Abstract
The hydrolysis of colominic acid via microwave irradiation was studied for the production of short chain oligomers with a degree of polymerization (DP) of 1–6. This method was compared to the traditional acid hydrolytic method for the production of preparative quantities of short colominic acid oligomers. The oligomers were purified by size exclusion chromatography and characterized by 1H NMR. Optimal conditions for producing the dimer were found to be 12 minutes at 10% power in a 1000 Watt domestic microwave. This method is advantageous over the traditional technique in that the hydrolysis can be completed in just a few minutes, rather than hours, it is reproducible, and yields large quantities of the desirable short chain oligomers of colominic acid.
Keywords: Colominic acid, Polysialic acid, Acid hydrolysis, Microwave irradiation
Colominic acid is a capsular bacterial polysaccharide that was originally isolated from the culture filtrate of Escherichia coli K-235 by Barry and Goebel.1 It is released from the cell wall surface into the culture medium by the acidic conditions that occur during the late growth phase of the bacteria.2 These oligo-polymers consist of polydisperse linear chains of sialic acid linked α2,8 (Scheme 1).3–5 The chain length of the colominic acid polymer is reported to range from a degree of polymerization (DP) of 60–80 to up to approximately 100.6,7 Colominic acid is known more generally as polysialic acid (polySia) and is present on the surface of other neuroinvasive bacteria such as Neisseria meningitidis Group B.8 Structurally and immunologically identical polySia chains are found in the neural tissues of vertebrates covalently attached to N-CAM (neural cell adhesion molecules). Due to the structural identity between the neuropathogenic bacterial polySia capsules and the α2,8-linked polySia on mammalian N-CAM, polySia is non-immunogenic, and acts as a cloaking device for neurotropic bacteria to evade immune detection and facilitate meningeal infections.8 These bacteria are readily spread from person-to-person, and thus, can lead to epidemics of bacterial meningitis that can be debilitating, and fatal, when not diagnosed and treated early. Meningitis caused by the group B meningococci is especially dangerous for infants and young children, and there is currently no vaccine to prevent it.8
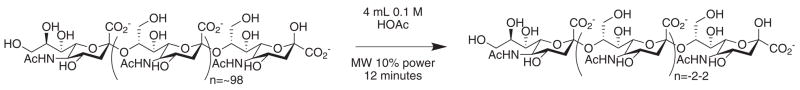
Scheme 1.
Microwave acid hydrolysis of colominic acid.
In mammals, polySia is found attached to N-CAM extending tri- and tetraantennary N-linked glycan chains.9 It lends a high negative charge density and hydrated volume to the cell surface. This can lead to diminution in cell-cell and cell-matrix interactions.9,10 The extended polySia chains on N-CAM are expressed primarily during embryonic development, and give plasticity to the cells, allowing for cell migration, axonal growth and synaptogenesis.10 PolySia, therefore, serves as an anti-adhesive glycotope during development.9 PolySia chain lengths peak during development, then get shorter as the organism progresses towards adulthood. 8,10,11 Some regions of the brain, including the hippocampus, hypothalamus, dentate gyrus and olfactory bulb express the embryonic, or extended polySia chains into adulthood, thus allowing for plasticity in these regions of the brain throughout life. This long-term neural plasticity is associated with neuronal functions including neural cell migration and generation, neurite outgrowth, cytokine response and reorganization of neural connections.8,10 For example, in mice, the loss of polysialylated N-CAM is associated with developmental and behavioral defects, and impaired learning.12 Alteration of polySia expression on N-CAM is also important in the progression of certain diseases, including many human cancers principally of neuroendocrine origin, schizophrenia and Alzheimer’s disease.9,13,11
PolySia has the potential of being used in the preparation of therapeutic agents. Due to its low toxicity and lack of immunogenicity, it may be possible to extend peptide drug half lives through conjugation of the peptide with polySia. PolySia may also serve to minimize immune response to these drugs.14,15 Because of the biological importance of polySia, there is potential for this molecule to be used in the development of synthetic and semi-synthetic therapeutics. These applications may necessitate the generation of preparative quantities of short chain oligomers in pure form. Colominic acid oligomers purchased commercially can be prohibitively expensive (25 mg of hexamer for appx. $1200)7 in the quantities needed for synthetic studies. Therefore, by starting with the commercially available form of polySia, colominic acid, preparative quantities of the necessary oligomers can be achieved at much less cost.
