Abstract
This study presents, for the first time, sequences of five beta-keratin cDNAs from turtle epidermis obtained by means of 5′- and 3′-rapid amplification of cDNA ends (RACE) analyses. The deduced amino acid sequences correspond to distinct glycine-proline-serine-tyrosine rich proteins containing 122–174 amino acids. In situ hybridization shows that beta-keratin mRNAs are expressed in cells of the differentiating beta-layers of the shell scutes. Southern blotting analysis reveals that turtle beta-keratins belong to a well-conserved multigene family. This result was confirmed by the amplification and sequencing of 13 genomic fragments corresponding to beta-keratin genes. Like snake, crocodile and avian beta-keratin genes, turtle beta-keratins contain an intron that interrupts the 5′-untranslated region. The length of the intron is variable, ranging from 0.35 to 1.00 kb. One of the sequences obtained from genomic amplifications corresponds to one of the five sequences obtained from cDNA cloning; thus, sequences of a total of 17 turtle beta-keratins were determined in the present study. The predicted molecular weight of the 17 different deduced proteins range from 11.9 to 17.0 kDa with a predicted isoelectric point of 6.8–8.4; therefore, they are neutral to basic proteins. A central region rich in proline and with beta-strand conformation shows high conservation with other reptilian and avian beta-keratins, and it is likely involved in their polymerization. Glycine repeat regions, often containing tyrosine, are localized toward the C-terminus. Phylogenetic analysis shows that turtle beta-keratins are more similar to crocodilian and avian beta-keratins than to those of lizards and snakes.
Keywords: beta-keratins, epidermis, mRNAs, protein evolution, turtle
Introduction
The epidermis of turtles exhibits two types of keratinocyte differentiation: softer keratinization in the limbs, neck and tail, and harder keratinization in the shell (Parakkal &Alexander, 1972; Maderson, 1985; Landmann, 1986). The hard keratins, known as beta-keratins, are produced in the precorneous layers of carapace and plastron epidermis, and in the tough scales of various terrestrial tortoises (Baden & Maderson, 1970; Alibardi, 2002, 2005). Immunological studies have indicated, however, that small amounts of beta-keratins are also produced in the soft epidermis of turtles, suggesting that these proteins are ubiquitous in reptilian epidermis (Alibardi et al. 2004; Alibardi & Toni, 2006, 2007). These results suggest that both the quality and amount of protein synthesized in the epidermis determine the degree of hardness of the scales.
Chemical-physical studies on beta-keratins in both birds and reptiles have found that they contain amino acid regions organized in beta-pleated sheets or strand conformation, aggregated into a densely packed lattice (Baden & Maderson, 1970; Fraser et al. 1972; Gregg & Rogers, 1986; Fraser & Parry, 1996, 2008). Their microfibrils show a typical X-ray and ultrastructural pattern of 3–4-nm electron pale filaments. The beta-keratin proteins (10–22 kDa) are smaller than alpha-(soft) keratins (40–68 kDa), which are found in the narrow hinge-regions of reptilian epidermis (see Wyld & Brush, 1979, 1983; Fuchs & Marchuk, 1983; Sawyer et al. 2000).
The nucleotide and amino acid sequences of beta-keratins, and their predicted secondary structures, have recently become available for three species of lizards, a snake species, and a crocodile species (Dalla Valle et al. 2005, 2007a,b, 2008). These studies show that lepidosaurian scales possess beta-keratins containing high amounts of glycine, serine, and proline. Beta-keratins from crocodile epidermis contain high amounts of tyrosine, in addition to glycine, serine, and proline (Dalla Valle et al. 2008). Another group of beta-keratins from some lizard and snake scales have recently been shown to contain high levels of cysteine (Hallahan et al., unpublished; Dalla Valle et al., unpublished).
Chelonians (turtles and tortoises) are the only group of reptiles of which the primary amino acid sequences and gene structure for beta-keratins are not known. Available data on the amino acid composition of chelonian hard keratinization (Baden & Maderson, 1970; Baden et al. 1974; Homer et al. 2001) have indicated that chelonian beta-keratins contain high amounts of glycine, proline, and cysteine. In comparison with the beta-keratins in the hard epidermis of other reptiles, chelonian beta-keratins show a characteristic high amount of tyrosine (over 10%). Two-dimensional electrophoretic and immunoblotting studies have indicated that chelonian beta-keratins of 13–14 and 22–24 kDa are present. Some chelonian beta-keratins have shown a basic pI (7.5–8.5), like those of the other reptiles (Alibardi & Toni, 2006, 2007; Toni et al. 2007b).
While the available data on chelonian beta-keratins provide a basic understanding of the composition of these proteins, the primary sequences of chelonian beta-keratins would facilitate a study of the evolution of these structural proteins in reptiles. Thus, the aim of this research was to determine the primary sequence and gene organization of turtle beta-keratins. This was achieved using nucleotide sequences previously determined in lizards, snakes and crocodiles for primer design (Dalla Valle et al., 2005, 2007a,b, 2008). We collected the shell epidermis during the season in which beta-cells are proliferating and forming new corneous layers. The presence of the mRNAs in turtle scutes was studied by in situ hybridization to evaluate the histological sites of beta-keratin expression. Finally, the phylogenetic relationships of chelonian and other sauropsid beta-keratins was inferred, yielding insights into the evolution of these proteins in birds and reptiles.
Materials and methods
Tissue preparation
Freshwater yellow ringed turtles (P. nelsonii), about 3–5 months old, were purchased in authorized pet shops. When the shell showed some scute growth in both carapace and plastron, the animals were sacrificed by decapitation and their tissue immediately frozen in liquid nitrogen or fixed for histological study. The skin of four animals was collected from shelled (carapace and plastron) and non-shelled (limbs, neck, and tail) parts of the body, and immediately stored in liquid nitrogen. The plastron, especially marginal scutes, was reduced to small pieces (2–3 mm or less) and used for RNA extraction (see later).
Two additional specimens were injected with tritiated thymidine (3–4 µCi g−1 body weight, Amersham, UK) (see details in Alibardi, 2005) for detecting the main sites of cell proliferation in scutes. Finally, two more specimens were injected with tritiated proline (2–3 µCi g−1 body weight, Amersham, UK) for determining the main sites of protein synthesis. The animals were sacrificed 2–3 h later, and tissues injected with tritiated tracers were studied by autoradiography on histological sections as previously detailed for Crysemys picta (see details in Alibardi et al. 2004; Alibardi, 2005; Alibardi & Toni, 2006).
Immunocytochemistry
Shell fragments of 2–5 mm in length (both carapace and plastron) were fixed for 4–5 h in 4% paraformaldehyde in 0.1 m phosphate buffer at pH 7.4. Tissues were dehydrated and embedded in the hydrophilic resin Bioacryl (Scala et al. 1992). Sections of 2–4 µm or 40–90 nm thickness were collected, respectively, for light and ultrastructural cytochemistry, as previously reported (Alibardi et al. 2004). For the histological study, sections were stained in 0.5% toluidine blue or by haematoxylin and eosin stain. A rabbit polyclonal antibody (antiserum, termed β-1), produced against a chick scale beta-keratin (Sawyer et al. 2000) was used at 1 : 100 dilution in 0.05 mTris buffer with 1% BSA. The antibody–antigen complex was detected by a goat anti-rabbit FITC-conjugated secondary antibody (1 : 50 dilution) under a UV epifluorescence microscope. In controls, the primary antibody was omitted or incubated with non-immune serum.
For ultrastructural immunocytochemistry, thin sections collected on nickel grids were incubated overnight with the above primary antibody, rinsed, and incubated with goat anti-rabbit IgG conjugated with 10-nm gold particles. After 5 min of staining with uranyl acetate, grids were observed under a Philips CM-100 electron microscope.
Tissues collected as above from animals injected with tritiated proline were fixed for 8 h in 2.5% glutaraldehyde, rinsed in buffer as above, post-fixed in 2% osmium tetroxide, dehydrated, and embedded in Durcupan resin. For autoradiography, after sectioning with an ultramicrotome, semi-thin sections were coated with K5-Ilford autoradiographic nuclear emulsion, exposed for 2–3 months, and developed with Kodak D19 and fixed in Ilford fixer.
