Abstract
14-3-3 proteins via binding serine/threonine-phosphorylated proteins regulate diverse intracellular processes in all eukaryotic organisms. Here, we examine the role of 14-3-3 self-dimerization in target binding, and in the susceptibility of 14-3-3 to undergo phosphorylation. Using a phospho-specific antibody developed against a degenerated mode-1 14-3-3 binding motif (RSxpSxP), we demonstrate that most of the 14-3-3-associated proteins in COS-7 cells are phosphorylated on sites that react with this antibody. The binding of these phosphoproteins depends on 14-3-3 dimerization, inasmuch as proteins associated in vivo with a monomeric 14-3-3 form are not recognized by the phospho-specific antibody. The role of 14-3-3 dimerization in the phosphorylation-dependent target binding is further exemplified with two well-defined 14-3-3 targets, Raf and DAF-16. Raf and DAF-16 can bind both monomeric and dimeric 14-3-3; however, whereas phosphorylation of specific Raf and DAF-16 sites is required for binding to dimeric 14-3-3, binding to monomeric 14-3-3 forms is entirely independent of Raf and DAF-16 phosphorylation. We also find that dimerization diminishes 14-3-3 susceptibility to phosphorylation. These findings establish a significant role of 14-3-3 dimerization in its ability to bind targets in a phosphorylation-dependent manner and point to a mechanism in which 14-3-3 phosphorylation and dimerization counterregulate each other.
INTRODUCTION
14-3-3 proteins are small acidic proteins naturally found in a dimeric form with a subunit mass of 28-33 kDa, first purified as abundant proteins present in brain tissue extracts (Moore and Perez, 1967). 14-3-3 proteins are expressed in all eukaryotic cells and are highly conserved in protein sequence from yeast to mammals (for review, see Fu et al., 2000; Tzivion et al., 2001; Tzivion and Avruch, 2002). Seven isoforms, encoded by seven distinct genes have been identified in mammals, >10 in plants, and two in yeast, Drosophila, and Caenorhabditis elegans. The high level of functional conservation of the 14-3-3 gene products is indicated by the finding that the yeast 14-3-3 proteins are functionally interchangeable with plant and mammalian isoforms. The primary function of the 14-3-3 proteins is to bind phosphoserine/threonine-containing motifs in a sequence-specific manner, e.g., RSxpSxP, mode-1 and RxxxpSxP, mode-2 (where pS represents phospho-serine), in a manner analogous to Src-homology 2 and phospho-tyrosine-binding domains that bind phosphotyrosine-containing motifs (Muslin et al., 1996; Yaffe et al., 1997). 14-3-3 proteins are necessary elements in the regulation of a variety of biological systems. For example, 14-3-3 proteins participate in regulation of cell cycle arrest in response to DNA damage, cell cycle timing, and intracellular signaling in response to stress and mating pheromone in yeast (van Hemert et al., 2001), photoreceptor development and learning in Drosophila (Skoulakis and Davis, 1998), cellular response to stress and survival factors in mammals, and the Ras/Raf signaling pathway in various organisms (Fu et al., 2000; Tzivion et al., 2001; Tzivion and Avruch, 2002). Nearly 100 proteins have been reported to associate with 14-3-3 proteins in vivo. Among these are proteins involved in cell cycle control, such as Cdc25, Wee1, p53, CDC2, and CDK2; cellular signaling and stress responses, such as Raf, IGF-I receptor, IRS-1, phosphatidylinositol-3 kinase (PI-3 kinase), protein kinase C, Cbl, Bcr, polyoma middle T antigen, MEKK-1 and 4, MLK2, BAD, and ASK-1; transcriptional regulation, such as FKHRL1, DAF-16, TAZ, TLX-2, and histone deacetylase; and cytoskeletal proteins such as keratin K18 and vimentin (for review, see Tzivion et al., 2001; Tzivion and Avruch, 2002). 14-3-3 binding regulates its partners through a variety of mechanisms, such as altering their catalytic activity, cellular localization, incorporation into protein complexes, or their susceptibility to proteases and phosphatases (Muslin and Xing, 2000; Tzivion and Avruch, 2002).
Structural analysis of 14-3-3, crystallized alone or together with bound synthetic short peptides, revealed the dimeric structure of 14-3-3 and pointed to the residues involved in dimerization and target binding (for review, see Fu et al., 2000; Tzivion and Avruch, 2002). These findings are supported by 14-3-3 mutational analysis that identified specific residues and regions in the amino terminus critical for dimerization and residues both in the carboxy and amino terminus critical for target binding (Fu et al., 2000; Tzivion and Avruch, 2002). Combined, these studies indicated that each 14-3-3-half dimer can bind a phosphopeptide independently and that dimerization may not be necessary for 14-3-3-target protein binding. Several findings indicated, however, that this conclusion does not apply to all 14-3-3-target protein interactions. For example, Wee1 (Honda et al., 1997; Wang et al., 2000), keratin K18 (Ku et al., 1998), Cbl (Liu et al., 1997), IGF-I receptor (Craparo et al., 1997), IRS-1 (Ogihara et al., 1997), and DAF-16 (Cahill et al., 2001) all require more than one phosphorylation site for stable 14-3-3 binding, suggesting that simultaneous binding of a dimeric 14-3-3 to two distinct sites is necessary to achieve stable association with these target proteins (Tzivion et al., 2001; Tzivion and Avruch, 2002). This apparent discrepancy can be explained by the finding of Yaffe et al. (1997) showing that the presence of two tandem 14-3-3 binding sites on a single peptide increases 14-3-3 binding affinity 30-fold compared with that of a single site, thus indicating that proteins that have two binding sites may bind 14-3-3 with much higher affinity than proteins containing a single site. With this in mind, monomeric 14-3-3 forms would bind only to proteins containing a high-affinity site, whereas dimeric 14-3-3 forms would also be able to associate with targets containing two or more low-affinity 14-3-3 binding sites. In agreement with this notion, dimeric, but not monomeric 14-3-3 forms bind vimentin, which contains several potential 14-3-3 binding sites; none of which matching an identified high-affinity 14-3-3 binding site (Tzivion et al., 2000).
14-3-3 binding to most of its targets has been shown to depend on the phosphorylation of the target protein; nevertheless, several reports indicate that 14-3-3 can bind some of its targets in a phosphorylation-independent manner (Zhai et al., 2001; Borch et al., 2002; Hallberg, 2002; Tzivion and Avruch, 2002). It is not established, however, what fraction of the in vivo 14-3-3 association is phosphorylation independent.
To further explore the importance of dimerization in 14-3-3 function and to determine the in vivo role of target protein phosphorylation in 14-3-3 binding, we generated a pan antibody that reacts with the peptide motif RSxpSxP, a mode-1, high-affinity 14-3-3 binding site. We used this antibody in combination with metabolic 32P and 35S-Met labeling to examine the basis of endogenous cellular protein binding in vivo to native, dimeric and mutant, monomeric 14-3-3 proteins. We find that most of the 35S-Met-labeled proteins that specifically bind dimeric 14-3-3 in vivo react with the pan-phospho-specific 14-3-3 binding site antibody and can be displaced from 14-3-3 by a competing, mode-1 phosphopeptide. Many cellular proteins associate also with a mutant, monomeric 14-3-3; however, none of these reacts with the phospho-specific antibody. In addition, although monomeric 14-3-3 forms bind several targets (e.g., Raf and DAF-16) with comparable affinity as wild-type dimeric 14-3-3, this binding is independent of Raf and DAF-16 phosphorylation. This is in contrast to the binding of dimeric 14-3-3, which depends entirely on Raf and DAF-16 phosphorylation. We also find that dimerization essentially abolishes 14-3-3 susceptibility to phosphorylation in vivo in response to various cellular treatments. Our results demonstrate an important role of 14-3-3 dimerization in the phosphorylation-dependent target binding and point to a counterregulatory mechanism of 14-3-3 dimerization and phosphorylation.
