Summary
The transcriptional status of a gene can be maintained through multiple rounds of cell division during development. This epigenetic effect is believed to reflect heritable changes in chromatin folding and histone modifications or variants at target genes, but little is known about how these chromatin features are inherited through cell division. A particular challenge for maintaining transcription states is DNA replication, which disrupts or dilutes chromatin associated proteins and histone modifications. PRC1-class Polycomb Group protein complexes are essential for development, and are thought to heritably silence transcription by altering chromatin folding and histone modifications. It is not known whether these complexes and their effects are maintained during DNA replication or subsequently re-established. We find that when PRC1-class Polycomb complex-bound chromatin or DNA is replicated in vitro, Polycomb complexes remain bound to replicated templates. Retention of Polycomb proteins through DNA replication may contribute to maintenance of transcriptional silencing through cell division.
Introduction
The hypothesis that chromatin structure is the basis for many epigenetic phenomena implies that it can be propagated through cell division. Transfer of chromatin based gene regulatory information through DNA replication would require that specific chromatin features (chromatin folding, chromatin associated proteins, histone variants, and histone modifications) are inherited or recreated with fidelity (Ng and Gurdon, 2008; Nightingale et al., 2006). Pioneering in vitro studies demonstrated that histones remain associated with DNA through replication, even in the absence of cellular factors (Bonne-Andrea et al., 1990); reviewed in (Annunziato, 2005). In vivo, parental histones and at least some histone modifications are inherited by daughter chromatin during DNA replication, although the mechanistic details of this process are not known (Annunziato, 2005; Benson et al., 2006). Extensive data linking histone modifications and their recognition by regulatory proteins including histone modifying enzyme complexes have led to a model for propagation of gene regulatory information through cell division. In this model, modified histones are transferred from parent to daughter DNA, where they recruit enzyme complexes that propagate the same modifications to newly added histones (Dodd et al., 2007; Ng and Gurdon, 2008; Nightingale et al., 2006). However, there is no direct evidence yet for this model in the context of DNA replication and the fate of most non-histone chromatin-associated epigenetic factors during DNA replication has not been determined.
Drosophila Polycomb Group (PcG) proteins maintain transcriptional silencing and are believed to act through epigenetic mechanisms (Grimaud et al., 2006). The best understood PcG target genes are the homeotic (Hox) genes, which control segmental identities during development. Transcriptional repression of Hox genes in specific segments early in development is essential to segmental identity and is initially carried out by transiently expressed transcription factors. Once established, Hox gene repression is maintained by PcG proteins. This regulation is believed to be epigenetic in that it is stable through multiple rounds of cell division despite the decay of early acting transcription factors (Ringrose and Paro, 2004; Simon and Tamkun, 2002).
Recent work in Drosophila and mammals implicates PcG proteins in biological functions beyond stable regulation of Hox genes. Genome-wide studies of PcG protein binding indicate that PcG regulation can be dynamic according to cell type and differentiation stage (reviewed in Pietersen and van Lohuizen, 2008; Ringrose, 2007; Schwartz and Pirrotta, 2008) (Kwong et al., 2008). PcG proteins also direct a range of gene expression levels not restricted to silencing. Early studies of Polycomb Response Elements (PREs), the DNA recognition elements that target PcG proteins in Drosophila, indicated that PcG repression involves recognition of both PREs and transcriptional status (Poux et al., 2001; Ringrose and Paro, 2004). PcG proteins maintain silencing of genes that are transcriptionally silenced early in development, but do not silence genes that are active early in development. More recent studies suggest that PcG proteins may be targeted to genes that are poised for activation, and that PcG proteins play a central role in ES cell pluripotency and differentiation by maintaining this potentiated state (Kwong et al., 2008; Pietersen and van Lohuizen, 2008).
Biochemical studies suggest that PcG proteins function in multiprotein complexes, several of which have been characterized. Polycomb Repressive Complex 1 (PRC1) includes four core PcG subunits: Polyhomeotic (Ph), Posterior Sex Combs (PSC), dRING1, and Polycomb (Pc) (Francis et al., 2001; Lavigne et al., 2004; Levine et al., 2002; Saurin et al., 2001; Shao et al., 1999). A reconstituted complex of these four proteins or three of the four can non-covalently alter chromatin and DNA structure, and inhibit chromatin remodeling and transcription of both naked DNA and chromatin (Francis et al., 2004; Francis et al., 2001; King et al., 2002; Mohd-Sarip et al., 2006; Shao et al., 1999). Components of this complex also can function as an E3 ligase for ubiquitylation of histone H2A (Cao et al., 2005; de Napoles et al., 2004; Elderkin et al., 2007; Wang et al., 2004a) probably in the context of a distinct complex (Lagarou et al., 2008). All of these activities correlate with silencing in vivo (Cao et al., 2005; Cao and Zhang, 2004; Grimaud et al., 2006; King et al., 2005; Lagarou et al., 2008; Ringrose and Paro, 2004; Wang et al., 2004a).
A second PcG complex, PRC2, whose components are also essential for gene silencing, can methylate histone H3 on lysine 27 (H3K27me). A component of PRC1, Pc, can recognize this methylation via its chromodomain (KD of the Pc chromodomain for an H3K27me3 histone peptide is 5 μM (Fischle et al., 2003)). This observation has led to the hypothesis that PRC2 is initially recruited to PREs, perhaps in part by the PRE-binding protein PHO, or the PHO-containing PHO Repressive Complex (Klymenko et al., 2006). PRC2 methylation of H3K27 leads to recruitment of PRC1 and silencing (Cao and Zhang, 2004; Wang et al., 2004b). If the H3K27me3 mark is transferred to daughter DNA during replication, PRC1-class complexes may be recruited back to their targets if they are disrupted by passage of the DNA replication fork. This model is supported by the presence of high levels of H3K27me3 around PREs and the loss of binding of Pc at PREs when expression or function of E(Z), the catalytic subunit of PRC2, is reduced (Cao and Zhang, 2004; Wang et al., 2004b). Recent work in mammalian cells indicates that PRC2 may also recognize H3K27me3, suggesting a mechanism for propagation of the methylation mark (Hansen et al., 2008).
PRC1 may also function independently of PRC2 and H3K27me3 in some cases. PHO, a protein that binds to many PREs (Brown et al., 1998; Fritsch et al., 1999; Mihaly et al., 1998) can directly contact and recruit a recombinant PRC1-class complex in vitro, and binds synergistically with the complex (Mohd-Sarip et al., 2005). Some PcG binding sites lose PRC1 components when PRC2 components are reduced, but others do not (Wang et al., 2004b), and H3K27me3 does not always co-localize with PcG binding (Ringrose et al., 2004). In early murine embryos, PRC1 binds to heterochromatin in the paternal genome independent of PRC2 and H3K27me3 (Puschendorf et al., 2008) and PRC1 components are targeted to the imprinted locus Kcnq1 independent of Ezh2 (Terranova et al., 2008). In an ES cell model for steps in X inactivation, at least one PRC1 component (Ring1B) can be recruited in the absence of PRC2 and H3K27me3 (Schoeftner et al., 2006).
