mitotic replication initiation proteins are not required for S. pombe pre-meiotic S phase. Nat. Genet. 25, 263-268. [DOI] [PubMed] [Google Scholar]
- Forsburg, S.L., and Sherman, D.A. (1997). General purpose tagging vectors for fission yeast. Gene 191, 191-195. [DOI] [PubMed] [Google Scholar]
- Fukushima, K., Tanaka, Y., Nabeshima, K., Yoneki, T., Tougan, T., Tanaka, S., and Nojima, H. (2000). Dmc1 of Schizosaccharomyces pombe plays a role in meiotic recombination. Nucleic Acids Res. 28, 2709-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov, D.V., and Kadonaga, J.T. (2001). The many faces of chromatin remodeling: switching beyond transcription. Cell 106, 523-525. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., Fink, G.R., Simon, M.I. and Abelson, J.N. (eds.) (1991). Guide to Yeast Genetics and Molecular Biology: Methods in Enzymology, vol. 194, San Diego, CA: Academic Press. Methods Enzymol. 194.
- Gutz, H., Heslot, H., Leupold, U., and Lopreno, N. (1974). Schizosaccharomyces pombe. In: Handbook of Genetics, ed. R.D. King, New York: Plenum Press, 395-446.
- Hiramoto, T., et al. (1999). Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene 18, 3422-3426. [DOI] [PubMed] [Google Scholar]
- Hiraoka, Y., Ding, D.Q., Yamamoto, A., Tsutsumi, C., and Chikashige, Y. (2000). Characterization of fission yeast meiotic mutants based on live observation of meiotic prophase nuclear movement. Chromosoma 109, 103-109. [DOI] [PubMed] [Google Scholar]
- Horie, S., Watanabe, Y., Tanaka, K., Nishiwaki, S., Fujioka, H., Abe, H., Yamamoto, M., and Shimoda, C. (1998). The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell. Biol. 18, 2118-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino, Y., and Yamamoto, M. (1985). Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol. Gen. Genet. 198, 416-421. [DOI] [PubMed] [Google Scholar]
- Jaskelioff, M., Van Komen, S., Krebs, J.E., Sung, P., and Peterson, C.L. (2003). Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 278, 9212-9218. [DOI] [PubMed] [Google Scholar]
- Keeney, J.B., and Boeke, J.D. (1994). Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136, 849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, T.J., Martin, G.S., Forsburg, S.L., Stephen, R.J., Russo, A., and Nurse, P. (1993). The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell 74, 371-382. [DOI] [PubMed] [Google Scholar]
- Kim, W.J., Park, E.J., Lee, H., Seong, R.H., and Park, S.D. (2002). Physical interaction between recombinational proteins Rhp51 and Rad22 in Schizosaccharomyces pombe. J. Biol. Chem. 277, 30264-30270. [DOI] [PubMed] [Google Scholar]
- Klein, H.L. (1997). RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair for meiosis. Genetics 147, 1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu, J.Y., Chua, P.R., and Roeder, G.S. (1998). The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell 94, 375-386. [DOI] [PubMed] [Google Scholar]
- Lin, Y., and Smith, G.R. (1994). Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics 136, 769-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa, K., Tsuruga, T., Kinomura, A., Usui, K., Katsura, M., Tashiro, S., Mishima, H., and Tanaka, K. (2002). A role for RAD54B in homologous recombination in human cells. EMBO J. 21, 175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar, M., Bahler, J., Kohli, J., and Hiraoka, Y. (2001). Live observation of fission yeast meiosis in recombination-deficient mutants: a study on achiasmate chromosome segregation. J. Cell Sci. 114, 2843-2853. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Muris, D.F., Vreeken, K., Schmidt, H., Ostermann, K., Clever, B., Lohman, P.H., and Pastink, A. (1997). Homologous recombination in the fission yeast Schizosaccharomyces pombe: different requirements for the rhp51+, rhp54+ and rad22+ genes. Curr. Genet. 31, 248-254. [DOI] [PubMed] [Google Scholar]
- Muris, D.F.R., Vreeken, K., Carr, A.M., Broughton, B.C., Lehmann, A.R., Lohman, P.H.M., and Pastink, A. (1993). Cloning the rad51 homolog of Schizosaccharomyces pombe. Nucleic Acids Res. 21, 4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muris, D.F.R., Vreeken, K., Carr, A.M., Murray, J.M., Smit, C., Lohman, P.H.M., and Pastink, A. (1996). Isolation of the Schizosaccharomyces pombe RAD54 homologue, rhp54+, a gene involved in the repair of radiation damage and replication fidelity. J. Cell Sci. 109, 73-81. [DOI] [PubMed] [Google Scholar]
- Nabeshima, K., Kakihara, Y., Hiraoka, Y., and Nojima, H. (2001). A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J. 20, 3871-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, F., Adriance, M., and McCready, S. (2000). The genetic control of spontaneous and UV-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Curr. Genet. 38, 113-125. [DOI] [PubMed] [Google Scholar]