MATERIALS AND METHODS
cDNA Constructs, Antibodies, and Peptides
Wild-type glutathione S-transferase (GST)-14-3-3 ζ (wt, 1-245), amino-terminal GST-14-3-3 ζ (nt, 1-140), carboxy-terminal GST-14-3-3 ζ (139-245), and dimerization-deficient GST-14-3-3 ζ (dm, E5K, L12AE to Q12QR, Y82Q, K85N, E87Q) were in the pEBG mammalian expression vector (see Figure 3A). myc-Epitope-tagged c-Raf-1, Raf S259A, and S259/621A and myc-14-3-3 were in pMT2 vector. m2-FLAG-epitope tagged wild-type DAF-16 (wt), m2-DAF-16 T54A, m2-DAF-16 S240A/T242A (2A), and m2-DAF-16 T54A/S240A/T242A (3A) were in pcDNA3 vector (see Figure 6A). The construction of these vectors has been described previously (Luo et al., 1995; Tzivion et al., 1998; Cahill et al., 2001). Constitutive and dominant negative forms of GST-AMP-activated kinase were a kind gift from Dr. Lee Witters (Dartmouth, Hanover, NH). The phospho-site-specific antibody to Raf pS259 and the pan phospho-specific 14-3-3 binding site antibody were produced by Cell Signaling Technology (Beverly, MA) against the phosphopeptides LSQRQRSTpST-PNVH and xxxxRSxpSxPxxxxC, respectively (pS represents phospho-serine and x any amino acid besides Cys). Briefly, rabbits were injected with the above-mentioned phosphopeptides, and the IgG fraction from the rabbit bleeds was first preabsorbed to a respective nonphosphorylated peptide to remove the antibody fraction reacting with the nonphosphorylated peptide, followed by affinity purification be using the respective phosphopeptide (Zhang et al., 2002). Specificity tests performed by Cell Signaling Technology show a signal-to-noise ratio (phospho versus nonphospho 14-3-3 binding motif peptides) of ∼40 in enzyme-linked immunosorbent assay tests. In addition, the enzyme-linked immunosorbent assay tests show specificity for phosphopeptides derived from 14-3-3 binding proteins in comparison with nonrelevant phosphopeptides (the complete specificity information can be found in the company data sheets). m2-FLAG and GST antibodies were from Sigma-Aldrich (St. Louis, MO) and UBI (Charlottesville, VA), respectively. The myc antibody was purified from mice ascitis fluids injected with the 9E10 hybridoma (American Type Culture Collection, Manassas, VA). Peptides corresponding to c-Raf-1 AA 613-627 and its phosphorylated version at a position corresponding to S621 (613LPKINRSApSEPSLHR627), generating a canonical high-affinity 14-3-3 binding site and a control, doubly phosphorylated peptide corresponding to c-Raf-1 AA 461-486 (AKNIIHRDMKpSNNIFLHEGLpTVKIGD), not containing a 14-3-3 binding site, were synthesized by Quality Controlled Biochemicals (Boston, MA).
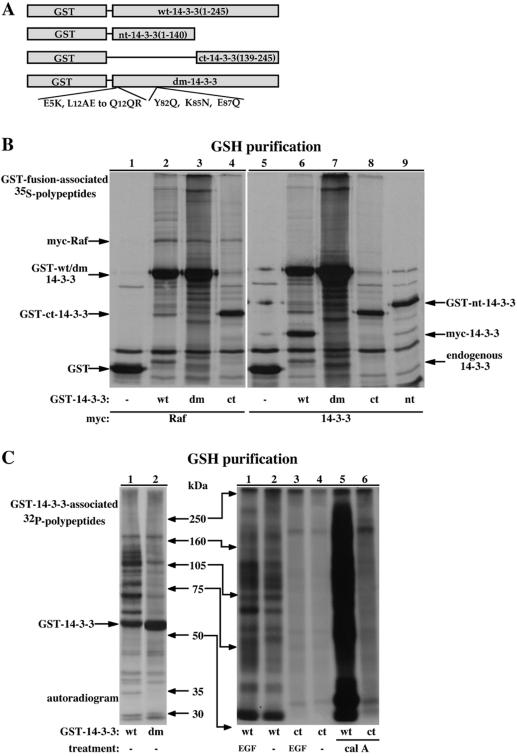
Figure 3.
Dimerization-deficient 14-3-3 forms present reduced phosphoproteins binding. (A) Schematic representation of 14-3-3 ζ GST-fusion variants used in the study. (B) GST-fusion proteins were purified from 35S-Met-labeled COS-7 cells coexpressing the indicated GST-fusion proteins and either myc-Raf (lanes 1-4) or myc-14-3-3 (lanes 5-9) and resolved using 10% SDS-PAGE. An autoradiogram showing the 35S-labeled proteins is presented. Indicated are the migrations of the GST-fusion proteins, endogenous 14-3-3 and the myc-epitope tagged Raf and 14-3-3. (C) GST-fused wild-type 14-3-3 (wt), dimerization-deficient 14-3-3 (dm) or amino-terminally truncated 14-3-3 (ct) were purified from 32P-labeled COS-7 cells treated as indicated with 100 nM EGF or 150 nM calyculin A (cal A) for 15 min. The recovered proteins were separated using a continuous 6-15% gradient (left) or a 6.5% (right) SDS-PAGE and analyzed by autoradiography. Indicated are the positions of the molecular weight markers and of the GST-14-3-3 fusion protein (left only).
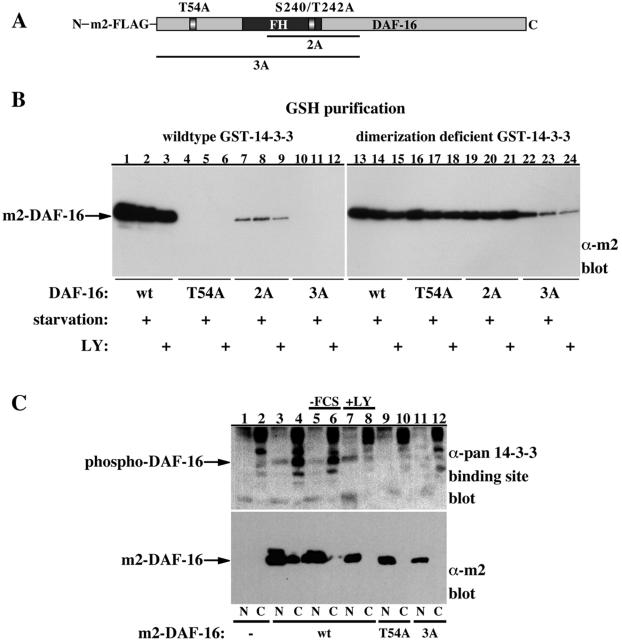
Figure 6.
Binding of dimerization-deficient 14-3-3 to DAF-16 is independent of DAF-16 phosphorylation. (A) Schematic representation of DAF-16 mutants used in the study. (B) HEK-293 cells were transfected with GST-14-3-3 (lanes 1-12) or dimerization-deficient GST-14-3-3 (lanes 13-24) together with m2-FLAG-epitope-tagged DAF-16 (wt, lanes 1-3 and 13-15), T54A DAF-16 (lanes 4-6 and 16-18), S240A/T242A DAF-16 (2A, lanes 7-9 and 19-21), or T54A/S240A/T242A DAF-16 (3A, lanes 10-12 and 22-24). The transfected cells were either left untreated or were serum deprived for 18 h in the absence or presence of the PI-3 kinase inhibitor LY294002 (LY, 10 μM) as indicated. GSH-bead-pulled-down proteins were resolved using 10% SDS-PAGE and immunoblotted for m2-DAF-16 (Sigma-Aldrich). The migration position of m2-DAF-16 is indicated. (C) Nuclear/cytoplasmic extracts of HEK-293 cells expressing control vector (lanes 1 and 2), m2-FLAG-DAF-16 (lanes 3-8), T54A DAF-16 (lanes 9 and 10), or T54A/S240A/T242A DAF-16 (3A, lanes 11 and 12) grown in serum (lanes 1-4 and 9-12) or deprived of serum in the presence of vehicle (lanes 5 and 6) or LY294002 (10 μM, lanes 7 and 8) were separated on 8.5% SDS-PAGE and were immunoblotted with the pan phospho-specific 14-3-3 binding site antibody (top) or with m2-FLAG antibody (bottom). The location of DAF-16 is indicated.
Cell Culture and Transfection
COS-7 and human embryonic kidney (HEK)-293 cells were maintained in DMEM supplemented with 10% fetal calf serum. For transient expression of proteins cells were transfected using LipofectAMINE (Invitrogen, Carlsbad, CA) as detailed in the figure legends according to the manufacturer's instructions. For cell stimulation, 24 h after transfection, cells were serum deprived for 18 h before the addition of the agonist.
Metabolic Labeling
Serum-deprived cells were washed once with medium lacking either methionine/cysteine (for 35S-Met labeling) or phosphate (for 32P-labeling) followed by 30-min incubation in the same media for depleting intracellular methionine/cysteine or phosphate, respectively. Cells were radiolabeled by incubation in the presence of either 0.5 mCi/ml 35S-Met or 32P for 4 or 2 h, respectively.
Cell Extraction and Protein Purification
For regular cell extraction, cells were lysed for 30 min in ice-cold extraction buffer containing 50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 1% Triton X-100, 1 mM dithiothreitol, 1 mM EDTA, 1 mM EGTA, 2 mM Na3VO4, 50 mM β-glycerophosphate, and a protease inhibitor cocktail (Pharmacia, Peapack, NJ). Cleared cell lysates were incubated at 4°C for 90 min with the appropriate antibody precoupled to protein A/G-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) for immunoprecipitation or with glutathione (GSH)-Sepharose beads (Pharmacia) for GSH-affinity purification. The beads were washed twice with extraction buffer, twice with extraction buffer containing 0.5 M LiCl, and twice with buffer A (40 mM Tris-Cl, pH 7.5, 0.1 mM EDTA, 5 mM MgCl2, and 2 mM dithiothreitol). The proteins were eluted directly in SDS-sample buffer and subjected to SDS-PAGE.