The focus on H3K27me3 and histone modification in general as the principle means by which chromatin may transmit epigenetic memory through replication implicitly assumes that chromatin proteins other than histones dissociate during the biochemical process of DNA replication. To test this idea, we examined the effect of DNA replication on the association of PRC1 class PcG protein complexes with chromatin and naked DNA in vitro.
Results
Inhibition of chromatin remodeling by PCC is preserved through chromatin replication in vitro
To test the effect of DNA replication on PCC-bound chromatin, PCC was bound to chromatin templates, followed by DNA replication in the presence of α32P-dATP to radiolabel the DNA (Fig. 1A). We used the well characterized cell free Simian Virus 40 (SV40) DNA replication system (Li and Kelly, 1984; Stillman and Gluzman, 1985). The SV40 protein large T-Antigen (TAg) binds specifically to the replication origin of SV40 viral DNA or to plasmids containing this sequence to initiate bidirectional DNA replication. SV40 origin-containing plasmids were assembled into chromatin using the salt gradient dialysis method with histones purified from HeLa cells (templates and proteins are described in sFig. 1). TAg was next bound to the template, and S100 cytoplasmic extracts were added to initiate DNA replication.
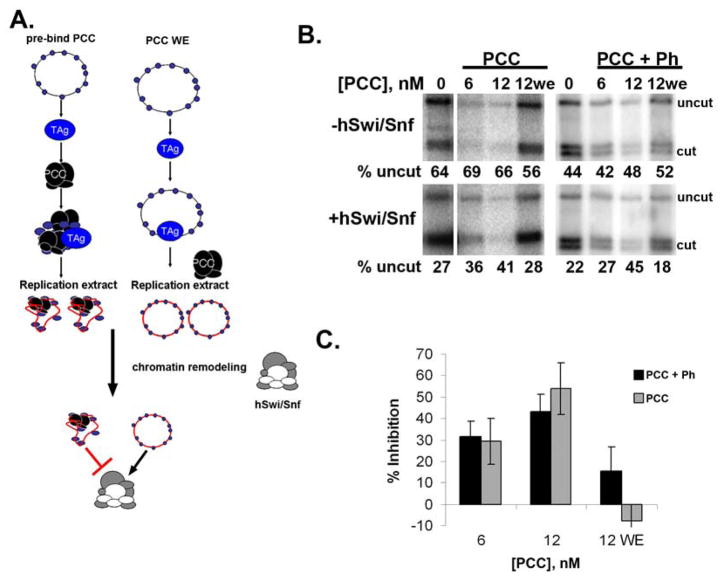
Figure 1. PCC inhibits chromatin remodeling of replicated templates.
A. Reaction scheme for in vitro replication followed by chromatin remodeling. The scheme on the right is the control in which PCC is added with the replication extract (WE). B. Example of restriction enzyme accessibility assay carried out after in vitro replication of chromatin. ATP-dependent chromatin remodeling by hSwi/Snf decreases the fraction of the template that is uncut (compare top and bottom panels); this reaction is inhibited when PCC is bound to chromatin prior to in vitro replication. Panel shows phosphorimager scan of chromatin remodeling reactions so that only replicated templates (that incorporated radiolabeled dATP during replication) are visible. PCC+Ph contains PSC, Pc, dRING, and Ph, while PCC has only PSC, Pc, dRING (see sFig. 1E). C. Summary of inhibition of chromatin remodeling of replicated templates. Error bars in all Figures are SEM unless otherwise indicated.
An aliquot of each replication reaction was used in a restriction enzyme accessibility assay (Francis et al., 2001; Logie and Peterson, 1997; Polach and Widom, 1995). In this assay, chromatin remodeling by hSwi/Snf increases restriction enzyme accessibility and this effect is blocked by PCC (Francis et al., 2001). All of the experiments reported here were carried out with Drosophila PRC1 core complex (PCC) containing Posterior Sex Combs (PSC), dRING, and Polycomb (Pc); in some experiments, Polyhomeotic (Ph) was included (which we refer to as PCC+ Ph). Although the original studies with PCC were carried out with the four protein complex, in our previous work and in these studies, complexes with and without Ph behaved identically (Francis et al., 2004; Francis et al., 2001; Lavigne et al., 2004).
When PCC was bound to chromatin before replication, less chromatin remodeling occurred on replicated templates than in reactions without PCC (Fig. 1B, C). This effect is dose dependent, and occurs over a similar range as previously described inhibition of chromatin remodeling (partial inhibition is observed at one complex for 3–5 nucleosomes, or ~5 PCCs per plasmid). In these experiments, both PCC and nucleosomes are above their KD for interaction, and nucleosomes are in excess of PCC (see supplemental material and sFig. 2 for further explanation). When PCC is added with the replication extracts instead of pre-bound to chromatin (indicated as “WE” in Figures), inhibition of chromatin remodeling is not observed, suggesting binding of PCC to chromatin is prevented in extracts. Thus, chromatin templates bound to PCC prior to DNA replication behave as though PCC is bound after DNA replication in that they are refractory to chromatin remodeling.
PCC-bound templates are completely replicated
The PCC-like effects observed on newly replicated chromatin could indicate that PCC, or its effect on chromatin, are preserved through DNA replication. Alternatively, PCC-bound templates may be partially replicated and PCC bound to unreplicated segments. This possibility was of particular concern because PCC inhibits replication in a dose dependent manner (sFig. 3). If PCC blocks passage of DNA replication forks, then PCC bound templates should generate partially replicated products. In contrast, if PCC prevents replication initiation, then full-length products should be generated in reactions with PCC bound to the template. We used three assays to determine whether full-length or partial replication products are produced from PCC-bound templates (Fig. 2). Under our replication conditions, replication of chromatin is about 3 times less efficient than replication of naked DNA. To increase replication of chromatin, we included a fraction from Xenopus extracts that contains high levels of the histone chaperone nucleoplasmin (referred to as NPE-1000) (sFig. 4, 5) in some of our experiments.