Nucleic acid extraction
Samples of turtle skin were collected, immediately frozen in liquid nitrogen, and stored at −80 °C until nucleic acid extraction. Total RNA was extracted using the commercial product Trizol (Invitrogen, Milan, Italy) according to the manufacturer's instructions. The RNA samples were kept at −80 °C until use. Genomic DNA was extracted from the liver, with the Genomic DNA Purification Kit (Fermentas, M-Medical, Milan, Italy) according to the manufacturer's instructions.
Cloning of turtle keratin cDNAs
The turtle beta-keratin cDNAs were cloned and sequenced using a two-step protocol based on 5′- and 3′-rapid amplification of cDNA ends (RACE) analyses. In the first step we used a 5′-RACE protocol with two primers whose sequence was designed from a conserved region of lizard and gecko beta-keratins (Dalla Valle et al. 2005, 2007a). In the second step, we performed a 3′-RACE analysis using a specific sense primer designed from the 5′ RACE sequence to obtain the complete cDNA sequence. The cloning progression is described in the Results section and the oligonucleotide sequences are reported in Table 1. RNA ligase-mediated rapid amplifications of cDNA 5′-ends (RLM-5′-RACE) and 3′-RACE were performed using the FirstChoice RLM-RACE kit (Ambion, Milan, Italy) following the manufacturer's instructions and methods described previously (Dalla Valle et al. 2007b).
Table 1.
Primers used in RT-PCR and 5′- and 3′-RACE analyses
| Primer | Sequences | Position* | Accession number |
|---|---|---|---|
| Gec1R | 5′-GAGGATGGGTCCAGGGATGGT-3′ | +360 →+340 | AM162665 |
| Gec3R | 5′-CATGGAGTGTTGCCTCCCAC-3′ | +371 →+352 | AM263206 |
| Tu1F | 5′-ctcACACTTCTCCGGACTTC-3′ | −56 →−40 | AM765814 |
| Tu2F | 5′-TTACCTCCATCACAGAAAGATG-3′ | −19 →−1 | AM765816 |
| Tu1R | 5′-GTTCCTGCTGGGTTTAGCATG-3′ | +451 →+431 | AM765815 |
| Tu2R | 5′-TGGCAGGTTTAACATGGCCCA-3′ | +377 →+357 | AM765814 |
| Tu3R | 5′-CTGTGCTGAGGGAAATTGCT-3′ | +232 →+213 | AM765814 |
| Tu4R | 5′-CTTGAGGGCAGGTAGCGAG-3′ | +809 →+791 | FM163397 |
Tu1F was lengthened with three additional random nucleotides (lowercase letters).
Nucleotide position in the reported sequence.
The resultant amplicons of 5′- and 3′-RACE analyses were purified from the sliced gel bands, directly sequenced or ligated into a pGEM-T vector using the pGEM-T Vector System I according to the supplier's recommendations (Promega, Milan, Italy). Plasmids from positive colonies were purified and sequenced.
To perform expression analyses, cDNAs produced by reverse transcription with random hexamers and the ThermoScript RT-PCR System kit (Invitrogen) were amplified with different sets of primers (see Results).
cRNA and cDNA probe synthesis
To prepare sense and antisense cRNA probes, a recombinant plasmid containing the whole coding region of a turtle beta-keratin, obtained by RT-PCR amplification with primers Tu2F and Tu1R (Table 1) and corresponding to sequence Tu-gptrp-2 (see Results), was linearized by restriction cleavage and used as a template. The cRNA transcripts were digoxigenin-labeled by in vitro transcription using a DIG RNA Labeling kit (Roche Diagnostics, Milan, Italy) and T7 and SP6 polymerase. Using the same plasmid, a double-stranded digoxigenin-labeled cDNA probe was prepared by PCR amplification of the fragment with M13F and M13R primers, with digoxigenin-labeled dNTPs (Roche).
Northern blot analysis
Three samples of total RNA from turtle epidermis were used. One sample derived from the apical region of limbs and included softer skin and claws, the second sample mainly comprised softer skin from the neck and non-apical regions of limbs (no claws were present in the latter sample). The third sample consisted in hard skin of scutes (large scales) from the carapace. The RNA samples were electrophoresed through a 1.1% formaldehyde-denaturing gel, blotted onto a positively charged nylon membrane (Roche), and baked at 80 °C for 2 h. The High Range RNA Ladder (M-Medical) was used as a size standard. The 28S and 18S rRNA genes were visualized using methylene blue staining to check RNA loading and integrity. The membranes were hybridized overnight at 68 °C with the DIG-labeled cRNA antisense probe in 5X SSC containing 50% formamide, 0.02% SDS, 0.1% lauroylsarcosine, 1% blocking reagent, and 100 µg mL−1 of transfer RNA. After incubation with an anti-DIG antibody, the signals were detected using the CPD-Star DIG Luminescent reagent according to the manufacturer's instructions. The signals were revealed following exposure of the membranes to an X-ray film for different intervals of time (5–60 min). The analysis was performed twice.
Genomic Southern blotting analysis
Genomic DNA samples (12 µg) were individually digested with Bg1II and XbaI for 3 h at 37 °C. The restriction enzymes were purchased from Promega. Digested samples were fractionated on 1% agarose gels in TBE buffer and blotted onto a positively charged nylon membrane (Roche) using 20X SSC (3 mNaCl, 300 mm sodium citrate, pH 7.0) as transfer buffer. Membranes were hybridized with a digoxigenin-labeled cDNA probe corresponding to the whole coding region of turtle beta-keratin Tu-gptrp-2. Digoxigenin-labeled bands were visualized as described above (exposure time 30 min to 2 h).
Nucleotide sequencing
Sequencing was performed on double-stranded DNA using the ABI PRISM Dye Terminator Cycle Sequencing Core Kit (Applied Biosystems, Monza, Italy). Electrophoresis of sequencing reactions was completed on the abi prism model 377, version 2.1.1 automated sequencer. The homology searches were carried out using the basic blast program version 2.0 at http://www.ncbi.nlm.nih.gov/BLAST/.
In situ hybridization
Pieces of skin, 2–4 mm in length, were collected from soft regions (tail, limbs and neck) and from the shell (carapace and plastron) of two turtles. They were immediately fixed in 4% formaldehyde as described above. Tissues were dehydrated in ethanol at increasing concentration, infiltrated in xylene, and embedded in wax.
Tissues 6–8 µm thick were collected using a rotary microtome, dewaxed with xylene, and hybridized using the antisense-cRNA probes. As controls, a sense cRNA probe was used, as well as negative controls omitting the probes, following a hybridization protocol previously described (Dalla Valle et al. 2005). The hybridizing medium contained 50% formamide, 4X SSC, Tween-20 (1 µL mL−1), tRNA (200 µg mL−1), 0.1% Chaps, 0.5 mm EDTA, heparin (50 µg mL−1), and 2 % blocking reagent (Roche).
For hybridization, an overnight incubation at 60 °C in the hybridization buffer containing 0.5–1.5 ng µL−1 of digoxigenin-labeled probe was carried out. Hybridization was followed by successive washes at decreasing concentrations of standard saline citrate (2X SSC, 0.5X SSC, 0.2X SSC, 0.1X SSC, increasing stringency) until the final phosphate-saline-Tween-buffer (PBT-buffer). Sections were incubated for 2 h at room temperature with anti-digoxigenin-fluorescein Fab fragment antibodies diluted 1 : 15 in Tris Buffer (Roche). The labeling was detected using fluorescence microscopy. Other sections were incubated with anti-digoxigenin alkaline phosphatase-conjugated antibody (Roche) diluted 1 : 500 in PBT buffer. Detection was performed with PBT buffer containing 4-nitroblue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) as substrates, as indicated by the manufacturer (Roche). These sections were mounted in permanent medium and studied under a bright field optical microscope.