For nuclear and cytoplasmic extraction, cell extracts were prepared using the NE-PER kit (Pierce Chemical, Rockford, IL) according to the manufacturer's instructions.
Raf Phosphopeptide Mapping
Two-dimensional phospho-peptide mapping was performed according to previously described protocols (Boyle et al., 1991; Luo et al., 1991). Briefly, 32P-labled myc-Raf was resolved using 7.5% SDS-PAGE, transferred to a polyvinylidene membrane, excised, and 32P incorporation in myc-Raf was determined by Cherenkov counting. After incubation with 0.5% polyvinylpyrrolidone in 100 mM acetic acid for 30 min at 37°C and extensive washes, the Raf protein was digested with 10 μg of sequencing grade-modified trypsin (Promega, Madison, WI) in 50 mM ammonium bicarbonate buffer for 2 h at 37°C and with additional 10 μg of trypsin for overnight. The eluted peptides were washed twice with 50 mM ammonium bicarbonate buffer and once with pH 1.9 thin layer chromatography-electrophoresis buffer (2.2% formic acid and 7.8% acetic acid). Samples were spotted on cellulose thin layer chromatography plates (Eastman Kodak, Rochester, NY) and separated using the Hunter thin-layer system (CBS Scientific, Del Mar, CA) in pH 1.9 buffer for 25 min at 1000 V. The plates were dried overnight and subjected to the second dimension of chromatographic separation in phospho-chromatography buffer (37.5% n-butanol, 25% pyridine, and 7.5% acetic acid). The plates were dried, and the phospho-peptide spots were visualized by autoradiography and phosphorimaging (this method routinely allowed recovery of 85-95% of the initial radioactivity in myc-Raf).
RESULTS
Development of Phospho-Site-specific Antibodies Recognizing Pan and Distinct 14-3-3 Binding Sites
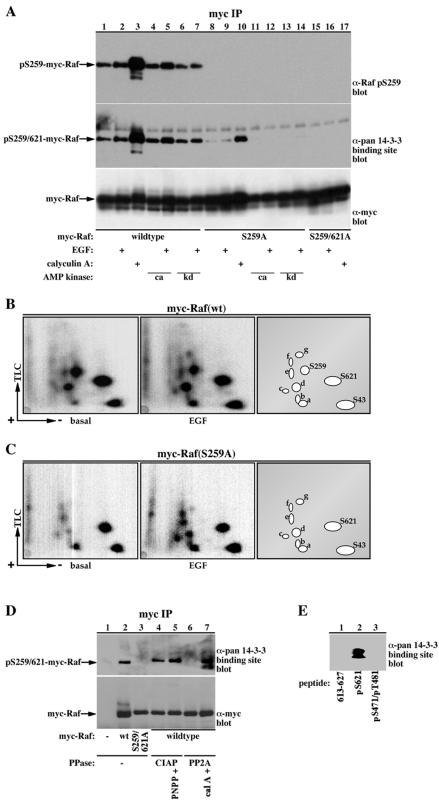
To aid in detecting phosphorylation of 14-3-3 target proteins at their high-affinity 14-3-3 binding sites, antibodies were generated against a Raf phosphopeptide corresponding to the 14-3-3 binding site of Raf at Ser-259 (e.g., LSQRQRSTp-STPNVH) and against a degenerated phosphopeptide sequence containing a mode-1 14-3-3 binding sequence, e.g., xxxxRSxpSxPxxxxC (where pS represents phospho-serine and x any amino acid beside Cys; for detailed information on antibody production and specificity, see MATERIALS AND METHODS). The high specificity of these antibodies is demonstrated in Figure 1. Raf contains two 14-3-3 binding sites (Tzivion et al., 1998), one surrounding Ser-259 (256RSTp-STP) and the second surrounding Ser-621 (618RSApSEP). The phospho-specific antibody to pS259 recognizes wild-type Raf (Figure 1A, lanes 1-7), which when expressed in COS-7 cells is phosphorylated on four major sites (S43, S259, S621, and a fourth unidentified site) and at several other minor sites (Figure 1B; Morrison et al., 1993). Conversely, the antibody is unable to recognize a Raf S259A mutant (Figure 1A, lanes 8-14), which lacks phosphorylation at the S259 site, but maintains phosphorylation at all the other sites, including S621 (Figure 1, compare B and C). These results demonstrate that despite the high similarity between the S259 and S621 sites, this antibody can successfully distinguish between these two distinct 14-3-3 binding sites.
Figure 1.
Raf phospho-serine 259 antibody and a pan phospho-specific 14-3-3 binding site antibody display high specificity for their phosphorylated targets. (A) Myc-epitope-tagged c-Raf-1 variants were immunoprecipitated from COS-7 cells expressing wild-type (lanes 1-7), S259A (lanes 8-14), or S259/621A (lanes 15-17) myc-Raf alone, or coexpressing constitutive (ca, lanes 4, 5, 11, and 12) or kinase-dead (KD, lanes 6, 7, 13, and 14) AMP kinase, treated with 100 ng/ml EGF or 100 nM calyculin A for 15 min as indicated. The resulting Raf proteins were separated with 10% SDS-PAGE and immunoblotted with either anti-myc antibody (clone 9E10, bottom), phospho-Raf S259 (top; Cell Signaling Technology) or with a pan phospho-specific 14-3-3 binding site antibody (middle; Cell Signaling Technology). The position of myc-Raf in the various blots is indicated. (B and C) Serum-deprived COS-7 cells expressing wild-type (B) or S259A (C) myc-Raf variants were metabolically labeled with 32P and stimulated with 100 ng/ml EGF for 15 min as indicated. myc-Raf proteins were immunoprecipitated with a myc-epitope tag antibody and subjected to phosphopeptide mapping as described in MATERIALS AND METHODS. Representative autoradiograms of the Raf phosphopeptide maps and a schematic representation of the phosphopeptide spots are presented. The locations of phospho-S43, -S621, and -S259 peptides are indicated. Note that the phospho-S259 spot is missing in the map of the Raf S259A mutant. The identity of the other phosphopeptide spots is unknown. (D) myc-Immunoprecipitates from vector control (lane 1), wild-type myc-Raf (lanes 2 and 4-6) or S259/621A myc Raf (lane 3)-transfected COS cells were treated with vehicle (lanes 1-3), alkaline phosphatase (CIAP, 20 U, lanes 4 and 5; Promega), or protein phosphatase 2A (UBI, PP2A, 0.5 U, lanes 6 and 7) for 30 min at 30°C. Lane 5 also included the alkaline phosphatase inhibitor PNPP (50 mM) and lane 7 the protein phosphatase inhibitor calyculin A (cal A, 10 nM). After dephosphorylation, the myc-immunoprecipitates were washed twice and analyzed as in A. Immunoblotting results with the pan phospho-specific 14-3-3 binding site antibody (top) and myc (bottom) are presented. (E) Twenty nanomoles of each synthetic peptide: Raf 613-627 (LPKINRSASEPSLHR, 613-627, lane 1), Raf 613-627 phosphopeptide phosphorylated on a site corresponding to Raf S621 (LPKINRSApSEPSLHR, pS621, lane 2), and Raf 461-486 phosphopeptide phosphorylated on two sites, corresponding to Raf S471 and T481 (AKNIIHRDMKpSNNIFLHEGLpTVKIGD, pS471/pT481, lane 3) were separated on a 17.5% SDS-PAGE and immunoblotted with the pan phospho-specific 14-3-3 binding site antibody.
The pan phospho-specific 14-3-3 binding site antibody, nevertheless, recognizes both wild-type Raf and the S259A Raf mutant. This antibody, however, binds wild-type Raf with a higher efficiency than the S259A mutant, indicating that the antibody is capable of binding to both 14-3-3 binding sites on Raf, pS259, and pS621 (Figure 1A, compare lanes 1-7 and 8-14). Conversion of both Ser-259 and Ser-621 to Ala abolishes reactivity with the pan antibody, demonstrating the dependency of the antibody on phosphorylation of the 14-3-3 binding sites of Raf (Figure 1A, lanes 15-17). Interestingly, expression of Raf together with a constitutively active (Figure 1A, lanes 4, 5, 11, and 12) or a kinase dead (Figure 1A, lanes 6, 7, 13, and 14) forms of AMP-activated kinase, a candidate Raf S621 kinase (Sprenkle et al., 1997), did not alter Raf S621 phosphorylation in COS-7 cells. On the other hand, treatment of the cells with the phosphatase inhibitor calyculin A strongly enhances phosphorylation of both the S259 and the S621 sites (Figure 1A, lanes 3 and 10). The specificity of the pan phospho-specific 14-3-3 binding site antibody was further demonstrated by showing that the reactivity of the antibody with Raf can be abolished by Raf dephosphorylation with protein phosphatase 2A (Figure 1D, compare lanes 2 and 6). In addition, the antibody reacts with a synthetic phosphopeptide corresponding to the 14-3-3 binding site on Raf at S621, but not with the unphosphorylated peptide or with a phosphopeptide from a non-14-3-3 binding region of Raf (Figure 1E, compare lane 2 with lanes 1 and 3). Also, the antibody reacts with cytoplasmic DAF-16, which is phosphorylated on 14-3-3 binding sites, but not with nuclear DAF-16, which is in an unphosphorylated form (Figure 6C, compare lanes 3 and 4). In addition, mutation of the AKT phosphorylation sites of DAF-16 or treatment of the cells with the PI-3 kinase inhibitor LY294002 abolishes DAF-16 reactivity with the antibody (Figure 6C, compare lane 4 with lanes 8, 10, and 12).