Short oligomers of colominic acid can be generated using traditional acid hydrolysis methods. Roy and Pon reported the use of an unbuffered acid hydrolysis using standard heating and various pH and temperature conditions to generate distributions of oligomers from colominic acid.16 Their reaction time required 30 minutes to four hours to run and was complicated by the unbuffered reaction solution because a constant pH had to be maintained over the course of the reaction. Later, Cheng and coworkers reported the use of acid hydrolysis via microwave irradiation to produce qualitative amounts (10–50 μg) of a wide range of oligomers with DPs ranging from monomer through unhydrolyzed products (DPs >200). These DPs were determined using MECC (micellar electrokinetic capillary chromatography).6
In our present study, we sought to generate quantitative amounts of a narrow DP distribution of colominic acid oligomers for use in the synthetic preparation of potential therapeutic agents. We initially targeted the dimer chain length to work with synthetically. Our experience with the traditional acid hydrolysis methods showed that it was time-consuming, difficult to maintain the proper conditions, and highly variable from batch to batch with respect to the amount of each DP that was generated (Table 1). Additionally, there was always a portion of high molecular weight sialyl oligomers that was not resolved by our preparative size exclusion chromatography column. This material was unusable for our synthetic studies, and thus resulted in an overall loss of expensive starting material (approximately $200 per gram).7 It was thought that the microwave hydrolytic method of Cheng and coworkers could be used to develop an efficient, reproducible, high yielding microwave hydrolytic method, whereby a narrow DP range of sialyl oligomers could be obtained in large quantities.
Table 1.
Summary of traditional and microwave acid hydrolysis results. Each number is reported as a percentage of the total product recovered. The column entry listed as unhydrolyzed refers to mixtures of chain lengths greater than hexamer that were not resolved by the size exclusion column.
