Abstract
Colorimetric uranium sensors based on uranyl (UO22+) specific DNAzyme and gold nanoparticles (AuNP) have been developed and demonstrated using both labeled and label-free methods. In the labeled method, a uranyl-specific DNAzyme was attached to AuNP, forming purple aggregates. The presence of uranyl induced disassembly of the DNAzyme functionalized AuNP aggregates, resulting in red individual AuNPs. Once assembled, such a “turn-on” sensor is highly stable and worked in a single step at room temperature and had detection limit of 50 nM after 30 min. of reaction time. The label-free method, on the other hand, utilizes the different adsorption properties of single stranded and double stranded DNA on AuNPs, which affects the stability of AuNPs in the presence of NaCl. The presence of uranyl resulted in cleavage of substrate by DNAzyme, releasing a single stranded DNA that can be adsorbed on AuNPs and protect them from aggregation. Taking advantage of this phenomenon, a “turn-off” sensor was developed, which is easy to control through reaction quenching, and has 1 nM detection limit after 6 min. of reaction at room temperature. Both sensors have excellent selectivity over other metal ions and have detection limits below the maximum contamination level of 130 nM for UO22+ in drinking water defined by the US Environmental Protection Agency (EPA). The study represents the first direct systematic comparison of these two types of sensor methods using the same DNAzyme and AuNPs, making it possible to reveal advantages, disadvantages, versatility, and limitations and potential applications of each method. The results obtained not only allow practical sensing application for uranyl, but also serve as a guide for choosing different method for designing colorimetric sensors for other targets.
Introduction
Uranium is a radioactive metal that exists ubiquitously in the environment.1 Since uranium is one of the main sources in nuclear energy generation and enriched uranium is a major component in nuclear weapons, human beings have high chance to be exposed to uranium, which can cause severe adverse effects to human health.2, 3 For these reasons, detection of uranium is very important. However, current analytical techniques, such as inductively coupled plasma, atomic absorption spectrometry, phosphorimetry all require expensive and complicate instruments, making on-site real-time sensing difficult.4-7 Toward portable metal ion sensors, remarkable progresses have been made on the design of sensors using various techniques including fluorescence,8-17 surface plasmon resonance,18 electrochemistry,19-21 and colorimetry.22-25 Despite the progress, there are only a few reported sensors specific for uranium26-30 and most of them cannot yet match instrument-based detection in terms of sensitivity and selectivity. A contributing factor in the difficulty of designing sensors for uranium is that uranium has many forms in aqueous solution, and the most soluble or bioavailable form is uranyl (UO22+). Unlike most metal ions such as Zn2+, the oxycationic uranyl poses a special challenge for designing a ligand to bind it specifically.
DNA is generally known as a passive genetic informational storage material. In 1994, however, it was reported that DNA with active catalytic functions can been obtained through in vitro selection process from a large DNA library, especially in the presence of metal cofactors, and it is thus called catalytic DNA, deoxyribozymes or DNAzyme.31 Since then, a number of DNAzymes have been selected that are highly specific for metal ions such as Pb2+,8, 31 Cu2+,32-34 Zn2+,35 Co2+,36, 37 and Mn2+.38 Such a selection method has also been shown to be generally applicable to other forms of metal ions, including the oxycation UO22+.39 Recognizing the potential of DNAzymes as a new class of molecules specific for a wide range of metal ions, we and others have converted the DNAzymes into highly sensitive and selective fluorescent sensors using catalytic beacon method.8, 40-43 For example, recently reported uranyl (UO22+) specific DNAzyme fluorescence sensor has 45 pM detection limit and million fold selectivity,39 which rivals those of analytical instruments such as inductively coupled plasma mass spectrometry.
While fluorescent sensors are applicable for accurate on-site and real-time detection of metal ions, they still require portable fluorimeters. Colorimetric sensors gain a lot of interest nowadays since they have the advantage of allowing simple on-site real-time detection without instruments.44 There are only a few colorimetric sensors reported for uranium,27,30 but most of them are not selective and have interference with other metal ions. Therefore developing colorimetric uranium specific sensors with high sensitivity and selectivity is very important and highly desirable.