Most of the 14-3-3-associated Proteins in COS-7 Cells Are Reactive with the Pan Phospho-specific 14-3-3 Binding Site Antibody
Many of the identified 14-3-3 target proteins have been demonstrated to require phosphorylation for 14-3-3 binding; and in several cases, it has been demonstrated that the 14-3-3 phosphopeptide binding pocket mediates the binding (Zhang et al., 1997; Wang et al., 1998; Xing et al., 2000). Nevertheless, several reports indicate that 14-3-3 proteins can also bind nonphosphorylated peptides and proteins through their phosphopeptide binding pocket (Masters et al., 1999; Wang et al., 1999; Mils et al., 2000). To examine what fraction of the 14-3-3-associated proteins in COS-7 cells contains phosphorylation sites recognized by the pan phospho-specific 14-3-3 binding site antibody, we compared the pattern of 14-3-3-associated proteins detected by metabolic 35S-Met labeling (Figure 2, top) with that detected by immunoblotting with the pan phospho-specific antibody (Figure 2, bottom). Recombinant GST-14-3-3 transiently expressed in COS-7 cells copurifies with a large number of proteins, both under basal conditions and after calyculin A-induced protein phosphatase inhibition (Figure 2, top, lanes 2-11). The specificity of 14-3-3 protein binding is exemplified by the GST control (Figure 2, top, compare lanes 1 and 6). This binding specificity is also manifested in the ability of a competitor phosphopeptide corresponding to the 14-3-3 binding site at Raf S621 to displace all the specific 14-3-3-bound proteins (Figure 2, top, compare lanes 7 and 11 with lanes 12 and 13). The residual 35S-Met-labeled proteins (Figure 2, top, lanes 12 and 13) represent the GST-14-3-3 protein and its degradation products (35-55 kDa), endogenous 14-3-3 that dimerizes with the GST-14-3-3 (29-30 kDa) and proteins that associate with GST and GSH beads (20-32 kDa), which are also present in the control GST lane (Figure 2, top, lane 1). The ability of the competitor phosphopeptide to dissociate all the specifically 14-3-3-associated proteins demonstrates that 14-3-3 binds all its targets in COS-7 cells through the conserved amphipathic phosphopeptide binding pocket. This conclusion is further supported by the finding that most of the specifically 14-3-3-bound 35S-Met-labeled proteins are recognized by the pan phospho-specific 14-3-3 binding site antibody (Figure 2, bottom). Importantly, none of the nonspecifically GST-14-3-3-associated proteins, including the GST-14-3-3 itself, cross-react with the antibody. In addition, all proteins that react with the pan phospho-specific antibody are displaced from 14-3-3 by the competitor phosphopeptide, demonstrating the high specificity of the antibody toward 14-3-3 targets (Figure 2, compare lanes 7 and 11 with lanes 12 and 13).
Figure 2.
14-3-3 binds all of its target proteins in COS-7 cells via the phosphopeptide binding pocket, and most of the associated proteins contain a phosphopeptide recognized by a pan phospho-specific 14-3-3 binding site antibody. Serum-deprived COS-7 cells transiently expressing GST (50 ng of DNA/10-cm plate, lane 1) or GST-fused 14-3-3 ζ (10 ng/plate, lanes 2-6 or 50 ng/plate lanes 7-14) were 35S-Met labeled and treated with either carrier (lanes 2, 7, and 12) for 60 min or calyculin A (100 nM, lanes 3-6, 8-11, 13, and 14) for the indicated times. Cells were extracted in the absence (lanes 1-11 and 14) or presence (lanes 12 and 13) of 300 μM synthetic competitor phosphopeptide corresponding to the Raf S621 14-3-3 binding site. GST-fusion proteins were precipitated using GSH-Sepharose beads, and the resulting proteins were separated using a continuous 6-15% gradient SDS-PAGE and visualized using either autoradiography (top) or by immunoblotting with the pan phospho-specific 14-3-3 binding site antibody (bottom). The migration positions of the molecular weight protein markers, GST-fusion proteins, and coassociated cellular vimentin are indicated. It is important to note that presented are exposure levels that most optimally represent the general result; however, our experiments used multiple exposures of both the 35S-Labeling and immunoblotting experiments to make a comprehensive comparison of the two.
Combined, these results demonstrate that in COS-7 cells the majority of the 14-3-3 target proteins bind 14-3-3 through the 14-3-3 phosphopeptide binding pocket and that these proteins bind 14-3-3 via a specific phosphorylated site, or at least contain a specific phosphorylated site recognized by the pan phospho-specific 14-3-3 binding site antibody. Thus, even though some peptides and proteins can bind 14-3-3 in a phosphorylation-independent manner, in vivo, the majority of proteins seem to bind 14-3-3 in a phosphorylation-dependent manner.
Self-Dimerization Is Important for 14-3-3 Ability to Bind Phosphoprotein Targets In Vivo
We and others previously showed that dimerization-deficient 14-3-3 forms can bind several targets with comparable efficiency as wild-type 14-3-3 (Ichimura et al., 1995, 1997; Luo et al., 1995; Gu and Du, 1998; Tzivion et al., 1998). Crystal structure analyses of 14-3-3, either alone or with bound peptides, indicate that each 14-3-3 half dimer can bind a target peptide independently (Yaffe et al., 1997; Rittinger et al., 1999). Combined, these findings suggest that dimerization may not be necessary for 14-3-3 to bind its phosphorylated targets. To examine this issue, we compared the pattern of cellular protein binding to wild-type and dimerization-deficient 14-3-3 forms, visualized using either 35S/32P-labeling (Figures 3 and 4, top) or immunoblotting with the pan phospho-specific 14-3-3 binding site antibody (Figure 4, bottom).
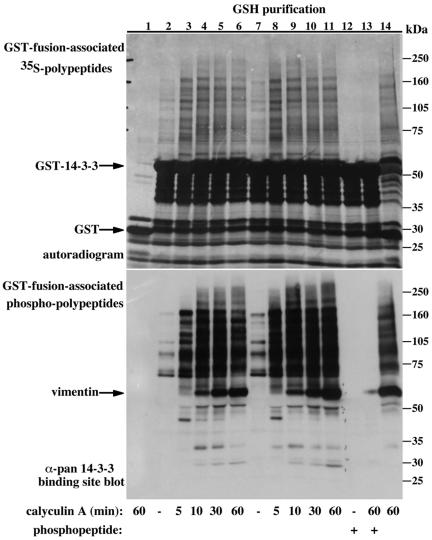
Figure 4.
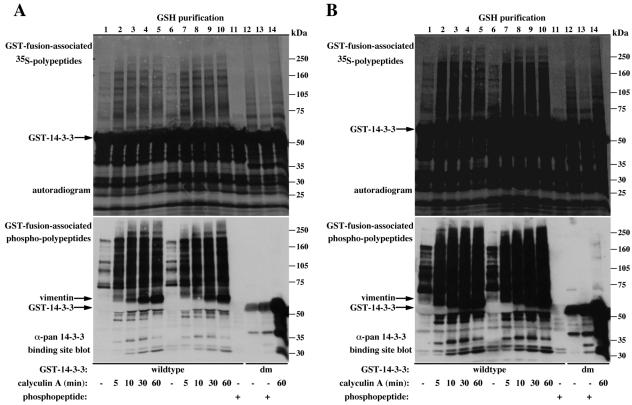
Dimerization-deficient 14-3-3-associated proteins are not recognized by the pan phospho-specific 14-3-3 binding site antibody. (A and B) Serum-deprived COS-7 cells transiently expressing wild-type GST-14-3-3 (50 ng of DNA/10-cm plate, lanes 1-5 or 250 ng/plate, lanes 6-11) or dimerization-deficient GST 14-3-3 (250 ng/plate, lanes 12-14), were 35S-Met labeled and treated with either carrier (lanes 1, 6, 11, and 12) for 60 min or calyculin A (100 nM, lanes 2-5, 7-10, and 14) for the indicated times. Cells were extracted in the absence (lanes 1-10, 12, and 14) or presence (lanes 11 and 13) of 300 μM synthetic Raf competitor phosphopeptide, and the recovered GST-fusion proteins were analyzed as described in Figure 2. The migration position of the molecular weight protein markers, GST-14-3-3 and coassociated cellular vimentin are indicated. To stress the difference between wild-type and dimerization-deficient 14-3-3-associated proteins and to demonstrate the ability of the competitor phosphopeptide to completely dissociate all associated proteins from wild-type 14-3-3 provided are two exposure levels (A and B).