| Trial | % Monomer | % Dimer | % Trimer | % Tetramer | % Pentamer | % Hexamer | % Unhydrolyzed |
|---|---|---|---|---|---|---|---|
|
Traditional | |||||||
| Trial 1 | 17.1 | 24.1 | 9.2 | 0.0 | 49.6 | 0 | 0 |
| Trial 2 | 20.8 | 37.2 | 13.9 | 12.0 | 14.3 | 5 | 10.6 |
| Trial 3 | 25.6 | 30.5 | 12.4 | 28.1 | 0 | 0 | 3.41 |
| Std. Dev. | 4.24 | 6.55 | 2.40 | 14.10 | 25.50 | 2.89 | 5.41 |
|
Microwave | |||||||
|
6 Minutes | |||||||
| Trial 1 | 9.3 | 29.6 | 12.3 | 10.1 | 38.7 | 0 | 0 |
| Trial 2 | 17.1 | 41.1 | 21.8 | 7.1 | 12.9 | 0 | 0 |
| Std. Dev. | 5.54 | 8.15 | 6.71 | 2.16 | 18.20 | 0 | 0 |
|
8 Minutes | |||||||
| Trial 1 | 19.1 | 43.6 | 13.6 | 23.8 | 0 | 0 | 0 |
| Trial 2 | 6.9 | 44.94 | 24.5 | 17.8 | 6.7 | 0 | 0 |
| Trial 3 | 10 | 30.61 | 25.3 | 34.1 | 0 | 0 | 0 |
| Std. Dev. | 6.35 | 7.91 | 6.54 | 8.25 | 3.89 | 0 | 0 |
|
9 Minutes | |||||||
| Trial 1 | 17.7 | 47.9 | 17.7 | 15.9 | 0.9 | 0 | 0 |
| Trial 2 | 13.7 | 37 | 18.9 | 15.3 | 15.1 | 0 | 0 |
| Std. Dev. | 2.78 | 7.69 | 0.89 | 0.46 | 10.00 | 0 | 0 |
|
10 Minutes | |||||||
| Trial 1 | 17.8 | 46.1 | 16.2 | 19.9 | 0 | 0 | 0 |
| Trial 2 | 18.1 | 39.5 | 12.5 | 29.9 | 0 | 0 | 0 |
| Std. Dev. | 0.15 | 4.64 | 3.19 | 7.06 | 0 | 0 | 0 |
|
12 Minutes | |||||||
| Trial 1 | 17.0 | 50.4 | 17.3 | 15.4 | 0 | 0 | 0 |
| Trial 2 | 22.1 | 50.4 | 12.7 | 14.8 | 0 | 0 | 0 |
| Std. Dev. | 3.63 | 0.00 | 3.19 | 0.4 | 0 | 0 | 0 |
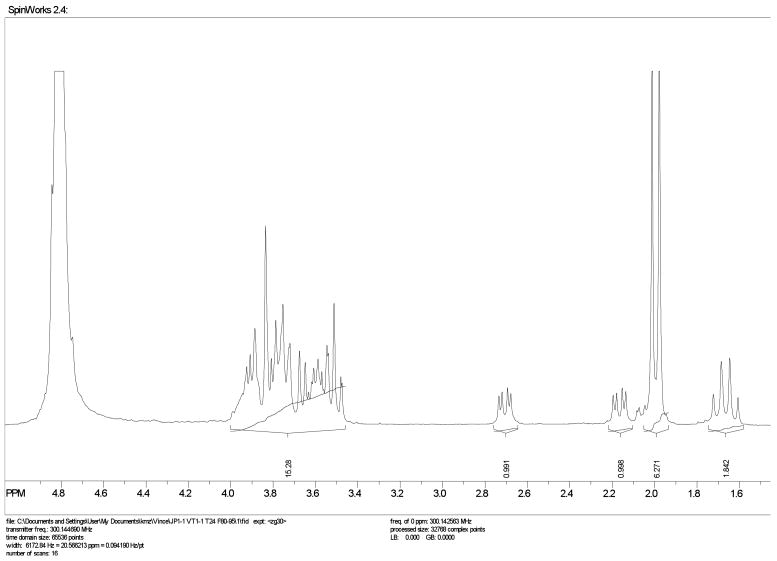
This method was simple to implement. Using a domestic1000 W microwave oven and simple reaction glassware consisting of an Erlenmeyer flask, several conditions were tested for the acetic acid-catalyzed reaction (Scheme 1, Table 1). The reaction was neutralized with sodium hydroxide, lyophilized and separated by preparative size exclusion chromatography. As all of the sialyl oligomers are known in the literature, confirmation of purity and oligomer length was conducted by proton NMR. In the NMR analysis, the integration of the protons corresponding to the methyl group of the N-acetate were compared to the non-reducing H3e protons. Additionally, the total proton count was used as a measure of the oligomer purity (Fig. 1).
Figure 1.
1H NMR of dimeric colominic acid in D2O.