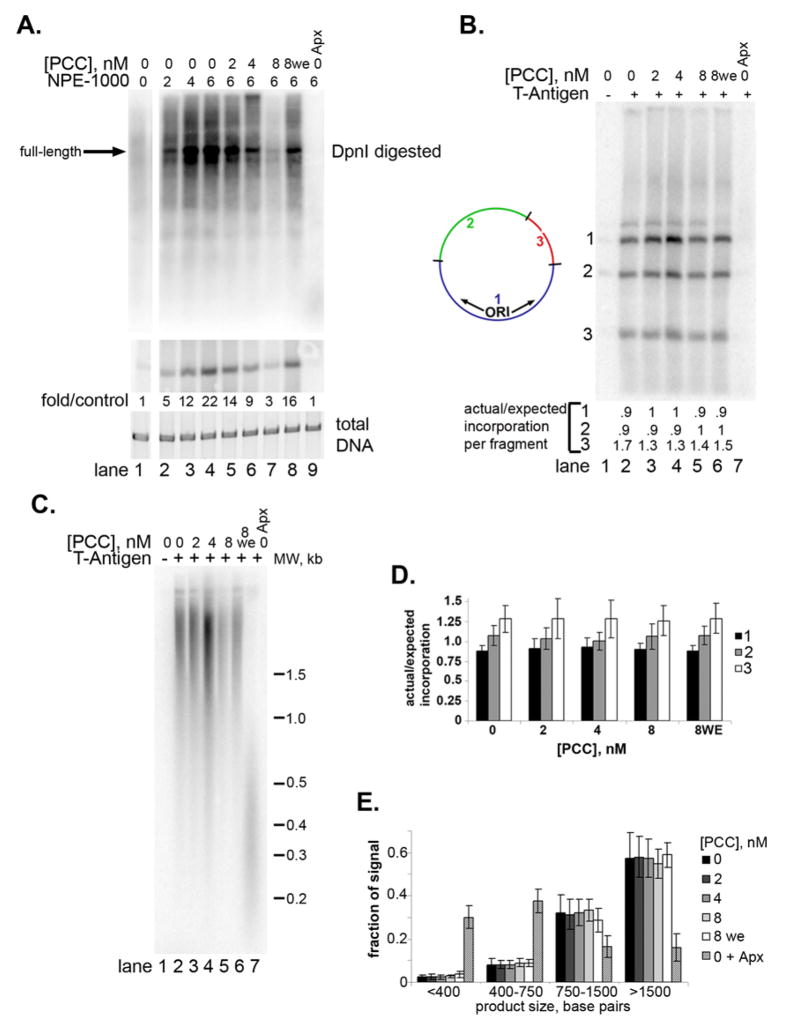
Figure 2. Full-length replication products are present after replication of PCC-bound chromatin and DNA.
A. Top panel is phosphorimager scan of chromatin replication reactions digested with DpnI. Middle panel is a phosphorimager scan of the same reaction products linearized with a restriction enzyme, and bottom panel shows total DNA (SYBR gold stain). NPE-1000 was added to stimulate replication (see sFig. 4, 5) B. Phosphorimager scan of replication products digested with three restriction enzymes as indicated. Replication should initiate in fragment 1. The ratio of actual/expected incorporation in each fragment is indicated below each lane. C. Denaturing gel of replication products to show length of nascent strand. D. Graph summarizing the actual/expected incorporation into each DNA fragment from experiments like the one in B. E. Graph summarizing the distribution of radioactivity in replication products analyzed by denaturing gel electrophoresis as in C. Each lane was divided into 4 segments and signals are expressed as a fraction of the total signal for the lane. Aphidicolin (Apx) was added after 25′ (lane 9 in A, 7 in B and C) to stall replication forks.
The restriction enzyme DpnI can distinguish replicated from unreplicated DNA. DpnI digests DNA that is methylated at adenine residues in its recognition site (GA′TC). Plasmids used in these experiments were propagated in bacteria and therefore are fully methylated at their 26 DpnI sites. Hemimethylated DNA in which one strand is methylated and the other is not is digested 60-fold more slowly than methylated DNA (Sanchez et al., 1992). Replicated DNA is hemimethylated and DpnI resistant. When replication products were digested with DpnI, DpnI-resistant, full-length products were observed (Fig. 2A, sFig. 6A) whether or not PCC was pre-bound to the chromatin. Full-length, DpnI-resistant products were also observed from naked DNA templates (sFig. 6D).
As a second means of determining whether full-length replication products were present, replicated plasmids were digested into three fragments, one of which contains the SV40 origin of replication. If replication initiates but fails to complete, then incorporation of radiolabeled nucleotide should occur preferentially near the origin (fragment 1 in Fig. 2B). The ratio of actual to expected incorporated radiolabeled nucleotide was close to 1 for each fragment for both naked DNA and chromatin, implying that the entire plasmid was replicated (Fig. 2B, D; sFig. 6B, C, E, F). We found that fragments 2 and 3 were consistently over-represented in reactions from chromatinized, but not naked DNA templates (compare 2D with sFig. 6F). We do not understand the reason for this discrepancy, but note that the ratio of incorporation into the three fragments is not altered by PCC. As a third means of detecting partial replication products, we used denaturing gels, which separate the nascent and template DNA strands (Fig. 2C, E). Large products were observed in both the presence and absence of PCC (Fig. 2C, lanes 3–6), but when aphidicolin was added to stall DNA polymerase, products less than 500 base pairs accumulate (Fig. 2C, lane 7; 2E). Both in the DpnI analysis and on denaturing gels, some partial replication products are detected, particularly from chromatin templates, but they are not increased when PCC is added.
These results demonstrate that PCC effects on chromatin persist through DNA replication in vitro. This could mean that PCC alters the template in a way that is stable through DNA replication, or that the complex itself is associated with the replicated templates. We therefore asked whether PCC is physically associated with replicated templates.
PCC remains bound to replicated chromatin and DNA
We first characterized the association of PCC with naked DNA and chromatin by sucrose gradient sedimentation. Chromatinized plasmids incubated with PCC sediment further into sucrose gradients than those incubated with buffer (sFig. 7A, B). Binding was not disrupted by competitor DNA, indicating that it is stable under these conditions. Sedimentation of labeled PCC in similar reactions indicates that it co-sediments with chromatin (sFig. 8A). Stable binding of PCC to naked DNA required a two-fold higher concentration of PCC (sFig. 7C, D). Thus, once PCC is bound to a template, it remains bound over the time scale relevant for in vitro DNA replication.
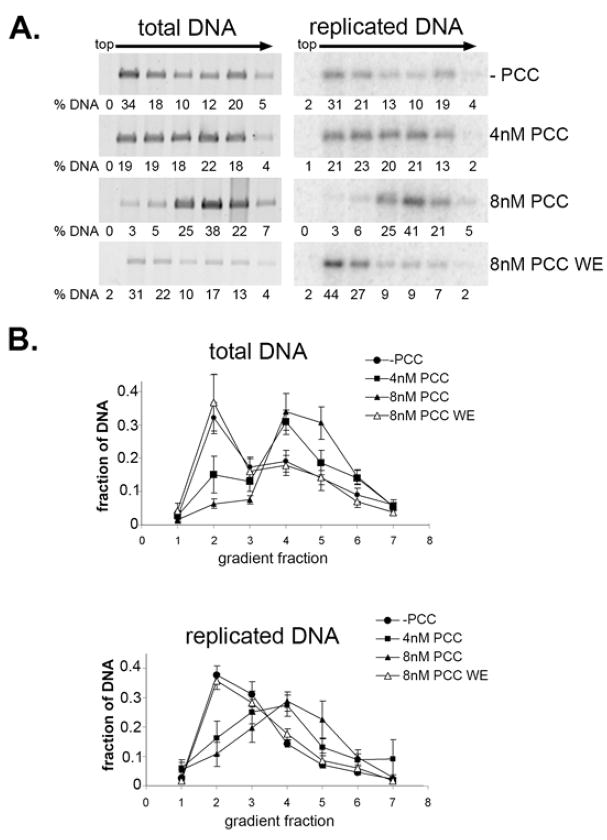
To determine whether replication dissociates PCC from chromatin, PCC-bound and unbound templates were separated by sucrose gradient sedimentation after DNA replication in vitro (Fig. 3). The DNA content of each fraction was monitored by agarose gel electrophoresis followed by SYBR gold staining, and replicated DNA was monitored by radiolabeled dATP incorporation. When PCC was pre-bound to chromatin before addition of replication extracts, both replicated and total chromatin sedimented further in the gradient than when PCC was added with replication extracts (Fig. 3A–C). Thus, PCC is likely physically associated with chromatin in replication extracts, and replication does not disrupt this association. This is further supported by the observation that labeled PCC co-sediments with chromatin templates (sFig. 8B). Little evidence for release of PCC from chromatin (which would cause more of the radiolabeled DNA to sediment in upper fractions) was observed. In contrast, when PCC is not pre-bound to chromatin but is added with the replication extract, both replicated and unreplicated chromatin sediments in the same position as chromatin in the absence of PCC (Fig. 3A–C).