The metabolic labeling experiments show that a variety of proteins associate with both wild-type and dimerization-deficient 14-3-3 forms (Figure 3B). This binding seems specific as far as recombinant GST and GST-fused to the amino-terminal part of 14-3-3, lacking the target protein-binding domain (GST-nt-14-3-3; Figure 3A) fail to bind with most of the proteins that associate with full-length 14-3-3 (Figure 3B, compare lanes 1, 5, and 9, nonspecific binding, and lanes 2-4, 6-8, specific 14-3-3 binding). The inability of the dimerization-deficient 14-3-3 forms to self-associate is demonstrated by their inability to bind endogenous 14-3-3 or a coexpressed myc-epitope-tagged 14-3-3 (Figure 3B, compare lanes 2 and 6, wild-type 14-3-3 with lanes 3, 4, 7, and 8, dimerization-deficient 14-3-3). Although the dimerization-deficient 14-3-3 forms bind the coexpressed myc-epitope-tagged Raf (Figure 3B, compare lane 2 with lanes 3 and 4), and display a pattern of bound proteins resembling the one observed with wild-type 14-3-3, a closer examination reveals large differences between the two patterns (Figure 3B, compare lanes 2 and 6 with lanes 3, 4, 7, and 8). This difference is much more apparent when using metabolic 32P-labeling experiments (Figure 3C). This figure also shows that while treatment of cells with agents that increase cellular protein phosphorylation (i.e., epidermal growth factor [EGF] and calyculin A) enhance phosphoprotein association with wild-type 14-3-3, they have negligible effect on protein association with dimerization-deficient 14-3-3 forms (Figure 3C, right, compare lanes 1 and 5 with lanes 3 and 6). These results pointed to a significant role of 14-3-3 dimerization in phosphotarget binding.
To further examine this possibility and to characterize the differences between wild-type and dimerization-deficient 14-3-3-associated proteins and their mode of binding to 14-3-3, we immunoblotted the variant 14-3-3-associated proteins with the pan phospho-specific 14-3-3 binding site antibody and tested the ability of the Raf-derived competitor phosphopeptide to displace the bound proteins (Figure 4). Similarly to results shown in Figure 2, the majority of proteins that associate with wild-type 14-3-3 react with the pan phospho-specific antibody; however, much to our surprise, none of the proteins that are bound to the dimerization-deficient 14-3-3 mutant is recognized by this antibody (Figure 4, A and B, bottom, compare lanes 1-10 with lanes 12-14). The synthetic phosphopeptide (corresponding to the 14-3-3 binding site on Raf at Ser 621) displaced all the specifically bound 35S-Met-labeled proteins from wild-type 14-3-3, and to a lesser extent, also from the dimerization-deficient 14-3-3 (Figure 4, A and B, top, compare lanes 11 and 13). These results indicate that the proteins that bind with monomeric 14-3-3 lack the mode-1, phosphorylated motif RSxpSxP. These proteins may nevertheless bind, at least in part, to the 14-3-3 phosphopeptide binding pocket, which may explain their displacement by the Raf phosphopeptide. The reduced ability of the phosphopeptide to displace bound proteins from the dimerization-deficient 14-3-3, compared with wild-type 14-3-3, may reflect a reduced affinity of monomeric 14-3-3 for the phosphopeptide, implicating a significant role of 14-3-3 dimerization in phosphopeptide binding. This notion is supported also by the observation that calyculin A treatment, which elevates the level of phosphoproteins in the cell, dramatically increases association of 35S-Met-labeled proteins with wild-type 14-3-3, but not with the dimerization-deficient 14-3-3 (Figure 4, A and B, top, compare lanes 6 and 10, wild-type 14-3-3 with lanes 12 and 14, monomeric 14-3-3). Combined, these findings indicate that 14-3-3 dimerization is important, not only to enable binding to proteins containing “low-affinity” motifs but also for binding proteins containing the mode-1, high-affinity motif.
Dimerization-deficient 14-3-3 Forms Can Bind Target Proteins in a Phosphorylation-independent Manner
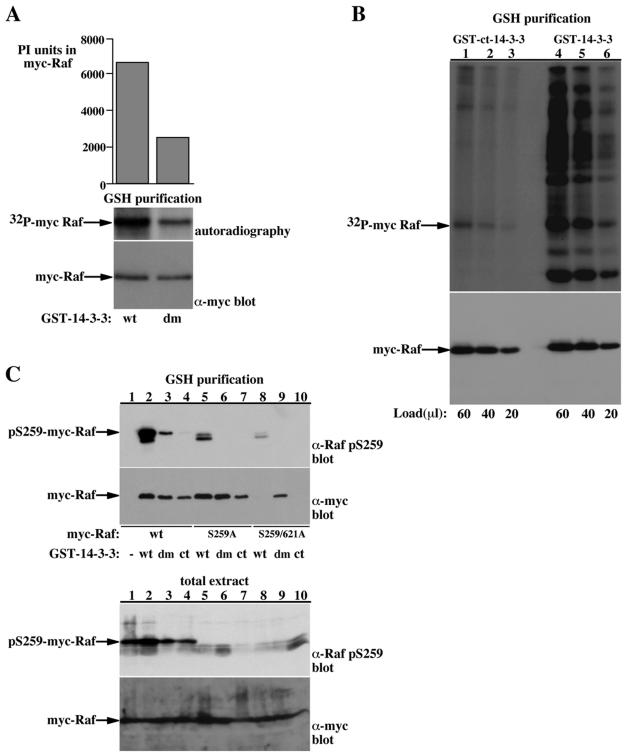
We next examined the dependency on target protein phosphorylation to wild-type and dimerization-deficient 14-3-3 binding to two well-characterized 14-3-3 targets, Raf and DAF-16 (Figures 5 and 6). Using the yeast two-hybrid interaction system and association analysis in mammalian cells, we previously demonstrated that Raf contains two phosphorylation sites that cooperatively mediate 14-3-3 binding, e.g., S259 and S621 (Tzivion et al., 1998). We and others also found that dimerization-deficient 14-3-3 forms maintain their ability to bind Raf (Luo et al., 1995; Ichimura et al., 1997; Tzivion et al., 1998); however, the necessity of Raf phosphorylation for binding to the dimerization-deficient 14-3-3 forms was not examined. To address this question, we first compared the in vivo phosphorylation level of Raf bound to wild-type and dimerization-deficient 14-3-3 forms (Figure 5, A and B). Raf bound to wild-type 14-3-3 contains up to threefold more total 32P than Raf bound to dimerization-deficient 14-3-3 forms (Figure 5, A and B). This difference in phosphate content is distributed equally on all Raf phosphorylation sites, because we did not find a significant difference between the 32P-tryptic peptide map of wild-type and dimerization-deficient 14-3-3-associated Raf (our unpublished data). Next, we used the pS259 phospho-site-specific antibody to compare between the phosphorylation of the 14-3-3 binding site of Raf bound to wild-type and dimerization-deficient 14-3-3 forms (Figure 5C). These experiments demonstrated that even though equal amounts of Raf protein are recovered with wild-type and dimerization-deficient 14-3-3 (as assessed by myc immunoblotting; Figure 5C, GSH purification, bottom, compare lane 2 with lanes 3 and 4), the myc-Raf recovered with wild-type 14-3-3 contains severalfold higher phosphorylation of Ser259 than the myc-Raf recovered with dimerization-deficient 14-3-3 forms (Figure 5C, GSH purification, top, compare lane 2 with lanes 3 and 4). In addition, although mutation of both Raf phosphorylation sites that mediate 14-3-3 binding abolishes Raf binding to wild-type 14-3-3 (Figure 5C, GSH purification, bottom, compare lanes 2 and 8), the binding to the dimerization-deficient 14-3-3 remains unchanged (Figure 5C, GSH purification, bottom, compare lanes 3 and 9). Combined, these results indicate that binding of dimerization-deficient 14-3-3 forms to Raf is largely independent of Raf phosphorylation.
Figure 5.