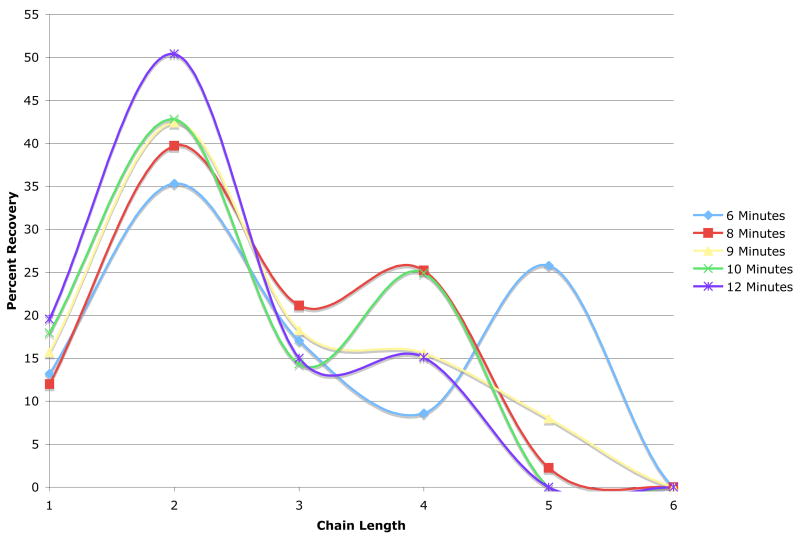
Using these methods, it was determined that higher power settings such as 20–30% did not work well, as the small volume of reaction solution boiled too quickly, and in some instances, either boiled over, resulting in a loss of material, or boiled dry during the irradiation. Because of these difficulties, the reaction would have to be stopped frequently, and allowed to cool before the reaction could commence, or additional solution would have to be added. This was not desirable, as it led to inconsistencies in the product distribution of oligomers and quantities produced. For example, a 6 minute reaction time at 20% power yielded monomer through pentamer for both trials, but in disparate trial to trial quantities (Table 1). The remainder of the trials were conducted at a lower power setting of 10%, and different reaction times were evaluated. Trials conducted at 6 minutes and 20% power, and 8 and 9 minutes at 10% power resulted in oligomer lengths of monomer through pentamer, but still suffered from some variability in quantities of each from trial to trial. The trials at 10 and 12 minutes gave the most consistent results with respect to low standard deviations, and also had the added benefit of generating a smaller oligomeric distribution, monomer through tetramer, with very high amounts of dimer produced. Optimal conditions were found to be 12 minutes at 10% power, based on the narrow product quantity distribution (as evidenced by low standard deviations), the highest amount of dimer produced (over 50% of recovered product), and a small DP range (monomer through tetramer) (Table 1). The superiority of these conditions compared to other microwave conditions tested can be further visualized in Figure 2.
Figure 2. Optimization of Microwave Hydrolysis Conditions for Generating Different DPs of PolySia.
Graph shows the percent recovery of DPs 1–5 as a function of time at 20% power (6 min) and 8,9,10, and 12 minutes at 10% power. Results are representative of the average of two to three trials (Refer to Table 1).
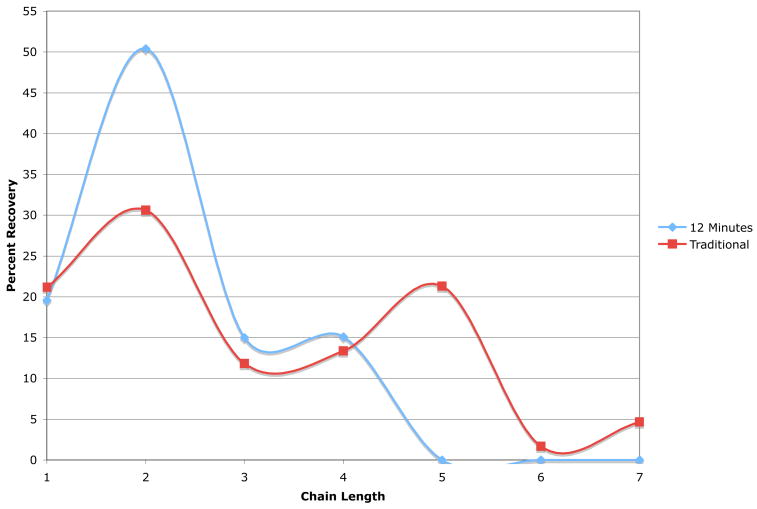
Comparison of the microwave conditions of 12 minutes at 10% power versus the traditional acid hydrolysis method by Roy and Pon showed significant differences in the product distribution and quantities.16 The optimized microwave conditions yielded approximately 20% more of dimer than the traditional method, and did not give any pentamer or hexamer chain lengths (Fig. 3). Additionally, no larger DPs were evident in the microwave results, whereas small quantities of extended DPs of unhydrolyzed colominic acid were produced in the traditional hydrolytic technique. Comparing our method to that of Cheng and coworkers, it is interesting to note that our method resulted in a much smaller DP distribution of products in a shorter microwave reaction time.6 In some of the conditions tested by Cheng et al., it was noted that either inter-residue lactonization of the C-9 of the non-reducing residue to the reducing end carboxylic acid, or incomplete hydrolysis of the polySia chains occurred at the shorter reaction times (1–3 minutes).6 Lactonized products were not isolated by our procedure, likely due to the neutralization of the microwave reaction via sodium hydroxide addition. Another advantage of our method is that it was not necessary to monitor the pH of the microwave reaction due to the very short reaction times. This further simplifies microwave hydrolysis and makes the procedure amenable to successful handling by relatively inexperienced student researchers in the laboratory.