Sequence alignment and phylogenetic analysis
Amino acid and nucleotide sequence alignments of various sauropsids were performed using ClustalX2.0. Lepidosaurian sequences include beta-keratins from lizard [lizard-glycine-proline-rich protein (Li-gprp)-1, AC CAI67601; Li-gprp-2, AC CAJ90482; Li-gprp-3, AC CAJ90483; Li-gprp-4, AC CAJ90484; Li-gprp-5, AC CAJ90485], gecko lizard [Gecko-glycine-proline-rich-protein (Ge-gprp)-1_2, AC CAJ44302; Ge-gprp-3, AC CAK19321; Ge-gprp-4, AC CAJ90467; Ge-gprp-5, AC CAK19322], and snake [snake-glycine-proline-rich-protein (Sn-gprp)-1, AC CAL49457; Sn-gprp-2_3, AC CAL49457; Sn-gprp-4, AC CAL49458; Sn-gprp-5, AC CAL51276]. Archosaurian sequences include beta-keratins from crocodile [Crocodile-glycine-proline-tyrosine-rich protein (Cr-gptrp)-1, AC CAO78748; Cr-gptrp-2, AC CAO78749; Cr-gptrp-3, AC CAP49205], chick keratinocyte (AC NP_001001310), chick beak (AC AA085139), chick claw (AC AAA62730), chick scale (AC p04459), and chick feathers I–IV (AC P02450, P04458, P20307, and P20308). Finally, six of the 17 turtle sequences obtained in this study were included in the phylogenetic analysis: turtle-glycine-proline-tyrosine-rich protein (Tu-gptrp)-1 (AC AM765814), Tu-gptrp-2 (AC AM765815), Tu-gptrp-3 (AC AM765816), Tu-gptrp-4 (AC AM765817), Tu-gptrp-5 (AC AM765818), and Tu-gptrp-17 (AC FM163397).
As these proteins contain many insertions/deletions, regions of ambiguous alignment were excluded and the alignment of 49 central amino acids was used for maximum likelihood phylogenetic inference of all beta-keratins listed above. Bayesian inference was implemented with the program Mr.Bayes2.1, using a mixed model of amino acid evolution (Huelsenbeck & Ronquist, 2001). Branch support is represented by the Bayesian posterior probability (BP). Maximum likelihood was implemented in the program PhyML (Guindon & Gascuel, 2003), using the LG model of amino acid evolution (Quang Le & Gascuel, 2008). Branch support in PhyML is estimated with 500 nonparametric bootstrap replicates.
For greater resolution and support among the turtles and archosaurians, which grouped together in the tree inferred by the methods described above, nucleotide sequences were aligned and a 210-bp region of unambiguous alignment was used for maximum likelihood phylogenetic inference. Bayesian inference was implemented with Mr.Bayes2.1, using the HKY model of evolution and a site-specific rate model, allowing each codon position to evolve at its own rate (Huelsenbeck & Ronquist, 2001). Maximum likelihood was implemented with paup*4.0 (Swofford, 1998) using the HKY model of evolution and a site-specific rate model. In all, 100 bootstrap pseudo-samples were generated using the baseml program in PAML (Yang, 2007), and the samples were analyzed using paup*4.0 to obtain bootstrap confidence values.
For the secondary structure prediction of proteins, the PSIPRED Protein Structure Prediction Server at http://bioinf.cs.ucl.ac.uk/psipred/ was used (Jones, 1999; McGuffin, 2000; Bryson et al. 2005).
Results
Histology, autoradiography and immunocytochemistry
As previously reported, we found that new beta-cells in turtle epidermis were produced for the growth of plastron and carapace near the hinge region of scutes of the shell (Alibardi, 2005, 2006). In these areas, new cells derived from the proliferation of germinal cells moved in the two to four suprabasal layers and became spindle-shaped to be incorporated in the corneous layer (Fig. 1A). In fact, thymidine-labeled cells were more frequently present in these regions than in more central areas of the scute (Fig. 1B). In the latter, a cubic layer with sparse suprabasal cells was present, whereas the stratum corneum appeared very thick (Fig. 1C). Therefore, in a random sampling of scutes, proliferating and differentiating beta-cells were more numerous around hinge regions than in more central areas of scutes. Pre-corneous (or transitional) cells took up most of the injected proline, indicating that they were metabolically very active (Fig. 1D). These same beta-cells were also labeled with the Beta-1 antibody, which also immunostained the compact corneous layer (Fig. 1E). This labeling was confirmed using an electron microscopic, which showed an intense immunogold decoration of both pale and clear areas of the corneous layer using the Beta-1 antibody (Fig. 1F).
Fig. 1.
(A) Schematic image of growing centers of turtle scutes. The arrows indicate the movement of neo-generated keratinocytes to expand the scutes. Asterisks show the main sites of proliferation. (B) Autoradiographic image of the hinge region of scute of the carapace showing tritiated thymidine-labeled nuclei (arrows). Bar, 10 µm. The inset shows a detail of the labeling in the transitional layer (arrow) after injection of tritiated histidine. Bar, 10 µm. (C) Thick corneous layer (arrow) of the central part of a carapace scute with a narrow epidermis. Toluidine blue stain. Bar, 20 µm. (D) Autoradiography after tritiated proline injection. Silver grains are concentrated in the pre-corneous layer occupied by transitional keratinocytes (arrows). Arrowheads point to the shedding layer between the outer and inner corneous layer. Bar, 10 µm. (E) Beta-1 immunofluorescent corneous layer of carapace scute. The square indicates the relative position of Fig. 1F. The arrow indicates the position of transitional cells. Bar, 10 µm. (F) Ultrastructural detail of the beta-1 immunolabeled corneous layer. The arrows indicate the remnants of the perimeter of single corneocytes, largely merged into the corneous layer. Bar, 150 nm. βi, inner beta-layer; βo, outer beta-layer; β1, beta-1 immunoreactivity; c, corneous layer; de, dermis; ds, dense areas of the corneous layer; e, epidermis; h, hinge region; HIS, tritiated histidine labeling; pa, pale areas of the corneous layer; PRO, tritiated proline labeling; sl, shedding line (along which the outer corneous layer detaches from the inner layer, often still forming); t, tip of scutes (bridge scutes); TB, toluidine blue stain; THY, tritiated thymidine (nuclear) labeling. Curved arrows indicate the direction of scute growth. Dashes underlie the basal layer of the epidermis.
Cloning of the turtle beta-keratins
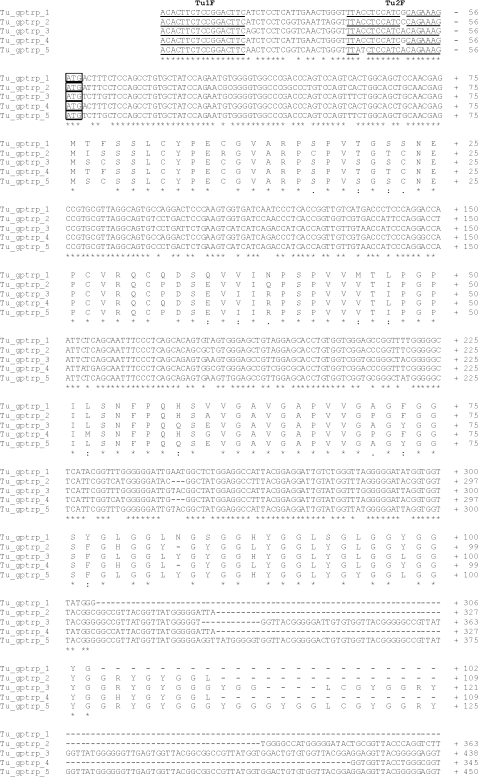
Cloning of turtle beta-keratins was initiated by performing 5′-RACE analysis using two antisense primers (Gec3R and Gec1R) selected from conserved regions of lizard and gecko beta-keratins (Table 1). A specific sense primer (Tu1F), located at the beginning of the 5′-UTR, was designed from the turtle 5′-RACE-amplified and -sequenced fragment, and used to carry out the 3′-RACE analysis. The cloning of the amplification products and the sequencing of eight clones yielded five beta-keratin transcripts named Tu-gptrp-1 to -5 (Fig. 2;AC AM765814, AM765815, AM765816, AM765817, AM765818, respectively). Of these, only Tu-gptrp-3 lacked the 3′-UTR as the 3′-RACE adaptor annealed to a stretch of adenines not corresponding to the polyadenylated region. A natural variant, ATTAAA of the poly-A signal, was present in the other four cDNAs. The transcript lengths ranged from 1.1 to 1.2 kb (except for Tu-gptrp-3, which was only 0.6 kb). The 5′-UTR region was well conserved and was composed of 56 nucleotides (nt) in each transcript. In all the transcripts, the ATG had a nucleotide context that only partially corresponded to the proposed consensus sequence for the initiation of translation (Kozak, 1986). Lower conservation was found in the 3′-UTR region, which differed in length from the other sequences (Fig. 2).
Fig. 2.