Binding of dimerization-deficient 14-3-3 to Raf is independent of Raf phosphorylation. (A and B) GST-fusion proteins purified from 32P-labeled COS-7 cells coexpressing myc-Raf and either wild-type GST-14-3-3 (wt) or dimerization-deficient GST-14-3-3 (dm) (A) or wild-type GST-14-3-3 or amino-terminally truncated GST-14-3-3 (ct) (B) were resolved using SDS-PAGE. The GST-14-3-3-associated myc-Raf was analyzed by myc immunoblotting (A and B, bottom) to quantify Raf recovery and by phosphorimaging to quantify 32P incorporation in Raf (A, middle; B, top). Indicated are the migration positions of myc Raf and the 32P-myc-Raf. Top, A, graphic representation of the phosphorimaging quantification of 32P in myc-Raf presented as PhosphorImager (PI) units. (C) COS-7 cells were transfected with myc-epitope tagged wild-type Raf (lanes 1-4), S259A Raf (lanes 5-7), or S259/621A Raf (lanes 8-10) together with either GST (lane 1), GST-14-3-3 (wt, lanes 2, 5, and 8), dimerization-deficient GST-14-3-3 (dm, lanes 3, 6, and 9), or carboxy-terminal GST-14-3-3 (ct, lanes 4, 7, and 10). Total cell extracts and GSH-bead-pulled-down proteins were resolved using 8.5% SDS-PAGE and immunoblotted for myc-Raf (bottom) or phospho-S259 Raf (top). The migration position of myc-Raf and phospho-S259-myc-Raf are indicated. The lower double band in the phospho-S259 Raf blots corresponds probably to the endogenous phospho-S259 Raf protein.
Similar results were obtained with DAF-16 (Figure 6). DAF-16 contains four putative AKT phosphorylation sites: T54, S240, T242, and S314. We recently showed that phosphorylation of DAF-16 with AKT induces 14-3-3 binding in vitro and that the PI-3 kinase pathway regulates 14-3-3-DAF-16 association in vivo (Cahill et al., 2001). This association is abolished by substituting the T54 site with Ala or by a combined substitution of the T54 site with the other AKT phosphorylation sites (Cahill et al., 2001; Figure 6B, compare lanes 1-3 with lanes 4-12). Binding of the dimerization-deficient 14-3-3 to DAF-16 is, however, independent of DAF-16 phosphorylation, as demonstrated by the ability of the dimerization-deficient 14-3-3 to bind with comparable efficiency wild-type and mutant DAF-16 forms lacking the AKT phosphorylation sites (Figure 6B, lanes 13-24). Because we were unable to detect any 32P incorporation in the DAF-16 3A mutant in vivo by using metabolic labeling experiments (our unpublished data), we infer that the dimerization-deficient 14-3-3 binding to DAF-16 is entirely phosphorylation independent. The role of the phosphorylation-dependent 14-3-3 association with DAF-16 is emphasized by the specific localization of phospho-DAF-16 in the cytoplasm, whereas only the nonphosphorylated DAF-16 is present in the nucleus (Figure 6C, compare lanes 3 and 4). This experiment, as mentioned above, also serves to demonstrate the specificity of the pan phospho-specific 14-3-3 binding site antibody to cytoplasmic, phospho-DAF-16 versus the nuclear, unphosphorylated DAF-16.
Dimerization Diminishes 14-3-3 Susceptibility to Undergo Phosphorylation
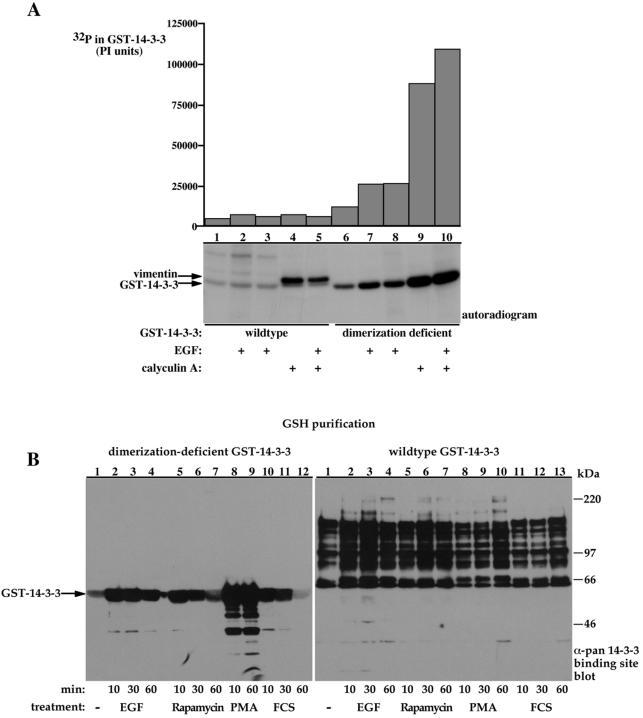
Several studies demonstrated phosphorylation of 14-3-3 at two sites, e.g., 14-3-3 ζ S58 and 14-3-3 ζ T233, and proposed a regulatory role of these phosphorylations in 14-3-3 dimerization and target binding, respectively (Megidish et al., 1995; Dubois et al., 1997a,b; Megidish et al., 1998). While conducting the metabolic 32P-labling experiments, we noticed that the dimerization-deficient 14-3-3 mutant contained significantly more 32P than the wild-type 14-3-3; this difference was further enhanced by treatment of cells with EGF or the phosphatase inhibitor calyculin A, reaching up to 50-fold higher 32P incorporation (Figure 7A, compare lanes 1 and 6, basal; lanes 2 and 7, EGF; and lanes 4 and 5 with 9 and 10, calyculin A). These results suggest that dimerization may mask 14-3-3 phosphorylation sites or change its conformation in a way that reduces its accessibility to kinases.
Figure 7.
Dimerization diminishes 14-3-3 susceptibility to phosphorylation. (A) Serum-deprived COS-7 cells expressing either wild-type GST-14-3-3 (lanes 1-5) or dimerization-deficient GST-14-3-3 (lanes 6-10) were metabolically labeled with 32P for 2 h and stimulated with 100 ng/ml EGF and/or 300 nM calyculin A for 15 min as indicated. GSH-bead-pulled-down proteins were resolved using 8.5% SDS-PAGE and analyzed using autoradiography (bottom) and phosphorimaging (PI units, top). The migration positions of GST-14-3-3 and coassociated cellular vimentin are indicated. (B) COS-7 cells expressing dimerization-deficient GST-14-3-3 (left) or wild-type GST-14-3-3 (right) were deprived of serum for 18 h and were either left untreated (lane 1) or treated with 100 ng/ml EGF (lanes 2-4), 100 nM rapamycin (lanes 5-8), 200 nM phorbol 12-myristate 13-acetate (lanes 8 and 9, left; lanes 8-10, right) or 10% fetal calf serum (lanes 10-12, left; lanes 11-13, right) for the indicated periods. GSH-bead-pulled-down proteins were resolved using 8.5% SDS-PAGE and immunoblotted with the pan phospho-specific 14-3-3 binding site antibody. Indicated are the migration positions of the molecular weight protein markers and the GST-14-3-3 protein.
In accordance with these observations, we also noticed that the dimerization-deficient 14-3-3 reacts with the pan phospho-specific 14-3-3 binding site antibody (Figures 4, bottom, and 7B, left). This reactivity was observed under basal conditions and was augmented by up to 10-fold by cell treatment with various factors and by calyculin A-induced phosphatase inhibition. No reactivity with wild-type 14-3-3 was detected with this antibody under similar conditions (Figures 2 and 4, bottom, and 7B, right). This figure also demonstrates the striking difference between wild-type and dimerization-deficient 14-3-3-associated proteins in terms of their reactivity with the pan phospho-specific 14-3-3 binding site antibody (Figure 7B, compare left, dimerization-deficient 14-3-3-associated proteins with right, wild-type 14-3-3-associated proteins).
S58 is a 14-3-3 ζ phosphorylation site that was identified after cell treatment with sphingosine by a putative sphingosine-dependent kinase, SDK1 (Megidish et al., 1998). This site is also a putative protein kinase C site and the surrounding sequence, i.e., RRSpSWR resembles the sequence of the peptide used to generate the pan phospho-specific 14-3-3 binding site antibody. The reactivity of the pan phospho-specific antibody with the dimerization-deficient 14-3-3 and the ability of PMA to induce the highest increase in this reactivity, may indicate that phosphorylation of S58 could contribute to the increased reactivity with the pan phospho-specific antibody. Because S58 lies within the 14-3-3 dimerization domain, a probable explanation to the inability of phorbol 12-myristate 13-acetate and calyculin A to induce phosphorylation of wild-type 14-3-3 is that this site may be sequestered within the dimerization interface. Whether phosphorylation of Ser 58 is responsible for the increase in phosphate content of the dimerization-deficient 14-3-3 and increased reactivity with the pan phospho-specific antibody and whether it may serve as a physiological mechanism for regulating endogenous 14-3-3 dimer formation will require further study.