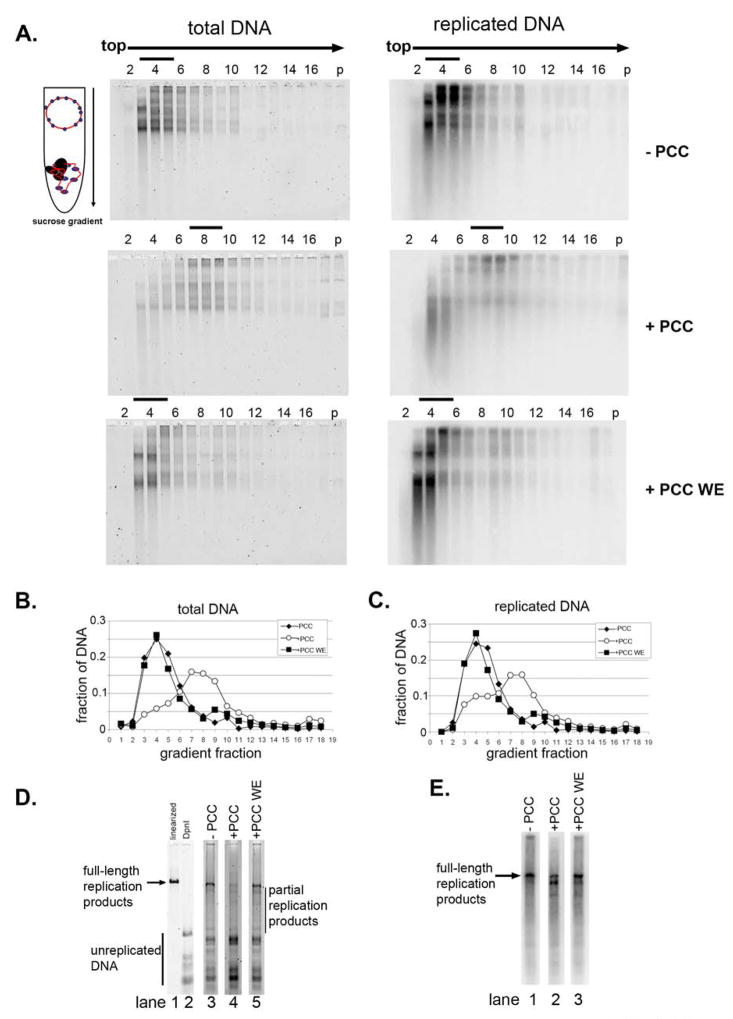
Figure 3. Association of PCC with chromatin following replication in vitro.
A. Agarose gels of sucrose gradient fractions from chromatin replication reactions. Left panel shows total DNA (SYBR gold) and right panels replicated DNA (phosphorimager scan). B, C. Quantification of gels shown in A. D. Fractions were pooled as indicated by black bars in A, and DNA was purified and digested with DpnI digestion; panel shows SYBR gold stained agarose gel. Lane 1 shows linearized template and lane 2 completely DpnI digested template. E. Phosphorimager scan of the reactions shown in D. Note that lane 2 is a longer exposure than 1 or 3 in D and E.
To determine whether the templates sedimenting in the position of PCC-bound chromatin were completely replicated, peak gradient fractions were pooled, and DNA was isolated from them and digested with DpnI. Full-length replication products were observed in fractions containing putative PCC- bound chromatin (lane 4 in Fig. 3D and lane 2 in Fig. 3E). These data suggest that when PCC is pre-bound to chromatin, it remains associated with chromatin after DNA replication. These observations were confirmed using mini sucrose gradients (sFig. 9A, B).
Replication reactions using naked DNA as a template were also fractionated by sucrose gradient sedimentation and again full-length, DpnI resistant replication reactions were observed in peak fractions from reactions without PCC, with PCC pre-bound, or with PCC added with the replication extract (sFig. 9C, D, and 10). In the experiments with naked DNA, we sometimes observed a small peak of replicated DNA sedimenting in the position of unbound templates, suggesting PCC is released from some templates during replication. From the sucrose gradient sedimentation experiments, we conclude that when PCC is pre-bound to chromatin or naked DNA before DNA replication, it is present on replicated chromatin or DNA.
PCC can be cross-linked to replicated templates
As a second means for testing whether PCC is bound to chromatin after DNA replication, we used biotinylated PCC to precipitate PCC-bound templates (sFig. 1E). Replication reactions were cross-linked with formaldehyde and biotinylated PCC- bound chromatin was recovered using streptavidin coated beads. A substantial fraction of both total and replicated chromatin was retained on the beads when templates were pre-bound with PCC (Figure 4A, B), implying that PCC remains bound to both replicated and unreplicated chromatin. A greater fraction of total than replicated chromatin was retained on the beads, suggesting PCC can dissociate during replication (Fig. 4B). This difference is significant for low but not high concentrations of PCC and may be overestimated since templates with fewer PCC bound are more likely to replicate, but less likely to be captured by streptavidin coated beads. Only a fraction of the PCC is captured under our conditions, which might explain capture of only part of the template (sFig. 11).
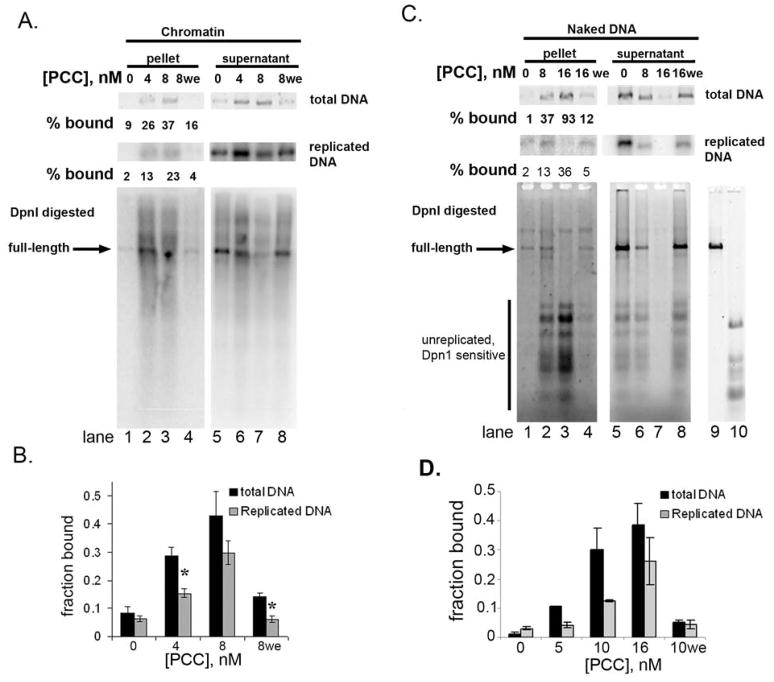
Figure 4. PCC can be cross-linked to replicated templates.