Nucleotide and deduced amino acid sequences of five turtle beta-keratins. One-letter symbols of encoded amino acids are shown below the DNA sequence. The numbers refer to the nucleotide and amino acid positions at the end of each line. Small lines indicate missing nucleotides or amino acids. The in-frame translation start codon, the stop codon, and the putative polyadenylation signal are boxed. The specific oligonucleotide primers used for the cloning are single or double underlined, and their specific names are reported in bold over the sequences. The sequences are available at the EMBL/GenBank/DDBJ database under Accession numbers AM765814, AM765815, AM765816, AM765817 and AM765818 for Tu-gptrp-1, 2, 3, 4, and 5, respectively.
Northern blotting and expression analysis
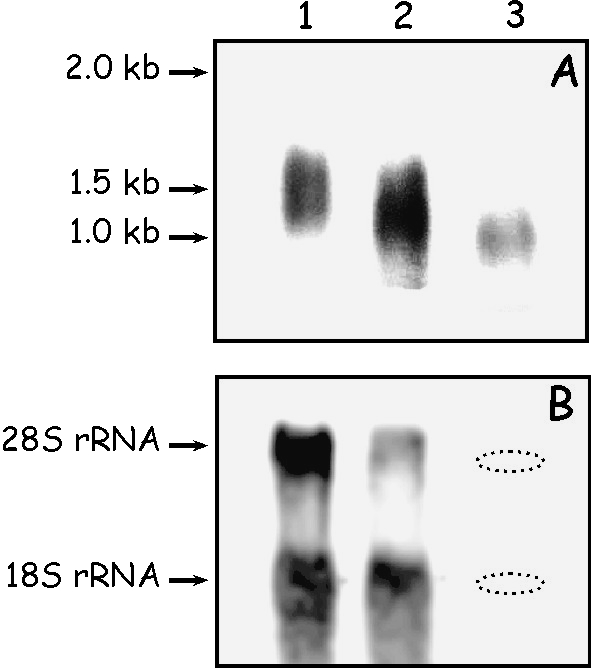
The expression of mRNA coding for turtle proteins was analyzed by Northern blotting using three samples of total RNA extracted from: (1) limb apical regions, including claws; (2) skin from neck and non-apical limbs; and (3) carapace scutes (Fig. 3). About 10 µg of RNA were loaded from samples 1 and 2, whereas, due to the difficulty of extracting RNA from scutes of sample 3, only 1 µg was loaded of this sample. The signal produced by methylene-blue staining was, in this case, very light and is undetectable in Fig. 3. The position of 28S and 18S rRNA genes of sample 3 is indicated in Fig. 3B. After 30 min of exposure, a transcript was detected in all the samples. The signal was of lower intensity in sample 3, containing much less total RNA (Fig. 3A). Based on comparison to the RNA ladder, the size of these transcripts was estimated to be about 1.5 kb. The size correlates well with the cDNA size of the cloned transcripts (between 1100 and 1200 bp), considering an average addition of 200–300 adenines in the mRNAs. The transcript of sample 3 appeared shorter than in the previous samples but this difference could be due to the different electrophoretic run.
Fig. 3.
(A) A representative Northern blot analysis of total RNAs extracted from turtle with a turtle DIG-labeled antisense cRNA beta-keratin probe. The arrows and reported numbers indicate the positions of RNA standards according to its migration. Exposure time: 30 min. 1, RNA from limb apical regions, including claws (10 µg); 2, RNA from skin from neck and non-apical limbs (10 µg); 3, RNA from carapace scutes (1 µg). (B) Methylene blue-stained 28S and 18S ribosomal RNA genes in each mRNA sample. The dotted ovals indicate the position of 28S and 18S.
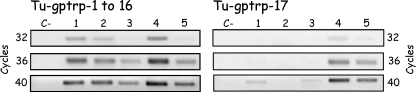
To analyse a possible different expression between turtle beta-keratin 1–16 and turtle beta-keratin 17, an RT-PCR analysis with the set of primers Tu1F-Tu3R (which amplifies the first 16 beta-keratins) and with the set of primers Tu1F-Tu4R (which amplifies turtle beta-keratin 17 and possibly other transcripts of the same group), was performed at 32–36 and 40 PCR cycles, starting with similar cDNA quantities. The normalization of cDNAs was performed on the basis of results obtained by PCR amplification with a set of primers for 18S rRNA gene (data not shown). The analysis included two samples of shell skin (1 and 2), a sample of soft skin from limbs and neck (3), a sample from claws and digit-tips skin (4), and a sample of digit-tips skin without claws (5). Although transcripts for beta keratins 1–16 were expressed in all five samples analysed, the level of expression was quite different in the analysed tissues. The highest levels were seen in samples from the skin of digit-tips containing claws and from the carapace skin, while the lowermost expression was observed from the sample of soft skin (Fig. 4).
Fig. 4.
RT-PCR analysis of β-keratin expression performed at different PCR cycles (32–36–40) with cDNA of shell skin (1 and 2); soft skin (3); claws and digit-tip skin (4); digit-tip skin only (5). C–, negative control (see text for more details).
As regards the expression of keratin 17, very low levels of expression were evident in samples from the carapace and from soft skin. A higher expression was seen in digit-tips including claws or digit tip without claws, suggesting a tissue specific-expression for this keratin.
The different levels of expression between the amplification performed for keratin 1–16 or for keratin 17 cannot be exclusively derived from the primers utilized. Whereas the first couple of primers amplify the numerous transcripts for beta-keratins 1–16, the second couple of primers amplify only transcripts for keratin 17. However, it appears that more transcripts for beta-keratin 17 are present as suggested from the second, weak amplified band seen in the samples from soft epidermis and from the sequencing of two amplification products obtained with this set of primers and genomic DNA (data not shown).
Southern blotting analysis and genomic DNA amplifications
Southern blotting was used to demonstrate that turtle beta-keratins belong to a family of highly related and conserved genes, as previously established for geckos, lizards, snakes, crocodiles, and birds. Southern blotting was performed with two restriction enzymes, Bg1II and XbaI, that had no restriction sites within the coding region sequences of the known turtle beta-keratins (Tu-gptrp-1 to -5). The probe was prepared using PCR by labeling the entire coding region corresponding to the sequence Tu-gptrp-2 that was obtained by RT-PCR with the primer pair Tu2F and Tu1R. Southern blotting yielded at least 10 bands with both enzymes, demonstrating the presence of multiple beta-keratin genes in the turtle genome (Fig. 5).
Fig. 5.

Southern blot analysis of turtle genomic DNA. The restriction enzymes used in the experiment are indicated on the top of the lanes. The sizes of the fragments that have hybridized with the probes are estimated from the 1-kb DNA ladder (Invitrogen).
This was confirmed by the amplification of genomic DNA fragments corresponding to beta-keratin genes. This amplification was performed to analyse the occurrence and position of possible introns inside turtle beta-keratin genes. The genomic DNA was PCR-amplified with the primer pairs Tu1F-Tu1R and Tu1F-Tu2R. The two antisense primers are located in conserved regions near the stop codon of the five beta-keratin transcripts isolated from cDNA cloning (Fig. 2). We obtained two different amplification products of about 1.6 and 1.2 kb, which were cloned and sequenced. All the positive clones (16 analysed) corresponded to beta-keratin genes. Each gene contained an intron localized to the 5′-UTR, 35 bp after the beginning of the sequence (Fig. 6; Table 2), and the position of the intron was deduced by alignment analysis. There is one exception (the Tu-gptrp-8 gene), in which the changing of a G to C (which could be due to a Taq polymerase mistake) determined the loss of a splice signal. However, another GT splice signal was present four nucleotides downstream. All introns displayed splice signals consistent with the GT/AG rule. The introns end 21 bp before the ATG codon, except in the Tu-gptrp-17 gene which ends 20 bp before the ATG codon. In Fig. 6 we present the organization of the Tu-gptrp-17 gene as representative of all the turtle beta-keratin genes isolated in the present work.
Fig. 6.
Nucleotide sequence of one of the clones obtained from PCR on turtle genomic DNA. One-letter symbols of encoded amino acids are shown below the DNA sequence. The in-frame translation start codon and the stop codon are boxed. The oligonucleotide primers used for the PCR are underlined. The intron is shown in bold lowercase and the splice signals are shaded. The sequence is available at the EMBL/GenBank/DDBJ database under AC FM163397.
Table 2.