DISCUSSION
14-3-3 proteins are key regulatory proteins involved in cellular processes such as cell cycle control, intracellular signaling, and cellular response to stress and survival factors. The dimeric nature of the 14-3-3 proteins was established soon with their discovery through biochemical means (Boston et al., 1982) and confirmed by crystallographic analysis (Liu et al., 1995; Xiao et al., 1995). The structure of the 14-3-3 protein crystal showed the existence of a binding groove in each 14-3-3 half-dimer and cocrystallization of 14-3-3 with short synthetic phosphopeptides showed the ability of each part of the dimer to bind a phosphopeptide independently (Yaffe et al., 1997; Rittinger et al., 1999). These findings suggested that the ability of 14-3-3 to bind its target proteins should be independent of 14-3-3 dimerization. This suggestion was supported by several reports demonstrating that dimerization-deficient 14-3-3 forms can bind target proteins at same efficiency as wild-type 14-3-3 (Ichimura et al., 1995; Luo et al., 1995; Tzivion et al., 1998). The present results, demonstrating that 14-3-3 dimerization is important for the phosphorylation-dependent binding of cellular proteins, necessitate a revision of this previous conclusion. Even though 14-3-3 binding to short synthetic phosphopeptides containing a single phosphoserine, at saturating concentrations, seems to be independent of 14-3-3 dimerization, the present results clearly demonstrate that the phosphorylation-dependent binding of cellular proteins in vivo is facilitated by 14-3-3 dimerization. The finding by Yaffe et al. (1997) that the presence of two, tandem 14-3-3 binding sites on a single peptide increases 14-3-3 binding by 30-fold indicates that targets having two 14-3-3 binding sites will bind with substantially higher affinity to dimeric 14-3-3 than to monomeric 14-3-3. This fact, however, cannot explain the present finding that monomeric 14-3-3 is greatly impaired in its ability to bind cellular phosphoproteins, including those containing the high-affinity, mode-1 motif RSxpSxP. Thus, although monomeric 14-3-3 forms bind some phosphorylated proteins through the “phosphopeptide” binding pocket, the phosphorylation of these partners is evidently nonessential for binding to monomeric 14-3-3. It is important to note that although we deliberately mutated regions in the dimerization interface that do not coincide with the phospho-binding pocket, the possibility exists that these mutations indirectly affect the phospho-binding pocket structure. The significance of this caveat is, however, lessened by the fact that other deletions that abrogate dimerization also show similar results as the dimerization-deficient mutant (Figures 3 and 5; our unpublished data).
A recent study involving cocrystallization of 14-3-3 with the enzyme serotonin N-acetyltransferase (AANAT) provides a more comprehensive understanding of the 14-3-3-target structure (Obsil et al., 2001). As the only existing example of a structure of 14-3-3 bound to a target protein, this study provides a somewhat different picture than that derived from studies with short peptides; full-length AANAT, which contains two potential 14-3-3 binding sites (e.g., T31 and S205), forms exclusively 1:2 stoichiometric complexes with 14-3-3 in vitro, suggesting that a dimeric 14-3-3 binds to a single AANAT polypeptide at two sites. Because the authors were unable to obtain crystals from this complex, they used an AANAT mutant lacking the carboxy-terminal 14-3-3 binding site, which formed 2:2 stoichiometric complexes with 14-3-3 in addition to the 1:2 complexes; however, only the 2:2 complex was successfully crystallized. This structure shows a 14-3-3 dimer binding two AANAT molecules (one by each half-dimer). Importantly, these results demonstrated that in addition to interactions with the phospho-residue-containing motif, 14-3-3 interacts with AANAT at other sites distant from the phosphorylated serine. A computer-based extrapolation of the 2:2 structure supported, however, the existence of a structure where a dimeric 14-3-3 binds a single AANAT molecule at two sites. Interestingly, the computer extrapolated structure points on an ability of the dimeric 14-3-3 to induce a conformational change of AANAT. This hypothesis was supported by enzymatic studies demonstrating the ability of a dimeric 14-3-3 to increase AANAT enzymatic activity by binding it at two sites. It will be of interest to determine whether the dimerization-deficient 14-3-3 can bind to AANAT, and if so, whether it can alter AANAT enzymatic activity.
The ability of 14-3-3 to bind nonphosphorylated peptides and proteins has been demonstrated by several groups; however, the physiological relevance of this potential has not been determined (Petosa et al., 1998; Masters et al., 1999; Wang et al., 1999). Our finding showing that the in vivo 14-3-3-associated proteins in COS-7 cells bind 14-3-3 through the phosphopeptide binding groove, as demonstrated by the ability of a 14-3-3 binding phosphopeptide to displace all specifically associated proteins, combined with the finding that most of these associated proteins are reactive with the pan phospho-specific 14-3-3 binding site antibody, raises questions about the physiological significance of 14-3-3 binding to nonphosphorylated targets. Nevertheless, it is likely that once bound to the phosphoserine-containing motif through its phosphopeptide binding pocket, the ability of other 14-3-3 residues and/or the second half of the dimer to interact with nonphosphorylated epitopes on the target surface may be significant for the physiological action of the 14-3-3 protein. Furthermore, the assembly of the 14-3-3 dimer may interfere with these secondary, phosphorylation-independent interactions unless their occurrence is facilitated by the presence of a high-affinity phosphopeptide-dependent binding. This may explain why the native 14-3-3 dimer cannot bind to the Raf S259/621A mutant, whereas this mutant exhibits considerable binding in vivo to monomeric 14-3-3. Impairment of the ability of 14-3-3 to form phosphopeptide-dependent interactions by interdicting 14-3-3 dimerization, may allow us to uncover the occurrence of these lower affinity, phosphorylation-independent interactions.
Few mechanisms underlying the regulation of 14-3-3 function have been proposed (Tzivion et al., 2001; Tzivion and Avruch, 2002). Among these, regulation of 14-3-3 function by its phosphorylation has attracted the most attention (Dubois et al., 1997a) and has been proposed to control both target binding and dimerization (Dubois et al., 1997b; Megidish et al., 1998). The enormous difference in the susceptibility of dimeric versus monomeric 14-3-3 to phosphorylation (Figure 7) indicates that regulation of 14-3-3 by phosphorylation probably has to occur at the monomeric state rather than after dimer assembly. Further examination of this mechanism will require assessment of the ratio of dimeric and monomeric 14-3-3 polypeptides under physiological conditions and in response to various treatments and their relative susceptibility to phosphorylation.
The significance of 14-3-3 dimerization is further magnified by the existence of at least seven isoforms in mammals capable of heterodimerization. These isoforms are encoded by seven separate genes, each displaying somewhat different target binding specificity and susceptibility to phosphorylation (Megidish et al., 1998). Thus, the differential tissue and development-dependent expression of the various 14-3-3 genes and the considerable tendency toward heterodimerization in addition to homodimerization suggest that small differences in binding specificity among isoforms, when combined with heterodimerization and differential regulation at the level of expression and phosphorylation of individual 14-3-3 isoforms, can combine to generate a robust regulatory apparatus.
Acknowledgments
This work was supported in part by National Institute of Health grant R01 GM-067134 (to G.T.).