A. Precipitation of replicated chromatin with biotinylated PCC after cross-linking. Top panels show total and replicated (radioactive) DNA and bottom panel phosphorimager scan of the same reactions digested with DpnI. Lanes 1, 4, 5 and 8 contain 6% of the total reaction, lanes 2 and 6 18%, and lanes 3 and 7 54%. B. Summary of fraction of replicated and total chromatin isolated with biotinylated PCC after replication of chromatin. Asterisks indicate that the fraction of replicated DNA precipitated is significantly lower than of total DNA (p<0.05, two-tailed Student’s t-test). C, D. Same as A,B except with naked DNA templates. Equal amounts of each reaction were loaded. Bottom panel is SYBR gold stain of DpnI-digested replication products. Lanes 9 shows linearized and 10 completely DpnI digested template. In both A and C, longer exposures are shown for the pellets than supernatants.
When naked DNA was used as the template for replication reactions, PCC could also be cross-linked to replicated and unreplicated templates (Fig. 4C, D). A lower fraction of replicated than total DNA was captured when naked DNA was used as the template (compare Fig. 4A and C), suggesting nucleosomes help stabilize PCC on replicated or replicating templates. When PCC is added with the replication extracts, near background levels of chromatin or DNA are retained on the beads, consistent with the conclusion that PCC does not bind chromatin or DNA under these conditions (Fig. 4A, C lane 4). Bound and unbound replicated DNA was also digested with DpnI to confirm that PCC-bound chromatin and DNA can be fully replicated (Fig. 4A, C, bottom panels).
Together, the sucrose gradient sedimentation and precipitation experiments indicate that PCC is physically associated with replicated templates.
PCC does not transfer among templates during DNA replication
Our results indicate that if PCC is bound to chromatin or naked DNA prior to replication, then it is present on the replication products, but if PCC is added with the replication extracts, it is not bound to the replication product. The simplest explanation for these results is that PCC is transferred to newly replicated chromatin without being released into solution, since if it was released even transiently, our data predict that it would not be able to rebind. To address if PCC can transfer among DNA molecules during DNA replication, we carried out two types of experiments.
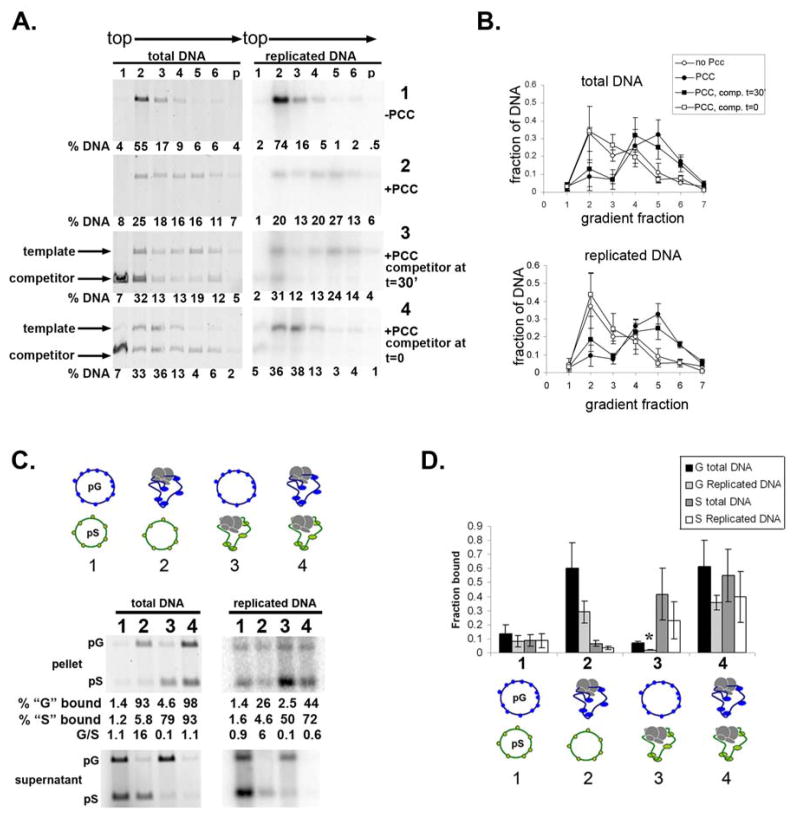
We used naked DNA without a replication origin as a competitor to capture any PCC that was destabilized during DNA replication. PCC was incubated with template or with template and competitor. Replication extract was added, either with or without competitor DNA. Replication was allowed to proceed, and products were analyzed by sucrose gradient sedimentation (Fig. 5A, B). We found that when PCC was pre-bound to the template, competitor added during DNA replication had no effect on the association of replicated templates with PCC (panel 3 of Fig. 5A). However, when the same amount of competitor was added during the binding reaction, PCC bound to the competitor and not detectably to the replicating template (panel 4).
Figure 5. PCC does not transfer to other DNA templates during replication.
A. Sucrose gradient sedimentation of replication reactions without competitor (panels 1 and PCC binding (t=0, panel 4). Replication products were linearized to distinguish template and competitor. B. Summary of sucrose gradient sedimentation experiments with competitor. C. Precipitation of chromatin following replication of mixtures of PCC-bound and unbound plasmids. In each reaction, one, both or neither plasmid was pre-bound with PCC as indicated. The plasmids were mixed when the replication extracts were added. The replicated DNA pellet samples were exposed for longer than the supernatant. For reaction 2, the amounts of replicated pG and pS in the bound fraction appear similar but the fraction of the replicated templates that is bound indicates preferential binding of pG. F. Summary of mixed plasmid experiments. Asterisks indicate cases where the fraction of replicated DNA that is precipitated with biotinylated PCC is significantly lower than the fraction of total DNA (p<0.05, two-tailed student’s t-test).
As a second means of assessing whether PCC can transfer among replicating templates, two plasmids that differ in size but both contain replication origins were used. Replication reactions in which both, one, or neither plasmid were pre-incubated with biotinylated PCC were carried out. We found that the plasmid that was pre-incubated with biotinylated PCC prior to replication is preferentially retained on the streptavidin coated beads (Fig. 5C, D). This was true both for replicated and unreplicated chromatin even though more replication is observed from the plasmid that was not pre-incubated with PCC. This suggests that significant transfer of PCC does not occur among replicating templates.