Length and identity values of the introns of 17 different turtle beta-keratin genes. The groups with the higher identity are reported in different colours
 |
Thirteen beta-keratin sequences were obtained by genomic DNA amplifications, but one (FM163398) corresponded to one of the five transcripts (Tu-gptrp-1) obtained from cloning cDNA. Thus, a total of 17 deduced beta-keratins have been identified in our analysis, and their sequences were deposited in GenBank with AC AM765814 to AM765818 for Tu-gptrp-1 to 5, and FM163386 to FM163397 for Tu-gptrp-6 to -17, respectively. The length of the intron in the sequenced genes ranged from 353 to 1069 nucleotides. The introns were divided into groups on the basis of sequence identity and length (Table 2). The introns with a length of 679 bp aligned well with the first portion of the intron sequences that were about 1 kb. In contrast, the 353-bp intron of Tu-gptrp-14 aligned well with the end of the 1-kb introns and partially overlapped (fewer than 100 bases) with the introns of 679 bp. This suggests that the shorter introns derive from the longer introns. Finally, the intron of Tu-gptrp-17 showed very low identity with the other intron sequences, reflecting the low levels of amino acid identity (42–48%).
In situ hybridization
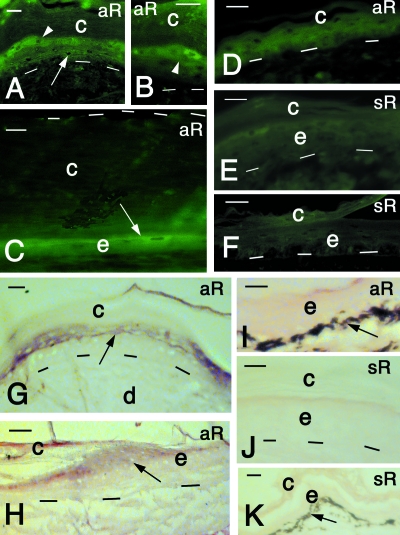
Single-stranded cRNA digoxigenin-labeled probes, targeting the complete coding sequence of Tu-gptrp-2 (470 bp), were used to study the expression of the isolated mRNA in the epidermis of carapace scutes and limb skin. The cRNA antisense probe (aR), detected by a fluorescent anti-digoxigenin antibody, exclusively labeled the spindle-shaped, differentiating cells of the beta-layer of scutes, especially in growing regions (Fig. 7A,B). In internal areas of scutes, where the corneous layer was thicker and the viable epidermis reduced to two to three layers, only fusiform suprabasal cells were fluorescent (Fig. 7C). A weaker labeling was also seen in pre-corneous cells of the softer epidermis of the neck and limbs (Fig. 7D). No labeling was detected in the undifferentiated cells of normal epidermis, or in the dermis.
Fig. 7.
In situ hybridization results on turtle skin after immunofluorescence (A–F) and alkaline phosphatase-colorimetric-based methods (G–K). (A) Reactive cells in the upper suprabasal (arrow) and pre-corneous (arrowhead) layers. Lateral area of carapace scute. Bar, 15 µm. (B) Close-up on reactive spindle-shaped cells (arrowhead) of the pre-corneous layer. Lateral areas of carapace scute. Bar, 20 µm. (C) Central area of carapace scute showing the thick corneous layer (delimited superiorly by dashes) and the thin, reactive cell of the epidermis (arrow). Bar, 10 µm. (D) Relatively soft epidermis of the limb showing some weak reactivity in the epidermis. Bar, 10 µm. (E) Sense control in the epidermis of a carapace scute. Bar, 15 µm. F, sense control of soft epidermis of limb. Bar, 15 m. (G) Reactive cells (arrow) of the precorneous layers (arrow) in carapace scute. Bar, 10 µm.(H) Reactive cells (arrow) near the hinge region (growing center) of carapace scute. Bar, 20 µm. (I) Non-reactive soft epidermis of the limb. Bar, 15 µm. (J) Sense control of carapace scute epidermis. No reactivity is present. Bar, 10 µm. (K) Sense control of soft epidermis of the limb. No reactivity is present. Bar, 10 m. aR, antisense RNA probe; c, corneous layer; d, dermis; e, epidermis; sR, sense RNA probe. Dashes underlie the basal layer of the epidermis.
The cornified layer was sometimes nonspecifically fluorescent, as confirmed by the controls. The negative control (omitting the probe) and the sense probe (sR) did not label or weakly labeled beta-cells of scutes (Fig. 7E,F). The initial observations were confirmed by detecting the hybridization product, with the anti-digoxigenin alkaline phosphatase antibody and the colorimetric reaction (see Materials and methods). The reddish-purple stain was seen over pre-corneous layers of the epidermis of scutes where spindle-shaped keratinocytes were expressing the specific mRNA (Fig. 7G). In particular, positive cells were seen in the growing areas near the hinge region of scutes (Fig. 7H). A very weak to absent reaction was present in the soft epidermis of tail and neck epidermis (Fig. 7I). The corneous layer was not stained in shell, limb or tail epidermis. Finally, no labeling was present in sense controls of shell scutes (Fig. 7J) or in the softer epidermis of limb or tail (Fig. 7K).
Amino acid content, secondary structure prediction and sequence analysis
On average, the deduced turtle beta-keratin sequences obtained in this study contain 145 amino acids, with a range of 122 to 174. The theoretical pI ranges from 6.8 (two sequences) to 8.4. The well-represented amino acids are glycine, proline and valine. Serine content is variable, but often high. Tyrosine is present in high concentrations (8.5–12.6%), except in Tu-gptrp-1 (4.9%). In general, these proteins contain low amounts of charged amino acids such as glutamate, aspartate, arginine, and lysine, and they contain almost no tryptophane.
The amino-acid alignment of the 17 turtle beta-keratins obtained in this study is presented in Fig. 8. The central sequence of 20 amino acids, which is well-conserved in all beta-keratins studied thus far, is indicated with the term ‘core-box’ (blue box in Fig. 8) for comparative purposes with homologous regions of other reptilian and bird beta-keratins (Fig. 9; Table 3). The prediction of secondary structure of two representative turtle beta-keratins (Tu-gptrp-2 and 17) was obtained using the PSIPRED Protein Structure Prediction Server at http://bioinf.cs.ucl.ac.uk/psipred/. It indicates that the region N-terminal to the core-box has a random-coiled configuration, whereas there are two beta-strands in the core box. Most of the remaining region has a random-coiled conformation.
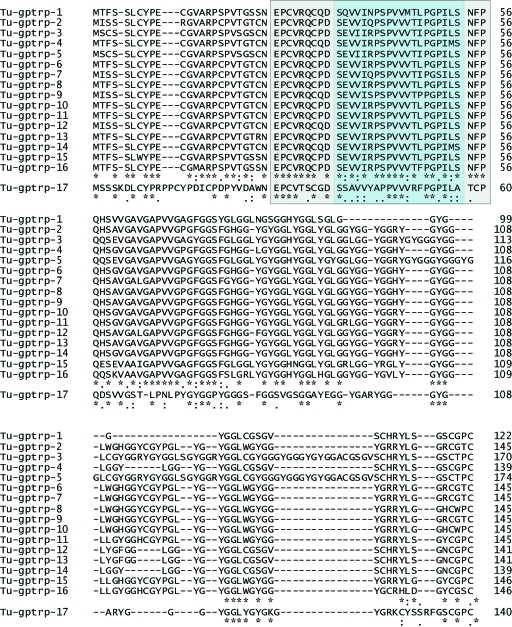
Fig. 8.
Amino acid alignment of the 17 deduced beta-keratins from turtle, using ClustalX2.0 software. The blue box indicates the core-box, and the surrounding grey-shaded box indicates the 32-aa extended beta-pleated region. Stars indicate identity; colons indicate conserved substitution; dots indicate semiconserved substitutions. Sequences are available at GenBank under: AC AM765814–AM765818 for Tu-gptrp-1 to -5, and FM163386–FM163397 for Tu-gptrp-6 to -17.
Fig. 9.
Comparison of various beta-keratins from turtles (Tu-gptrp-1 to Tu-gptrp-5, and Tu-gptrp-17) and other sauropsids (crocodiles: Cr-gptrp-1 and 2; chickens: chick-feather-I and -II; lizards: Li-gprp-2 and -4; geckos: Ge-gprp-3 and 5; snakes: Sn-gprp-1 and 2–3), that highlights the position of seven main amino acids found in turtle. (A) highlights glycine, proline, cysteine and serine. (B) highlights valine, leucine and tyrosine (see text for details). The sequences are centered on the core-box, indicated by the rectangle.