Figure 3. Comparison of the Traditional and Microwave Hydrolysis Methods for Generating Sialyl Oligomers with DPs 1–7.
Graph shows the DP and quantity of each chain length of colominic acid produced in 12 minutes at 10% power in the microwave method versus 3 hours at 70°C at pH = 3.0 by traditional acid hydrolytic method. Results are representative of the average of two to three trials (Refer to Table 1).
In summary, we have developed a simple, reproducible, and highly efficient microwave acid hydrolysis method for the generation of pure, short DPs of colominic acid oligomers in large isolable quantities. Comparison of the microwave technique to the traditional acid hydrolysis method showed that the microwave technique was more complete, simple and effective. Under optimal microwave conditions, no unhydrolyzed colominic acid was isolated, and the pH did not have to be monitored over several hours time. The optimum microwave conditions (12 minutes at 10% power) gave consistent product DP distribution (DP 1–4) and quantities as compared to the traditional acid hydrolysis method. When comparing our results with those of Cheng and coworkers, we noted a more complete hydrolysis in shorter times and with lower power settings, and again, a much smaller DP distribution of products in quantitative amounts.6 This method can be modified as necessary to yield whatever product distribution is desired, and is simple to implement. The potential cost savings achieved through starting with the large DP colominic acid as opposed to purchasing commercially available pure oligomers is significant, especially given the short turnaround time from start to finish of the reaction, purification and characterizations (3–4 days total).
1. Experimental
1.1 General
Colominic acid (approximate DP of 100) was purchased from Nacalai Tesque and was used without further purification. All microwave hydrolyses were carried out in a domestic microwave oven (Emerson, 1000 Watt, model number MW8111SS). 1H NMRs were conducted in D2O (Acros), at 300 MHz on a Bruker AVANCE NMR. The spectra were processed using Spinworks 2.4.
1.2 Microwave hydrolysis of colominic acid
Approximately 100 mg of colominic acid, was added to a 50 mL Erlenmeyer flask. The colominic acid was dissolved into 4 mL of 0.1 M acetic acid using a graduated pipet. The flask was then covered with plastic wrap and a rubber band, and several holes were added for ventilation. Microwave hydrolysis was carried out in a 1000 Watt conventional Emerson microwave oven, with the flask placed in an alumina bath. The reaction was run at either 10 or 20% power, with heating periods of 6, 8, 9, 10, or 12 minutes. The sample was neutralized to a pH of 7.0 using 0.1 M NaOH and freeze dried. The resultant white solid was weighed to determine an overall percent recovery.
1.3 Purification and characterization of colominic acid oligomers
The oligomeric mixtures generated by either the microwave or acid hydrolysis methods were dissolved in less than 2.0 mL of 0.03 M ammonium bicarbonate buffer, centrifuged, and injected into a Pharmacia Liquid Chromatography FPLC using a 5 mL syringe. A Biogel P-10 size exclusion column, 2.5 × 120 cm, was used to separate the oligomers. The column was run at a flow rate of 0.25 mL per minute, with 125 total fractions collected at a volume 3.5 mL per fraction. The fractions were pooled and freeze dried as individual peaks. Dialysis against pure distilled water was carried out using Spectrum Cellulose Ester 100 MWCO tubing and performed when necessary to remove excess salt impurities from the pure oligomers. 1H NMR analyses were conducted in D2O to confirm the chain lengths. The DP was confirmed by comparison of the integration of the N-acetyl methyl peak with the integration of the H3e non-reducing signal as well as the overall proton count.