H3Kc27me3 chromatin allows replication and transfer of PCC
Our data indicate that PCC can likely transfer to newly replicated chromatin without being released into solution. Because this is also observed with naked DNA, it does not require nucleosomes although nucleosomes make the interaction between chromatin and PCC more efficient. In vivo, the PcG proteins in PCC are bound to PREs, which tend to be depleted of nucleosomes (Mishra et al., 2001; Mito et al., 2007; Papp and Muller, 2006). Areas surrounding PREs also usually contain H3K27me3 (Papp and Muller, 2006; Schwartz et al., 2006), a modification deposited by PRC2, and recognized by Pc, a component of PCC (Fischle et al., 2003; Min et al., 2003). This modification has been suggested to play a central role in targeting and maintaining PRC1-class complexes in vivo. Our results indicate that once PCC is bound to chromatin (in this case binding is driven by mass action), H3K27me3 is not required to maintain the binding of PCC through DNA replication. However, given that the Pc-H3K27me3 interaction is likely to occur at or near PREs in vivo, we wondered if H3K27me3 would affect the replication of PCC-bound chromatin or the association of PCC with chromatin through DNA replication.
Histone octamers including histone H3 with a methyl-lysine analogue (MLA) (Simon et al., 2007) at residue 27 that mimics H3K27me3 (referred to as H3Kc27me3) were assembled onto replication templates. PCC inhibits replication of H3Kc27me3 chromatin at similar concentrations to chromatin assembled with histones purified from cells (sFig. 12A, B) or produced in bacteria (not shown). In binding assays, PCC proteins bound similarly to H3Kc27me3 and unmodified templates. We were able to detect preferential binding to H3Kc27me3 chromatin in the context of an excess of naked DNA competitor (sFig. 12C, D), although the effect is modest. The most likely explanation for the results of our binding assays is that both PCC and nucleosomal templates are many fold above their KD in these experiments. Thus, it remains possible that there is a larger effect of H3Kc27me3 on binding affinity for PCC but that different conditions will be required to detect it. It is also possible that the methyl-lysine analogue binds less tightly to Pc than H3K27me3 and thus that these experiments underestimate the influence of H3K27me3.
To determine whether PCC remains associated with replicated chromatin that includes H3Kc27me3 through DNA replication, sucrose gradient sedimentation was used to separate bound and unbound templates after replication (Fig. 6). The results of these assays indicate that PCC remains associated with H3Kc27me3 templates. Thus, histones, and H3K27me3 are not required for maintenance of PCC complexes during DNA replication in vitro. However, both DNA replication and maintenance of PCC through DNA replication can occur in the context of H3Kc27me3 modified chromatin, which likely represents a more physiological substrate.
Figure 6. Association of PCC with chromatin that has H3Kc27me3 through DNA replication in vitro.
A. Sucrose gradient sedimentation of replication reactions carried out with H3Kc27me3 chromatin. These results are directly comparable to sFig. 9. B. Summary of multiple experiments with H3Kc27me3 chromatin. The H3Kc27me3 templates show a tendency to aggregate so that a fraction of the template migrates near the bottom of the gradient in the absence of PCC.
PcG proteins are bound to chromatin during S-phase and to newly replicated DNA in vivo
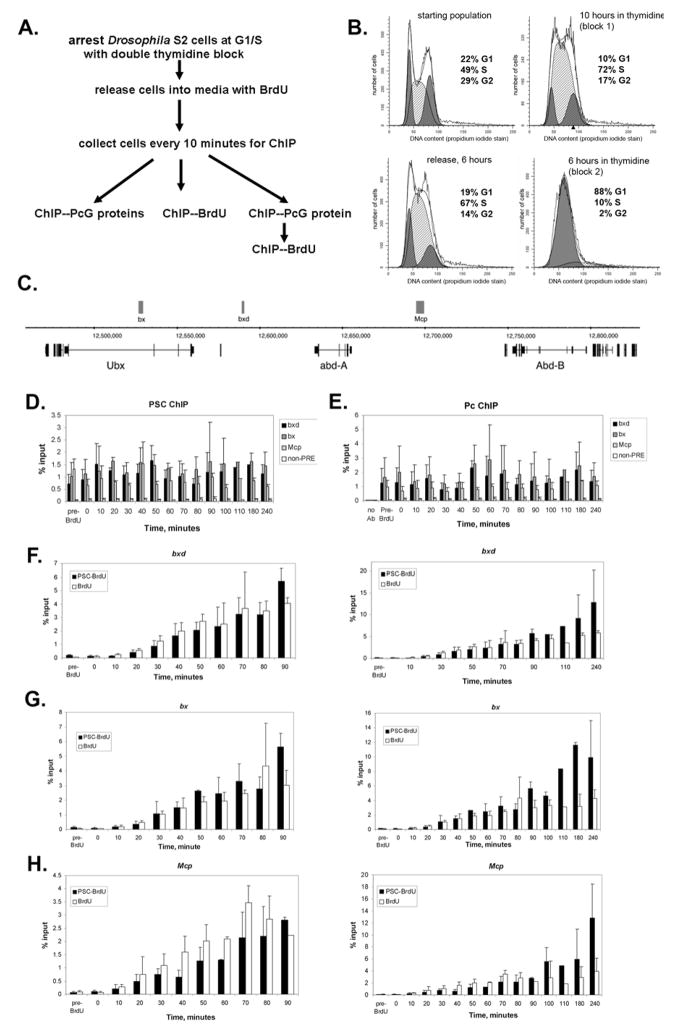
Our data predict that PcG proteins are present on chromatin throughout DNA replication. Although the methods to directly test this in vivo are not yet available, we wondered whether we could detect PcG proteins on newly replicated chromatin. To create a situation in which a large population of cells were replicating their DNA at the same time, we synchronized Drosophila S2 cells at G1/S using a double thymidine block (Jackman and O’Connor, 2003). Cells were released into media without thymidine but containing Bromodeoxyuridine (BrdU), and allowed to progress through S-phase. Aliquots of cells were collected every 10 minutes for two hours and used to analyze BrdU incorporation, PcG protein binding, and colocalization of the PcG protein PSC and BrdU at three PREs in the Bithorax complex (BX-C) homeotic gene cluster (Fig. 7C). Using antibodies to two PcG proteins in PRC1, Pc and PSC, we find that levels of PcG proteins at the three PREs are fairly constant over the entire time course, including when DNA replication is occurring (Fig. 7D, E). In control ChIPs without antibody, we recovered very little PRE DNA. Similarly, little DNA corresponding to a previously characterized PcG-negative heterochromatin region is recovered from ChIP experiments using antibodies to PSC or Pc (Fig. 7D,E) (Papp and Muller, 2006).
Figure 7. Association of PcG proteins from PRC1 with Polycomb Response Elements during DNA replication.