Table 3.
Percentage identity of the 20-amino-acid core-box region in reptilian beta-keratins (lizards, snakes, alligators) and chick scale, claw, and feather keratins, using ClustalX2.0 (Larkin et al. 2007). Li-gprp-1_5: core-box of lizard Podarcis siculabeta-keratins (CAI67601, CAJ90482, CAJ90483, CAJ90484, CAJ90485); Ge-gprp-1_2: core-box of gecko lizard Tarentola mauritanicabeta-keratins (CAJ44302 and CAK19320); Ge-gprp-3: core-box of gecko lizard Tarentola mauritanicabeta-keratin (CAK19321); Ge-gprp-4 and Ge-gprp-5: core-box of gecko lizard Hemidactylus turcicusbeta-keratins (CAJ90467 and CAK19322); Sn-gprp-1_4: core-box of snake Elaphe guttatabeta-keratins (CAL49457, CAL49458, CAL49459, CAL49460); Sn-gprp-5: core-box of snake Elaphe guttatabeta-keratin (CAL51276); Alligator: core-box of alligator beta-keratin (Sawyer et al. 2000); Chick-scale: core-box of chicken scale beta-keratin (P04459); Chick-claw: core-box of chicken claw beta-keratin (AAA62730); Chick-feather: core-box of chicken feather beta-keratin (P02450)
| Li-gprp-1_5 | Ge-gprp-1_2 | Ge-gprp-3 | Ge-gprp-4 | Ge-gprp-5 | Sn-gprp-1_4 | Sn-gprp-5 | Tu-gptrp-1 | Tu-gptrp-2 | Tu-gptrp-3–5 | Tu-gptrp-4 | Cr-gpyrp-1 | Cr-gpyrp-2 | Alligator | Chick Scale | Chick Claw | Chick feather | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Li-gprp-1_5 | 100 | 75 | 75 | 75 | 75 | 70 | 75 | 55 | 70 | 60 | 50 | 60 | 70 | 65 | 60 | 70 | 60 |
| Ge-gprp-1_2 | 100 | 90 | 100 | 90 | 70 | 75 | 80 | 80 | 70 | 70 | 80 | 80 | 70 | 75 | 70 | 80 | |
| Ge-gprp-3 | 100 | 90 | 85 | 70 | 75 | 70 | 80 | 70 | 70 | 80 | 80 | 70 | 75 | 70 | 80 | ||
| Ge-gprp-4 | 100 | 90 | 70 | 75 | 80 | 80 | 70 | 70 | 80 | 80 | 70 | 75 | 70 | 80 | |||
| Ge-gprp-5 | 100 | 65 | 70 | 70 | 75 | 65 | 70 | 75 | 75 | 65 | 70 | 65 | 75 | ||||
| Sn-gprp-1_4 | 100 | 95 | 60 | 80 | 75 | 60 | 70 | 80 | 70 | 70 | 75 | 70 | |||||
| Sn-gprp-5 | 100 | 65 | 85 | 80 | 65 | 75 | 85 | 75 | 75 | 80 | 75 | ||||||
| Tu-gptrp-1 | 100 | 80 | 75 | 75 | 75 | 75 | 65 | 70 | 65 | 85 | |||||||
| Tu-gptrp-2 | 100 | 90 | 80 | 80 | 90 | 80 | 80 | 75 | 90 | ||||||||
| Tu-gptrp-3–5 | 100 | 80 | 70 | 80 | 70 | 70 | 65 | 80 | |||||||||
| Tu-gptrp-4 | 100 | 70 | 70 | 60 | 65 | 60 | 80 | ||||||||||
| Cr-gpyrp-1 | 100 | 90 | 75 | 85 | 75 | 90 | |||||||||||
| Cr-gpyrp-2 | 100 | 85 | 85 | 80 | 90 | ||||||||||||
| Alligator | 100 | 80 | 80 | 75 | |||||||||||||
| Chick scale | 100 | 85 | 80 | ||||||||||||||
| Chick claw | 100 | 75 | |||||||||||||||
| Chick feather | 100 |
Many regions of Tu-gptrp 1–16 are well-conserved, with an identity score of 66.7% from amino acids 1–96 (Fig. 8). The alignment of the 17 turtle amino acid sequences shows that Tu-gptrp-17 differs greatly from the other 16 proteins (Fig. 8). However, secondary structure prediction indicates that two beta-strands are also present in the core box of Tu-gptrp-17. The alignment shows that from amino acid 70 (74 for Tu-gptrp-17) toward the C-terminus, the sequence consists mainly of gly-X (X is mainly tyrosine) and gly-gly-X (X is mainly leucine) repeats. Four of the total 11–13 prolines of these turtle beta-keratins are found in the core-box, and 4–5 of the 11–13 valines are also localized to the core-box.
The comparison of five representative turtle beta-keratins and some lepidosaurian and archosaurian (including chick) beta-keratins is presented in Fig. 9. This figure shows the proteins after alignment centered on the core-box. The core-box is localized toward the N-terminus of turtle and archosaurian beta-keratins, whereas it is found more toward the central region in lepidosaurians. To highlight the distribution of key amino acids in the beta-keratins presented, Fig. 9A highlights glycine, proline, cysteine and serine, and Fig. 9B highlights tyrosine, valine and leucine. Cysteine residues are present toward the N- and C-termini of beta-keratin proteins, whereas proline is concentrated in the core-box and surrounding regions. With the exception of feather keratins, glycine is found in glycine-rich repeats toward the C-terminus of these proteins. Serine is more randomly distributed. Whereas most valine is seen in the central region, including the core-box, leucine and tyrosine are present in lateral regions. Tyrosine is present in much higher amounts in turtle (and crocodilian) beta-keratins in comparison with those of lizards and snakes, and this aromatic amino acid is localized toward the N- and especially near the C-terminal region (Fig. 9). Within the turtle and reptilian core-box, the constant three-peptide PGP, a common VVV or VVM, and a common PPP or PSP, are present (and an APP in Tu-gptrp-17; Fig. 9).
Using Clustal-X2.0 we found that conservation between turtle and lepidosaurian beta-keratin sequences is quite low (17–29%) (Fig. 9,Table 3). Sequence identity rises to 50–85% when the core-box is compared (Table 3). In a comparison of turtle with crocodilian and avian beta-keratins, the conservation is much higher (25–36%), especially between amino acids 1 and 102 of the turtle proteins. This region includes the core-box and other amino acid sequences present around the core-box. The C-terminus of the molecule is more divergent between turtles and archosaurians.
Phylogenetic analysis
Maximum likelihood and Bayesian methods were used to infer the relationships between known lepidosaurian, archosaurian, and turtle beta-keratin sequences. Firstly, from an amino acid alignment of 49 central amino acids (including the core-box), Bayesian inference and maximum likelihood yielded an unrooted tree separating lepidosaurians (lizards, snakes, and gecko lizards) from turtles and archosaurians (crocodiles and birds) by a very long branch and 100% branch support for the lepidosaurian and turtle–archosaurian clades (tree not shown). Thus, turtle beta-keratins are more similar to archosaurian beta-keratins than to those of lepidosaurians.
From a nucleotide alignment of the first 210 nucleotides of turtle and archosaurian beta-keratin sequences, Bayesian inference and maximum likelihood methods were used to infer phylogenetic relationships within the turtle-archosaurian clade (Fig. 10). As turtle sequences Tu-gptrp-1 to -5 formed a clade that was separated from the archosaurians (and Tu-gptrp-17) by a long branch, these turtle sequences were designated as the outgroup in Fig. 10. For the most part, the gene tree in Fig. 10 reflects the well-accepted species tree for these taxa (Rest et al. 2003). Tu-gptrp-1 to -5 form a clade that is sister to the archosaurians, and the crocodile sequences form a clade that is sister to the bird beta-keratins. The one exception is Tu-gptrp-17, which groups with the bird beta-keratins and is most closely related to a chick-keratinocyte beta-keratin. While it is unclear why Tu-gptrp-17 and chick-keratinocyte sequences are grouping together, they do have the longest branches in the tree, leaving open the possibility that they are grouping together because of long-branch attraction.
Fig. 10.