References
- Borch, J., Bych, K., Roepstorff, P., Palmgren, M.G., and Fuglsang, A.T. (2002). Phosphorylation-independent interaction between 14-3-3 protein and the plant plasma membrane H+-ATPase. Biochem. Soc. Trans. 30, 411-415. [DOI] [PubMed] [Google Scholar]
- Boston, P.F., Jackson, P., Kynoch, P.A., and Thompson, R.J. (1982). Purification, properties, and immunohistochemical localisation of human brain 14-3-3 protein. J. Neurochem. 38, 1466-1474. [DOI] [PubMed] [Google Scholar]
- Boyle, W.J., van der Geer, P., and Hunter, T. (1991). Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201, 110-149. [DOI] [PubMed] [Google Scholar]
- Cahill, C.M., Tzivion, G., Nasrin, N., Ogg, S., Dore, J., Ruvkun, G., and Alexander-Bridges, M. (2001). Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J. Biol. Chem. 276, 13402-13410. [DOI] [PubMed] [Google Scholar]
- Craparo, A., Freund, R., and Gustafson, T.A. (1997). 14-3-3 (epsilon) interacts with the insulin-like growth factor I receptor and insulin receptor substrate I in a phosphoserine-dependent manner. J. Biol. Chem. 272, 11663-11669. [DOI] [PubMed] [Google Scholar]
- Dubois, T., et al. (1997a). Structure and sites of phosphorylation of 14-3-3 protein: role in coordinating signal transduction pathways. J Protein Chem. 16, 513-522. [DOI] [PubMed] [Google Scholar]
- Dubois, T., Rommel, C., Howell, S., Steinhussen, U., Soneji, Y., Morrice, N., Moelling, K., and Aitken, A. (1997b). 14-3-3 is phosphorylated by casein kinase I on residue 233. Phosphorylation at this site in vivo regulates Raf/14-3-3 interaction. J. Biol. Chem. 272, 28882-28888. [DOI] [PubMed] [Google Scholar]
- Fu, H., Subramanian, R.R., and Masters, S.C. (2000). 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40, 617-647. [DOI] [PubMed] [Google Scholar]
- Gu, M., and Du, X. (1998). A novel ligand-binding site in the zeta-form 14-3-3 protein recognizing the platelet glycoprotein Ibalpha and distinct from the c-Raf-binding site. J. Biol. Chem. 273, 33465-33471. [DOI] [PubMed] [Google Scholar]
- Hallberg, B. (2002). Exoenzyme S binds its cofactor 14-3-3 through a non-phosphorylated motif. Biochem. Soc. Trans. 30, 401-405. [DOI] [PubMed] [Google Scholar]
- Honda, R., Ohba, Y., and Yasuda, H. (1997). 14-3-3 zeta protein binds to the carboxyl half of mouse wee1 kinase. Biochem. Biophys. Res. Commun. 230, 262-265. [DOI] [PubMed] [Google Scholar]
- Ichimura, T., Ito, M., Itagaki, C., Takahashi, M., Horigome, T., Omata, S., Ohno, S., and Isobe, T. (1997). The 14-3-3 protein binds its target proteins with a common site located towards the C-terminus. FEBS Lett. 413, 273-276. [DOI] [PubMed] [Google Scholar]
- Ichimura, T., Uchiyama, J., Kunihiro, O., Ito, M., Horigome, T., Omata, S., Shinkai, F., Kaji, H., and Isobe, T. (1995). Identification of the site of interaction of the 14-3-3 protein with phosphorylated tryptophan hydroxylase. J. Biol. Chem. 270, 28515-28518. [DOI] [PubMed] [Google Scholar]
- Ku, N.O., Liao, J., and Omary, M.B. (1998). Phosphorylation of human keratin 18 serine 33 regulates binding to 14-3-3 proteins. EMBO J. 17, 1892-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., Bienkowska, J., Petosa, C., Collier, R.J., Fu, H., and Liddington, R. (1995). Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376, 191-194. [DOI] [PubMed] [Google Scholar]
- Liu, Y.C., Liu, Y., Elly, C., Yoshida, H., Lipkowitz, S., and Altman, A. (1997). Serine phosphorylation of Cbl induced by phorbol ester enhances its association with 14-3-3 proteins in T cells via a novel serine-rich 14-3-3-binding motif. J. Biol. Chem. 272, 9979-9985. [DOI] [PubMed] [Google Scholar]
- Luo, K.X., Hurley, T.R., and Sefton, B.M. (1991). Cyanogen bromide cleavage and proteolytic peptide mapping of proteins immobilized to membranes. Methods Enzymol. 201, 149-152. [DOI] [PubMed] [Google Scholar]
- Luo, Z.J., Zhang, X.F., Rapp, U., and Avruch, J. (1995). Identification of the 14.3.3 zeta domains important for self-association and Raf binding. J. Biol. Chem. 270, 23681-23687. [DOI] [PubMed] [Google Scholar]
- Masters, S.C., Pederson, K.J., Zhang, L., Barbieri, J.T., and Fu, H. (1999). Interaction of 14-3-3 with a nonphosphorylated protein ligand, exoenzyme S of Pseudomonas aeruginosa. Biochemistry 38, 5216-5221. [DOI] [PubMed] [Google Scholar]
- Megidish, T., Cooper, J., Zhang, L., Fu, H., and Hakomori, S. (1998). A novel sphingosine-dependent protein kinase (SDK1) specifically phosphorylates certain isoforms of 14-3-3 protein. J. Biol. Chem. 273, 21834-21845. [DOI] [PubMed] [Google Scholar]
- Megidish, T., White, T., Takio, K., Titani, K., Igarashi, Y., and Hakomori, S. (1995). The signal modulator protein 14-3-3 is a target of sphingosine- or N,N-dimethylsphingosine-dependent kinase in 3T3(A31) cells. Biochem. Biophys. Res. Commun. 216, 739-747. [DOI] [PubMed] [Google Scholar]
- Mils, V., Baldin, V., Goubin, F., Pinta, I., Papin, C., Waye, M., Eychene, A., and Ducommun, B. (2000). Specific interaction between 14-3-3 isoforms and the human CDC25B phosphatase. Oncogene 19, 1257-1265. [DOI] [PubMed] [Google Scholar]
- Moore, B.E., and Perez, V.J. (1967). Specific acidic proteins of the nervous system. In: Physiological and Biochemical Aspects of Nervous Integration, ed. F.D. Carlson, Englewood Cliffs, NJ: Prentice Hall, 343-359.
- Morrison, D.K., Heidecker, G., Rapp, U.R., and Copeland, T.D. (1993). Identification of the major phosphorylation sites of the Raf-1 kinase. J. Biol. Chem. 268, 17309-17316. [PubMed] [Google Scholar]
- Muslin, A.J., Tanner, J.W., Allen, P.M., and Shaw, A.S. (1996). Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889-897. [DOI] [PubMed] [Google Scholar]
- Muslin, A.J., and Xing, H. (2000). 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 12, 703-709. [DOI] [PubMed] [Google Scholar]
- Obsil, T., Ghirlando, R., Klein, D.C., Ganguly, S., and Dyda, F. (2001). Crystal structure of the 14-3-3zeta:serotonin n-acetyltransferase complex. A role for scaffolding in enzyme regulation. Cell 105, 257-267. [DOI] [PubMed] [Google Scholar]
- Ogihara, T., et al. (1997). 14-3-3 protein binds to insulin receptor substrate-1, one of the binding sites of which is in the phosphotyrosine binding domain. J. Biol. Chem. 272, 25267-25274. [DOI] [PubMed] [Google Scholar]
- Petosa, C., Masters, S.C., Bankston, L.A., Pohl, J., Wang, B., Fu, H., and Liddington, R.C. (1998). 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J. Biol. Chem. 273, 16305-16310. [DOI] [PubMed] [Google Scholar]
- Rittinger, K., Budman, J., Xu, J., Volinia, S., Cantley, L.C., Smerdon, S.J., Gamblin, S.J., and Yaffe, M.B. (1999). Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 4, 153-166. [DOI] [PubMed] [Google Scholar]
- Skoulakis, E.M., and Davis, R.L. (1998). 14-3-3 proteins in neuronal development and function. Mol. Neurobiol. 16, 269-284. [DOI] [PubMed] [Google Scholar]
- Sprenkle, A.B., Davies, S.P., Carling, D., Hardie, D.G., and Sturgill, T.W. (1997). Identification of Raf-1 Ser621 kinase activity from NIH 3T3 cells as AMP-activated protein kinase. FEBS Lett. 403, 254-258. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., and Avruch, J. (2002). 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277, 3061-3064. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., Luo, Z., and Avruch, J. (1998). A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature 394, 88-92. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., Luo, Z.J., and Avruch, J. (2000). Calyculin A-induced vimentin phosphorylation sequesters 14-3-3 and displaces other 14-3-3 partners in vivo. J. Biol. Chem. 275, 29772-29778. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., Shen, Y.H., and Zhu, J. (2001). 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene 20, 6331-6338. [DOI] [PubMed] [Google Scholar]
- van Hemert, M.J., van Heusden, G.P., and Steensma, H.Y. (2001). Yeast 14-3-3 proteins. Yeast 18, 889-895. [DOI] [PubMed] [Google Scholar]
- Wang, B., Yang, H., Liu, Y.C., Jelinek, T., Zhang, L., Ruoslahti, E., and Fu, H. (1999). Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38, 12499-12504. [DOI] [PubMed] [Google Scholar]
- Wang, H., Zhang, L., Liddington, R., and Fu, H. (1998). Mutations in the hydrophobic surface of an amphipathic groove of 14-3-3zeta disrupt its interaction with Raf-1 kinase. J. Biol. Chem. 273, 16297-16304. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Jacobs, C., Hook, K.E., Duan, H., Booher, R.N., and Sun, Y. (2000). Binding of 14-3-3beta to the carboxyl terminus of Wee1 increases Wee1 stability, kinase activity, and G2-M cell population. Cell Growth Differ. 11, 211-219. [PubMed] [Google Scholar]
- Xiao, B., Smerdon, S.J., Jones, D.H., Dodson, G.G., Soneji, Y., Aitken, A., and Gamblin, S.J. (1995). Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376, 188-191. [DOI] [PubMed] [Google Scholar]
- Xing, H., Zhang, S., Weinheimer, C., Kovacs, A., and Muslin, A.J. (2000). 14-3-3 proteins block apoptosis and differentially regulate MAPK cascades. EMBO J. 19, 349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M.B., Rittinger, K., Volinia, S., Caron, P.R., Aitken, A., Leffers, H., Gamblin, S.J., Smerdon, S.J., and Cantley, L.C. (1997). The structural basis for 14-3-3, phosphopeptide binding specificity. Cell 91, 961-971. [DOI] [PubMed] [Google Scholar]
- Zhai, J., Lin, H., Shamim, M., Schlaepfer, W.W., and Canete-Soler, R. (2001). Identification of a novel interaction of 14-3-3 with p190RhoGEF. J. Biol. Chem. 276, 41318-41324. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Zha, X., Tan, Y., Hornbeck, P.V., Mastrangelo, A.J., Alessi, D.R., Polakiewicz, R.D., and Comb, M.J. (2002). Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J. Biol. Chem. 277, 39379-39387. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Wang, H., Liu, D., Liddington, R., and Fu, H. (1997). Raf-1 kinase and exoenzyme S interact with 14-3-3zeta through a common site involving lysine 49. J. Biol. Chem. 272, 13717-13724. [DOI] [PubMed] [Google Scholar]