A. Strategy for analysis of PcG protein binding during DNA replication in S2 cells. B. Representative FACS analysis of S2 cells arrested at the G1/S boundary using a double thymidine block. The final panel shows cells after 6 hours of the final thymidine treatment and is representative of the cell population at the start of the time course (8 hours). C. Schematic diagram of part of the BX-C showing the position of three PREs used for this study, bx, bxd, and Mcp, and the position of the three homeotic genes in the complex (adapted from Fig. 4 of (Kwong et al., 2008). D, E. ChIP for PSC (D) and Pc (E) at PREs and a negative control site throughout the time course of DNA replication. F–H. Time course of BrdU incorporation (% input) and PSC-BrdU co-occupancy at three PREs over the time course of DNA replication. Data from sequential anti-PSC--anti-BrdU ChIPs are expressed as % of PSC-ChIP (since the PSC-ChIP elution is the input material for the BrdU IP in the sequential ChIP paradigm). The left panels show the first part of the time course, when replication is occurring (as demonstrated by BrdU incorporation). Right panels show the full four hour time course. Replication is largely completed by 90 minutes after release from the thymidine block for the sites analyzed; note that the broad peak of replication likely reflects imprecise synchronization of the cells, with some cells arrested in early S-phase and others in G1. Bars in all graphs show the average of three experiments and error bars show standard deviation.
The pattern of BrdU incorporation indicates that the BX-C is replicated within 90 minutes of release from the thymidine block, with the bulk of BrdU incorporation occurring between 20 and 60 minutes (Fig. 7F–H). To monitor PcG protein binding to replicated DNA, we first carried out ChIP for PSC, a component of PRC1. The PSC bound DNA was purified and antibodies to BrdU used to immunoprecipitate BrdU labeled DNA. We compared the fraction of DNA immunoprecipitated with anti-BrdU to the fraction of PSC-bound DNA immunoprecipitated with anti-BrdU and find that the enrichment of PSC-BrdU exactly follows the pattern of BrdU incorporation at the three PREs over the time course of replication (Fig. 7F–H, left panels). The specificity of the BrdU immunoprecipitation is indicated by the low level of DNA recovered from the chromatin harvested before addition of BrdU (“pre-BrdU”), or at the start of the time course (time =0). These data are consistent with PSC being bound to DNA during or shortly after replication of DNA, although the time scale of these experiments would not detect rapid dissociation and rebinding.
We continued to monitor PSC association with BrdU labeled DNA out to 4 hours after release from the block. Interestingly, we find that the level of PSC associated with BrdU labeled PRE DNA continues to increase after replication of these sequences is complete (Fig. 7F–H, right panels). These data are consistent with a model in which PSC bound before DNA replication is maintained on the daughter templates, and additional PSC is added after replication to restore the full complement required for silencing although higher resolution methods will be needed to confirm this.
Discussion
We examined the effect of DNA replication on the association of PRC1-class complexes with chromatin and DNA using a cell free system. Our principle finding is that PRC1 class complexes bound to chromatin or DNA remain associated during DNA replication in vitro. Our results suggest that transfer of chromatin regulatory proteins may be a mechanism for epigenetic inheritance through cell division.
How is PCC retained on replicating templates?
We do not know the mechanism by which PCC is retained through DNA replication. However, our data suggest that the complexes are not released into solution during passage of the DNA replication fork. Furthermore, they suggest that nucleosomes facilitate PCC binding to and retention on templates, but are not essential for either. The finding that PCC can be maintained on either chromatin or naked DNA is interesting in light of the finding that PREs are sites of rapid histone turnover and are depleted of nucleosomes (Mishra et al., 2001; Mito et al., 2007; Papp and Muller, 2006).
One model for the transfer of PCC during DNA replication is that the complexes remain in direct contact with DNA during passage of the DNA replication fork. Contacts between PcG proteins and nucleosomes or DNA could be disrupted in front of the replication fork, but replaced by contacts with nucleosomes or DNA behind the replication fork. This mechanism has been proposed for transfers of histone-DNA contacts during replication and transcription in vitro (Bonne-Andrea et al., 1990; Clark and Felsenfeld, 1992; Studitsky et al., 1994). PCC can likely contact multiple nucleosomes or a long stretch of DNA (Francis et al., 2004; Mohd-Sarip et al., 2006) which may allow the complex to remain on chromatin when some of these contacts are disrupted. A second model is that PCC interacts with the replication machinery, either directly, or through intermediary factors. These interactions would retain PCC near chromatin or DNA during replication, even if contacts between PCC and DNA are disrupted, allowing them to rapidly rebind newly replicated chromatin. Consistent with this idea, several chromatin modifying proteins can interact with components of the DNA replication machinery (Groth et al., 2007; Kohn et al., 2008).
PCC inhibits DNA replication in vitro
We observe strong inhibition of DNA and chromatin replication by PCC, raising the question of how PcG-bound regions are replicated if PRC1-class complexes are indeed continuously bound. If PCC inhibits replication initiation, but not elongation as our results suggest, then PRC1-class complexes would limit replication if they were bound near replication origins, but replication forks originating outside PcG-bound regions would be able to replicate through them. Consistent with this idea, targeting of Pc to a replication origin in Drosophila that mediates developmental chorion gene amplification in follicle cells decreased gene amplification (Aggarwal and Calvi, 2004) and PcG silenced regions of polytene chromosomes (such as Hox gene clusters) are under-replicated (Marchetti et al., 2003; Moshkin et al., 2001).
How might retention of PCC through DNA replication contribute to heritable transcriptional silencing?
The Polycomb Response Elements that recruit PcG proteins in vivo, like the plasmids used here, likely bind several copies of the complex. Some of these complexes may be inherited by each daughter, by the mechanism we demonstrate here. Reduction of PcG protein levels leads to reactivation of their target genes, suggesting that these genes are continuously susceptible to transcriptional activation (see for example (Beuchle et al., 2001; Breiling et al., 2001; Cao et al., 2002; Wang et al., 2004b). It may therefore be important that PRC1-class complexes, which may be directly involved in transcriptional silencing, maintain constant association with genes marked for silencing. Alternatively, the inherited complexes might be important for maintaining long range association between distal regulatory elements or trans interactions among PcG repressed genes (Bantignies et al., 2003; Lanzuolo et al., 2007). These associations, in turn, might prevent transcriptional activation after DNA replication by keeping newly replicated chromatin in PcG-rich nuclear compartments.
We were surprised to find that H3K27me3 is not essential for maintaining PRC1-class complexes through DNA replication in vitro. It is possible that retention of parental PRC1-class complexes and recruitment of new complexes are mechanistically distinct because we do not find evidence for recruitment of new PCC during replication and our in vivo data suggest PSC is present on newly replicated chromatin but that additional PSC is recruited after replication. This may be similar similar to histone proteins in that it is thought that parental histones are transferred randomly to the two daughter strands, followed by deposition of new histones by replication-coupled assembly complexes (reviewed in Groth et al., 2007). In the case of PRC1-class complexes, our in vivo data raise the possibility that addition of new complexes is not directly coupled to DNA replication. H3K27me3 might be important for recruitment of new PRC1-class complexes.
In our experiments, PCC interacts with chromatin through mass action, but in vivo, PRC1-class complexes are specifically targeted to PREs. This likely involves contacts between PcG proteins and multiple transcription factors (Mohd-Sarip et al., 2005; Ringrose and Paro, 2007), as well as H3K27me3. We hypothesize that the stable association of PCC with chromatin that we observe here reflects how the complex could behave once it is recruited to a PRE, but it will be important to test this mechanism in a system where PCC is targeted.