Phylogenetic inference of turtle and archosaurian beta-keratins using maximum likelihood (implemented in paup*4.0; Swofford, 1998) and Bayesian (implemented in Mr.Bayes2.1; Huelsenbeck & Ronquist, 2001) methods. For this figure, branch lengths were estimated in Mr.Bayes2.1 using the HKY model of evolution and a site-specific rate model. BP, Bayesian posterior probability; BS, bootstrap proportions.
Discussion
Tissue distribution and sequence determination
The autoradiographic and immunocytochemical data indicate that the keratinocytes near the hinge region and in the transitional layer of the scute epidermis are the main sites of production of beta-keratins (Alibardi, 2002, 2005, 2006; Alibardi et al. 2004). In fact, the observed tritiated thymidine and histidine uptake pattern suggests a higher proliferation and a higher protein metabolism in these regions, while the proline uptake correlates to the high percentage of this amino acid in beta-keratins (Dalla Valle et al. 2005, 2007a,b, 2008).
Although our initial attempts to obtain partial sequences of turtle beta-keratins using proteomic methods proved to be difficult (data not shown), obtaining complete sequences using molecular biology techniques presented fewer difficulties, especially when compared to recent attempts in lizards, snakes and crocodiles. After sequencing of the 5′-UTR and selection of a specific sense primer, we obtained the 3′-RACE fragment directly, yielding five different beta-keratin transcripts. This indicates that the 5′-UTR region is well conserved in turtle beta-keratins, which was confirmed with genomic amplification of 13 different genomic fragments containing beta-keratin genes. In sum, we have obtained 17 unique turtle beta-keratin amino acid sequences, with 16 of the 17 exhibiting high conservation, at least in the first 70 amino acids. Although beta-keratins may be present in turtle epidermis from scales of different body regions, our study suggests that most of these proteins are quite uniform. We do not known whether the 17 deduced proteins found in the present study represent the entire proteome for turtle beta-keratins. However, the number of deduced beta-keratins in P. nelsonii and their molecular weights determined in this study correspond to the 12 or more protein spots detected in the epidermis of the turtle C. picta (Alibardi & Toni, 2007). In the latter analysis, all the proteins were derived from the shell (carapace and plastron); thus, the additional sequences found in P. nelsonii may derive from other skin regions and from claws in particular. In fact, specific proteomic patterns from turtle claws have identified a few other protein spots (5–7), mainly basic, which are not present in the shell epidermis (L. Alibardi and M. Toni, unpublished data).
The main difference between amino acid sequences obtained by proteomic and molecular methods, although from different turtle species, is the pI values. Whereas sequences deduced in this study by cloning and genomic amplifications are generally basic (pI of 6.8–8.4), those proteins isolated from the epidermis are more acidic (pI 6.2– 7.2 in nine of 12 protein spots; see Alibardi & Toni, 2007). This difference is probably a result of post-translational modifications of beta-keratins, in particular phosphorylation, whose physiological function is unknown (Toni et al. 2007a,b).
Only the analysis of a whole turtle genome will make it possible to know the total number of beta-keratin genes in turtles. The only available genome of a reptile, the lizard Anolis carolinensis, has indicated that 40 different genes for beta-keratins are present, probably clustered in a single chromosomal region (Dalla Valle et al., unpublished results).
In situ hybridization shows that the mRNAs for turtle beta-keratins are present in spindle-shaped beta-cells that differentiate in upper layers or in the pre-corneous layer of the epidermis, well above the germinal layer. Also, the weak hybridization seen in pre-corneous alpha layers supports some immunological studies that have suggested low levels of expression in the soft epidermis (Alibardi & Toni, 2006). These results suggest that beta-keratins are widely expressed in turtle epidermis, but they are expressed in the highest levels in forming beta-layers of shell scutes (carapace and plastron), and likely of claws, for the production of mechanically proof layers. The synthesis of these proteins takes place after the cells have left the basal and first suprabasal layers, and this pattern of expression is also observed in lepidosaurian and crocodilian epidermis (Dalla Valle et al. 2005, 2007a,b, 2008). Ultrastructural in situ hybridization studies have indicated that beta-keratins are immediately polymerized into cytoplasmic bundles of beta-keratin (Dalla Valle et al. 2005; Alibardi et al. 2006, 2007).
Beta-keratin comparisons and relationships
While there is some variation in the amino acid composition between the deduced beta-keratins of P. nelsonii, the turtle beta-keratins as a whole are very different from those of lepidosaurian reptiles so far sequenced (Dalla Valle et al. 2005, 2007a,b). In contrast, turtle beta-keratins resemble those of archosaurian scales and claws, of both crocodilians and birds (Fig. 9; see Gregg & Rogers, 1986; Dalla Valle et al. 2008). Among shared characteristics are tyrosine richness, high conservation of the core-box and surrounding regions, and a relative richness in alpha-helix regions. Because of the relatively high percentage of tyrosine, turtle beta-keratins represent, like those isolated from the crocodile, a new class of proteins indicated as high-glycine-proline-tyrosine proteins (HGPTs). The deduced proteins (Tu-gptrp-1 to Tu-gptrp-17) possess molecular masses (11.9–17.0 kDa) in agreement with beta-keratins analysed by proteomic methods (Homer et al. 2001; Alibardi & Toni, 2006, 2007; Toni et al. 2007b).
The core-box of turtle beta-keratins is more similar to the core-box of crocodilian and avian beta-keratins (sequence identity of 60–90%) than to that of lizards and snakes (50–85%, see Table 3). Furthermore, turtle beta-keratins share a higher homology with archosaurian beta-keratins in a pre-core-box region of 17 amino acids. These observations were confirmed in a phylogenetic analysis of lepidosaurian, turtle, and archosaurian sequences: lepidosaurian proteins and turtle/archosaurian proteins clustered separately, reflecting the well-supported phylogenetic relationships of these species (tree not shown). In fact, with the exception of Tu-gptrp-17, all beta-keratin sequences grouped with other beta-keratins of the same species (see Fig. 10), suggesting that these proteins diversified independently in each sauropsid group.
Our secondary structure predictions indicate that turtle beta-keratins, like those of the crocodile, have a smaller percentage of beta-strand regions in comparison with those of lepidosaurian reptiles (4.8–8.3% in turtle, 4.4–8.4% in crocodile, 6.3–11.2% in lizards, and 9.4–15.3% in snakes). In the turtle proteins, two beta-sheets (strands) exist in tandem in the region with the highest conservation, the core-box. In general, beta-sheets are found near the core-box of beta-keratins, whereas in all core-boxes a double-strand region is present. However, the core-box is present within a more extended region of 32 amino acids (indicated by the grey-shaded area surrounding the blue-shaded area in Fig. 8) in which, as other studies have indicated, there are at least four strands (Fraser & Parry, 1996, 2008). The effects of these different secondary conformations on the properties of the various turtle proteins are not known.
The small core-box region is sufficient to produce the typical polymerization and filamentation of beta-keratins inside beta-cells of the epidermis (Fraser et al. 1972; Brush, 1983, 1993). Prolines and most valines are concentrated in the core-box region and probably contribute to the formation of the beta-pleated sheets (Fraser et al. 1972; Fraser & Parry, 1996).The effects of other amino acid regions on the chemical and physical properties of turtle beta-keratins are not known. In particular, the role of the glycine-rich tail of turtle and archosaurian proteins remains to be determined (Fig. 9). A proposed model of beta-keratins in feathers and scales (see Gregg & Rogers, 1986) indicated that the glycine-rich regions are allocated outside the framework of the beta-keratin filament, but their precise role was not explained. In general, glycine-rich sequences make the protein more insoluble (Gregg & Rogers, 1986), and may be involved in homophilic stabilizing interactions (glycine-loops), as shown in the protein loricrin in mammalian corneocytes of the stratum corneum (Kalinin et al. 2002).
In conclusion, this study has identified the nucleotide and deduced amino-acid sequences of 17 unique beta-keratins in P. nelsonii. As in the lizard A. carolinensis, turtle beta-keratins are members of a multigene family, although it is still unclear how many additional members of this gene family exist. In situ hybridization shows that beta-keratin mRNAs are expressed in cells of the differentiating beta-layers of the turtle shell scutes. The 17 turtle beta-keratin sequences are rich in glycine, proline, serine, and tyrosine, and are more similar to archosaurian beta-keratins than to those of the lepidosaurians.