1.4 Traditional acid hydrolysis of colominic acid
The traditional acid hydrolysis of colominic acid was carried out according to the procedure reported by Roy and Pon.16 Approximately 100 mg of colominic acid was added to a 50 mL round bottom flask and dissolved into 10 ml of deionized water and the pH was adjusted to approximately 3.0 using 1M hydrochloric acid. The hydrolysis was carried out at 70°C in an oil bath for 3 hours. Recordings of the temperature and the pH were taken every fifteen minutes and the pH was adjusted accordingly with acid and/or base to keep the pH constant at ~3.0. The sample was neutralized to pH 7.0 using 0.1 M sodium hydroxide then freeze dried. The resultant white solid was weighed to determine an overall percent recovery. Purification of the oligomeric mixture was performed as described above.
Acknowledgments
The authors appreciate the support of the California State University Department of Chemistry and funding received through a Research Corporation Cottrell College Science Award, NIH-NIAID AREA grant, and a CSUPERB (California State University Program for Education and Research in Biotechnology) Entrepreneurial Joint Venture Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barry GT, Goebel WF. Nature. 1957;179:206–206. doi: 10.1038/179206a0. [DOI] [PubMed] [Google Scholar]
- 2.Troy FAI, McCloskey MA. J Biol Chem. 1979;254:7377–7387. [PubMed] [Google Scholar]
- 3.Barry GT. J Exp Med. 1958;107:507–521. doi: 10.1084/jem.107.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGuire EJ, Binkley SB. Biochemistry. 1964;3:247–251. doi: 10.1021/bi00890a017. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Aparicio LB, Reglero A, Ortiz AIJML. Appl Microbiol Biotechnol. 1988;27:474–483. [Google Scholar]
- 6.Cheng MC, Wang KT, Inoue S, Inoue Y, Khoo KH, Wu SH. Anal Biochem. 1999;267:287–293. doi: 10.1006/abio.1998.2988. [DOI] [PubMed] [Google Scholar]
- 7.Nacalai USA Inc. [Accessed 01/06/09];2008 http://www.nacalaiusa.com/product.php?id=33.
- 8.Stein DM, Robbins J, Miller MA, Lin F-YC, Schneerson R. Vaccine. 2006;24:221–228. doi: 10.1016/j.vaccine.2005.07.084. [DOI] [PubMed] [Google Scholar]
- 9.Nakata D, Troy FAI. J Biol Chem. 2005;280:38305–38316. doi: 10.1074/jbc.M508762200. [DOI] [PubMed] [Google Scholar]
- 10.Gascon E, Vutskits L, Kiss JZ. Brain Res Rev. 2007;56:101–118. doi: 10.1016/j.brainresrev.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Easton EW, Schiphorst WECM, Koeleman CAM, Michalides RJAM, Van Den Eijnden DH. Glycoconj J. 1995;12:829–837. doi: 10.1007/BF00731245. [DOI] [PubMed] [Google Scholar]
- 12.Cremer H, Lange R, Christoph M, Plomann M, Vopper G, Roes J, Brown R, Baldwin S, Kraemer P, Scheff S, Barthels D, Rajewsky K, Wille W. Nature. 1994;367:455–459. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- 13.Angata K, Fukuda M. Biochimie. 2003;85:195–206. doi: 10.1016/s0300-9084(03)00051-8. [DOI] [PubMed] [Google Scholar]
- 14.Gregoriadis G, Fernandes A, Mital M, McCormack B. Cell Mol Life Sci. 2000;57:1964–1969. doi: 10.1007/PL00000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregoriadis G, Jain S, Papaioannou I, Laing P. Int J Pharm. 2005;300:125–130. doi: 10.1016/j.ijpharm.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Roy R, Pon RA. Glycoconj J. 1990;7:3–12. [Google Scholar]