In conclusion, the ability of parental PCC to be transferred to daughter chromatin may help explain how PcG-mediated repression established by transiently acting factors can be propagated through cell generations. They also raise the possibility that the maintenance of chromatin regulatory proteins, in addition to histones, through DNA replication might be an important mechanism of epigenetic inheritance.
Experimental Procedures
Detailed explanations of procedures, reagents, and buffer compositions and supporting Figures are presented in the supplemental materials.
Supplementary Material
Acknowledgments
We thank R. Kingston, T. Maniatis, W.M. Michael, N. Kleckner, A. Murray, A. Mazo, L. Kobrossy, B. Lengsfeld, and S. Lo for advice and support throughout the project and J. Müller, R. Jones, B. Stillman, and W. M. Michael for reagents. This work was supported by the NIH (GM078456-01), a Searle Scholar award and a March of Dimes Basil O’Connor Scholar award to N.J.F., and a Helen Hay Whitney fellowship to M.D.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 2003;17:2406–2420. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, Strack CL, Cook RG, Mizzen CA, Annunziato AT. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J Biol Chem. 2006;281:9287–9296. doi: 10.1074/jbc.M512956200. [DOI] [PubMed] [Google Scholar]
- Beuchle D, Struhl G, Muller J. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development. 2001;126:993–1004. doi: 10.1242/dev.128.6.993. [DOI] [PubMed] [Google Scholar]
- Bonne-Andrea C, Wong ML, Alberts BM. In vitro replication through nucleosomes without histone displacement. Nature. 1990;343:719–726. doi: 10.1038/343719a0. [DOI] [PubMed] [Google Scholar]
- Breiling A, Turner BM, Bianchi ME, Orlando V. General transcription factors bind promoters repressed by Polycomb group proteins. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Clark DJ, Felsenfeld G. A nucleosome core is transferred out of the path of a transcribing polymerase. Cell. 1992;71:11–22. doi: 10.1016/0092-8674(92)90262-b. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- Elderkin S, Maertens GN, Endoh M, Mallery DL, Morrice N, Koseki H, Peters G, Brockdorff N, Hiom K. A phosphorylated form of Mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol Cell. 2007;28:107–120. doi: 10.1016/j.molcel.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a Polycomb Group Protein Complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core Polycomb repressive complex. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- Fritsch C, Brown JL, Kassis JA, Muller J. The DNA-binding Polycomb group protein pleiohomeiotic mediates silencing of a Drosophila homeotic gene. Dev. 1999;126:3905. doi: 10.1242/dev.126.17.3905. [DOI] [PubMed] [Google Scholar]
- Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Jackman J, O’Connor PM. Methods for Synchronizing Cells at Specific Stages of the Cell Cycle. In: Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada KM, editors. Current Protocols in Cell Biology. Chapter 8.3. Hoboken, NJ: John Wiley & Sons, Inc; 2003. [DOI] [PubMed] [Google Scholar]
- King IF, Emmons RB, Francis NJ, Wild B, Muller J, Kingston RE, Wu CT. Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol Cell Biol. 2005;25:6578–6591. doi: 10.1128/MCB.25.15.6578-6591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IFG, Francis NJ, Kingston RE. Native and recombinant Polycomb-Group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol Cell Biol. 2002;22:7919–7928. doi: 10.1128/MCB.22.22.7919-7928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Muller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn KW, Aladjem MI, Weinstein JN, Pommier Y. Chromatin Challenges during DNA Replication: A Systems Representation. Mol Biol Cell. 2008;19:1–7. doi: 10.1091/mbc.E07-06-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong C, Adryan B, Bell I, Meadows L, Russell S, Manak JR, White R. Stability and dynamics of polycomb target sites in Drosophila development. PLoS Genet. 2008;4:e1000178. doi: 10.1371/journal.pgen.1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 2008;22:2799–2810. doi: 10.1101/gad.484208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Lavigne M, Francis NJ, King IF, Kingston RE. Propagation of silencing: recruitment and repression of naive chromatin in trans by Polycomb repressed chromatin. Mol Cell. 2004;13:415–425. doi: 10.1016/s1097-2765(04)00006-1. [DOI] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6080–6080. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Kelly TJ. Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logie C, Peterson CL. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 1997;16:6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti M, Fanti L, Berloco M, Pimpinelli S. Differential expression of the Drosophila BX-C in polytene chromosomes in cells of larval fat bodies: a cytological approach to identifying in vivo targets of the homeotic Ubx, Abd-A and Abd-B proteins. Development. 2003;130:3683–3689. doi: 10.1242/dev.00587. [DOI] [PubMed] [Google Scholar]
- Mihaly J, Mishra RK, Karch F. A conserved sequence motif in Polycomb-response elements. Mol Cell. 1998;1:1065–1066. doi: 10.1016/s1097-2765(00)80107-0. [DOI] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg SE, Schedl P. The iab-7 Polycomb Response Element maps to a nucleosome-free region of chromatin and requries both GAGA and Pleiohomeotic for silencing activity. Mol Cell Biol. 2001;21:1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- Mohd-Sarip A, Cleard F, Mishra RK, Karch F, Verrijzer CP. Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev. 2005;19:1755–1760. doi: 10.1101/gad.347005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer CP. Architecture of a polycomb nucleoprotein complex. Mol Cell. 2006;24:91–100. doi: 10.1016/j.molcel.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Moshkin YM, Alekseyenko AA, Semeshin VF, Spierer A, Spierer P, Makarevich GF, Belyaeva ES, Zhimulev IF. The Bithorax complex of Drosophile melanogaster: underreplication and morphology in polytene chromosomes. Proc Natl Acad Sci USA. 2001;98:570–574. doi: 10.1073/pnas.021353598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- Nightingale KP, O’Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev. 2006;16:125–136. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- Poux S, McCabe D, Pirrotta V. Recruitment of components of Polycomb Group chromatin complexes in Drosophila. Development. 2001;128:75–85. doi: 10.1242/dev.128.1.75. [DOI] [PubMed] [Google Scholar]
- Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 2008;40:411–420. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- Ringrose L. Polycomb comes of age: genome-wide profiling of target sites. Curr Opin Cell Biol. 2007;19:290–297. doi: 10.1016/j.ceb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Ehret H, Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell. 2004;16:641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Sanchez JA, Marek D, Wangh LJ. The efficiency and timing of plasmid DNA replication in Xenopus eggs: correlations to the extent of prior chromatin assembly. J Cell Sci. 1992;103(Pt 4):907–918. doi: 10.1242/jcs.103.4.907. [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. Embo J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu C-t, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Simon JA, Tamkun JW. Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr Opin Genet Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman BW, Gluzman Y. Replication and supercoiling of Simian Virus 40 DNA in cell extracts from human cells. Mol Cell Biol. 1985;5:2051–2060. doi: 10.1128/mcb.5.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studitsky VM, Clark DJ, Felsenfeld G. A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 1994;76:371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH. Polycomb Group Proteins Ezh2 and Rnf2 Direct Genomic Contraction and Imprinted Repression in Early Mouse Embryos. Dev Cell. 2008 doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004a;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004b;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.