Acknowledgments
The study was financed by 60% Grants from the Universities of Padova and Bologna, and by self support (L.A.). The beta-1 antibody was kindly provided by Dr R. H. Sawyer (University of South Carolina, Columbia, SC, USA).
References
- Alibardi L. Immunocytochemical observations on the cornification of soft and hard epidermis in the turtle Chrysemys picta. Zoology (Jena) 2002;105:31–44. doi: 10.1078/0944-2006-00048. [DOI] [PubMed] [Google Scholar]
- Alibardi L. Proliferation in the epidermis of chelonians and growth of the horny scutes. J Morphol. 2005;265:52–69. doi: 10.1002/jmor.10337. [DOI] [PubMed] [Google Scholar]
- Alibardi L. Ultrastructural and immunohistochemical observations on the process of horny growth in chelonian shells. Acta Histochem. 2006;108:149–162. doi: 10.1016/j.acthis.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Alibardi L, Toni M. Immunolocalization and characterization of beta-keratins in growing epidermis of chelonians. Tissue Cell. 2006;38:53–63. doi: 10.1016/j.tice.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Alibardi L, Toni M. Immunological characterization of a newly developed antibody for localization of a beta-keratin in turtle epidermis. J Exp Zool. 2007;308B:200–208. doi: 10.1002/jez.b.21138. [DOI] [PubMed] [Google Scholar]
- Alibardi L, Spisni E, Toni M. Differentiation of the epidermis in turtle: an immunocytochemical, autoradiographic and electrophoretic analysis. Acta Histochem. 2004;106:379–395. doi: 10.1016/j.acthis.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Alibardi L, Dalla Valle L, Toffolo V, Toni M. Scale keratin in lizard epidermis reveals amino acid regions homologous with avian and mammalian epidermal proteins. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:734–752. doi: 10.1002/ar.a.20342. [DOI] [PubMed] [Google Scholar]
- Alibardi L, Toni M, Dalla Valle L. Expression of beta-keratin mRNAs and proline-uptake in epidermal cells of growing scales and pad lamellae of gecko lizards. J Anat. 2007;211:104–116. doi: 10.1111/j.1469-7580.2007.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden HP, Maderson PF. Morphological and biophysical identification of fibrous proteins in the amniote epidermis. J Exp Zool. 1970;174:225–232. doi: 10.1002/jez.1401740211. [DOI] [PubMed] [Google Scholar]
- Baden H, Sviokla S, Roth I. The structural protein of reptilian scales. J Exp Zool. 1974;187:287–294. doi: 10.1002/jez.1401870212. [DOI] [PubMed] [Google Scholar]
- Brush AH. Self-assembly of avian φ-keratins. J Prot Chem. 1983;2:63–75. [Google Scholar]
- Brush AH. The origin of feather: a novel approach. In: Farner D, Kling J, Parker K, editors. Avian Biology. New York: Academic Press; 1993. pp. 121–162. [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Valle L, Toffolo V, Belvedere P, Alibardi L. Isolation of a mRNA encoding a glycine-proline-rich beta-keratin expressed in the regenerating epidermis of lizard. Dev Dyn. 2005;234:934–947. doi: 10.1002/dvdy.20581. [DOI] [PubMed] [Google Scholar]
- Dalla Valle L, Nardi A, Toffolo V, Niero C, Toni M, Alibardi L. Cloning and characterization of scale beta-keratins in the differentiating epidermis of geckoes show they are glycine-proline-serine-rich proteins with a central motif homologous to avian beta-keratins. Dev Dyn. 2007a;236:374–388. doi: 10.1002/dvdy.21022. [DOI] [PubMed] [Google Scholar]
- Dalla Valle L, Nardi A, Belvedere P, Toni M, Alibardi L. Beta-keratins of differentiating epidermis of snake show that they are glycine-proline-rich proteins with an avian-like gene organizaton. Dev Dyn. 2007b;236:1939–1953. doi: 10.1002/dvdy.21202. [DOI] [PubMed] [Google Scholar]
- Dalla Valle L, Nardi A, Gelmi C, Toni M, Alibardi L. Crocodilian epidermis contains glycine-proline-tyrosine rich beta-keratins and a gene structure more similar to that of birds than to lepidosaurian reptiles. J Exp Zool B. 2008 in press. [Google Scholar]
- Fraser RD, Parry DA. The molecular structure of reptilian keratin. Int J Biol Macromol. 1996;19:207–211. doi: 10.1016/0141-8130(96)01129-4. [DOI] [PubMed] [Google Scholar]
- Fraser RD, Parry DA. Molecular packing in the feather keratin filament. J Struct Biol. 2008;162:1–13. doi: 10.1016/j.jsb.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Fraser RD, MacRae TP, Rogers GE. Keratins: Their Composition, Structure and Biosynthesis. Springfield, IL: Thomas; 1972. [Google Scholar]
- Fuchs E, Marchuk D. Type I and type II keratins have evolved from lower eukaryotes to form the epidermal intermediate filaments in mammalian skin. Proc Natl Acad Sci U S A. 1983;80:5857–5861. doi: 10.1073/pnas.80.19.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg K, Rogers G. Feather keratins: composition, structure and biogenesis. In: Bereither-Hahn J, Matoltsy G, Sylvia-Richards K, editors. Biology of the Integument, Vertebrates. New York: Springer-Verlag; 1986. pp. 666–694. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Homer BL, Li C, Berry KH, et al. Soluble scute proteins of healthy and ill desert tortoises Gopherus agassizii. Am J Vet Res. 2001;62:104–110. doi: 10.2460/ajvr.2001.62.104. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kalinin AE, Kajava AV, Steinert PM. Epithelial barrier function: assembly and structural features of the cornified cell envelope. Bioessays. 2002;24:789–800. doi: 10.1002/bies.10144. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Landmann L. The skin of Reptiles: epidermis and dermis. In: Bereither-Hahn J, Matoltsy G, Sylvia-Richards K, editors. Biology of the Integument, Vertebrate. Berlin: Springer Verlag; 1986. pp. 150–187. [Google Scholar]
- Maderson PF. Some developmental problems of the reptilian integument. In: Gans C, Billett F, Maderson PF, editors. Biology of Reptilia. New York: John Wiley & Sons; 1985. pp. 525–598. [Google Scholar]
- McGuffin L. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Parakkal PF, Alexander NJ. Keratinization: A Survey of Vertebrate Epithelia. New York: Academic Press; 1972. [Google Scholar]
- Quang Le S, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Rest JS, Ast JC, Austin CC, Waddell PJ, Tibbetts EA, Hay JM, Mindell DP. Molecular systematics of primary reptilian lineages and the tuatara mitochondrian genome. Mol Phylogenet Evol. 2003;29:289–297. doi: 10.1016/s1055-7903(03)00108-8. [DOI] [PubMed] [Google Scholar]
- Sawyer RH, Glenn TC, French JO, et al. The expression of beta (β) keratins in the epidermal appendages of reptiles and birds. Am Zool. 2000;40:530–539. [Google Scholar]
- Scala C, Cenacchi G, Ferrari C, Pasquinelli G, Preda P, Manara GC. A new acrylic resin formulation: a useful tool for histological, ultrastructural, and immunocytochemical investigations. J Histochem Cytochem. 1992;40:1799–1804. doi: 10.1177/40.11.1431065. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland, MA: Sinauer Associates; 1998. Version 4. [Google Scholar]
- Toni M, Dalla Valle L, Alibardi L. The epidermis of scales in gecko lizards contains multiple forms of beta-keratins including basic glycine-proline-serine-rich proteins. J Proteome Res. 2007a;6:1792–1805. doi: 10.1021/pr060626+. [DOI] [PubMed] [Google Scholar]
- Toni M, Dalla Valle L, Alibardi L. Hard (beta-)keratins in the epidermis of reptiles: composition, sequence, and molecular organization. J Proteome Res. 2007b;6:3377–3392. doi: 10.1021/pr0702619. [DOI] [PubMed] [Google Scholar]
- Wyld JA, Brush AH. The molecular heterogeneity and diversity of reptilian keratins. J Mol Evol. 1979;12:331–347. doi: 10.1007/BF01732028. [DOI] [PubMed] [Google Scholar]
- Wyld JA, Brush AH. Keratin diversity in the reptilian epidermis. J Exp Zool. 1983;225:387–396. [Google Scholar]
- Yang Z. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]