- Ostermann, K., Lorentz, A., and Schmidt, H. (1993). The fission yeast rad22 gene, having a function in mating-type switching and repair of DNA damages, encodes a protein homolog to rad52 of saccharomyces-cerevisiae. Nucleic Acids Res. 21, 5940-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F., and Haber, J.E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasion, S.G., and Forsburg, S.L. (1999). Nuclear localization of Schizosaccharomyces pombe Mcm2/Cdc19p requires MCM complex assembly. Mol. Biol. Cell 10, 4043-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Hidalgo, L., Moreno, S., and San-Segundo, P.A. (2003). Regulation of meiotic progression by the meiosis-specific checkpoint kinase Mek1 in fission yeast. J. Cell Sci. 116, 259-271. [DOI] [PubMed] [Google Scholar]
- Petukhova, G., Stratton, S., and Sung, P. (1998). Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393, 91-94. [DOI] [PubMed] [Google Scholar]
- Petukhova, G., Sung, P., and Klein, H. (2000). Promotion of Rad51-dependent d-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 14, 2206-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill, B., and Roeder, G.S. (1994). The yeast med1 mutant undergoes both meiotic homolog nondisjunction and precocious separation of sister chromatids. Genetics 136, 65-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuckli-Maurer, J., and Heyer, W.D. (2000). Meiotic recombination in RAD54 mutants of Saccharomyces cerevisiae. Chromosoma 109, 86-93. [DOI] [PubMed] [Google Scholar]
- Schmuckli-Maurer, J., Rolfsmeier, M., Nguyen, H., and Heyer, W.D. (2003). Genome instability in rad54 mutants of Saccharomyces cerevisiae. Nucleic Acids Res. 31, 1013-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha, A., and Kleckner, N. (1997). Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90, 1123-1135. [DOI] [PubMed] [Google Scholar]
- Shimada, M., Nabeshima, K., Tougan, T., and Nojima, H. (2002). The meiotic recombination checkpoint is regulated by checkpoint rad(+) genes in fission yeast. EMBO J. 21, 2807-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara, M., Shita-Yamaguchi, E., Buerstedde, J.M., Shinagawa, H., Ogawa, H., and Shinohara, A. (1997). Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147, 1545-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith, H., Brown, G., and Forsburg, S. (2000). Schizosaccharomyces pombe Hsk1p is a potential Cds1p target required for genome integrity. Mol. Cell. Biol. 20, 7922-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, A., Tanaka, K., Okazaki, K., Nohima, H., and Okayama, H. (1994). A zinc finger protein controls the onset of premeiotic DNA synthesis of fission yeast in a mei2-independent cascade. EMBO J. 13, 1881-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, P., and Robberson, D.L. (1995). DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82, 453-461. [DOI] [PubMed] [Google Scholar]
- Suto, K., Nagata, A., Murakami, H., and Okayama, H. (1999). A double-strand break repair component is essential for S phase completion in fission yeast cell cycling. Mol. Biol. Cell 10, 3331-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington, L.S. (2002). Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66, 630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi, P., Heyer, W.-D., Schuchert, P., and Kohli, J. (1988). DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. J. Mol. Biol. 204, 917-925. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., Kagawa, W., Kinebuchi, T., Kurumizaka, H., and Miyagawa, K. (2002). Human Rad54B is a double-stranded DNA-dependent ATPase and has biochemical properties different from its structural homolog in yeast, Tid1/Rdh54. Nucleic Acids Res. 30, 1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui, Y., Khasanov, F.K., Shinagawa, H., Iwasaki, H., and Bashkirov, V.I. (2001). Multiple interactions among the components of the recombinational DNA repair system in Schizosaccharomyces pombe. Genetics 159, 91-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch, M., Zonneveld, J.B., Vreeken, K., de Vries, F.A., Lohman, P.H., and Pastink, A. (2002). Differential expression and requirements for Schizosaccharomyces pombe RAD52 homologs in DNA repair and recombination. Nucleic Acids Res. 30, 1316-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Komen, S., Petukhova, G., Sigurdsson, S., Stratton, S., and Sung, P. (2000). Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell 6, 563-572. [DOI] [PubMed] [Google Scholar]
- Wood, V., et al. (2002). The genome sequence of the eukaryote fission yeast Schizosaccharomyces pombe. Nature 415, 871-880. [DOI] [PubMed] [Google Scholar]
- Xu, L., Weiner, B.M., and Kleckner, N. (1997). Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 11, 106-118. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., and Hiraoka, Y. (2001). How do meiotic chromosomes meet their homologous partners?: lessons from fission yeast. Bioessays 23, 526-533. [DOI] [PubMed] [Google Scholar]
- Young, J.A., Schreckhise, R.W., Steiner, W.W., and Smith, G.R. (2002). Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9, 253-263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.