Metallic nanoparticles, especially gold nanoparticles [AuNPs], have emerged as a new class of reporters and have received much attention for colorimetric sensing44-49 due to their high extinction and strong size- and distance-dependent optical properties.44 The color of the AuNPs is red in dispersed state but changes to blue upon aggregation due to the shift of surface plasmon absorption to longer wavelength.47, 50 Since the plasmon peak shift of 13 nm AuNP can be directly observed by naked eye even with the concentration as low as a few nM, the AuNP can be ideally used as a reporter in colorimetric sensing. Especially when combined with DNA, AuNPs have been shown to be very useful for detecting a broad range of molecules, because DNA not only have molecular recognition functions31, 51 but also can control the assembly and disassembly status of AuNPs, in result tuning their optical properties.46, 47, 52
The DNA based AuNPs colorimetric sensors can be generally classified as either labeled or label-free sensors. The labeled method attaches DNA, DNAzymes or aptamers onto AuNPs before sensing operation, and relies on directed disassembly (or assembly) of AuNPs due to analyte-specific cleavage or conformational change of the DNA molecules. Since AuNPs are highly negatively charged due to the phosphate backbone of DNA, AuNP aggregation can be prevented even in 2.5 M NaCl, while the bare AuNPs are relatively less stable and form aggregates due to the salt induced screening effect.49 However, in the presence of the bridging DNA which is complementary to both DNA strands functionalized to AuNPs, AuNPs can be aggregated and the color of AuNPs changes from red to purple due to the plasmon peak shift.47 Using this phenomenon, DNA labeled sensors have been reported that can detect DNA,22, 24 protein,53, 54 small molecules,25 and metal ions. 23, 55-58 Such a method has been converted into dip stick format,59 making it even more simple and straightforward to use.
The label-free colorimetric sensors do not require any attachment of DNA onto AuNPs for the method to work, and utilizes different adsorption properties of single stranded [ssDNA] and double stranded [dsDNA] on citrate-coated AuNPs that affects salt (often NaCl)-induced aggregation of AuNPs.60 AuNPs are inherently not stable and will aggregate out of solution. Most AuNPs are stabilized by a small molecule such as citrate. The presence of salt, however, will decrease citrate’s stabilization effects and cause AuNP aggregation. Since ssDNA is flexible and can partially uncoil its bases, it can easily be adsorbed on AuNPs and enhance electrostatic repulsion between AuNPs, which in turn stabilizes AuNPs even in the presence of NaCl.52, 60 On the other hand, as dsDNA is stiff and exposed by negatively charged phosphate backbone,61 the strong repulsion between dsDNA and AuNPs makes their binding negligible, causing salt-induced aggregation. Based on these phenomena, several designs of label-free based sensors have been developed to detect specific DNA52 and RNA62 sequences and potassium ions,63 mercury ions,64-66 adenosine,67 cocaine,68 and thrombin69 using aptamers. Detection of Pb2+ using DNAzyme based on label free method has also been reported.70, 71 Recently, a combination of the two methods, salt induced aggregation method using AuNPs chemically functionalized with DNA has also been applied to detect adenosine.67, 72
Because of its high sensitivity and broad applicability, colorimetric sensors based on DNA and AuNPs have been reported recently numerous times, most of which use either labeled22-25, 53, 54, 56, 57 or label free method.52, 62-65, 67, 69-71 Even though both methods are based on the dispersed and aggregated states of AuNPs, the principles of these two methods are different, so are their general properties. It would be very interesting to compare and contrast the two methods to find out their differences in terms of advantages and disadvantages, versatility, limitations, and potential applications. In addition to offering critical guidance for researchers who want to use the methods, fundamental insight gained from such a comparison is important to advancing both the field of bioanalytical chemistry and the broad field of nanobiotechnology. Since previous publications used different DNA system, it has been difficult to compare the two methods directly in order to understand the difference between the two. It is therefore desirable to use the same DNA and AuNPs systems to provide a direct comparison of the two methods. Here we report the development and optimization of uranyl colorimetric sensors using both labeled and label-free methods by combining DNAzyme and AuNPs in different ways. Both methods results in highly sensitive and selective colorimetric sensors, with detection limits below that of maximum contamination level of uranium defined by the US Environmental Protection Agency (EPA). Through the process, the general properties of the two colorimetric sensors are compared in various aspects, making this work the first systematic comparison of these two methods using uranium sensing as an example.
Results and Discussion
Labeled DNAzyme-AuNP colorimetric sensor
A uranyl specific DNAzyme was used to assemble DNA functionalized AuNPs to form purple colored aggregates as shown in Fig. 1A and B. The substrate strand (39S-L) is elongated on both 5′ and 3′ ends to hybridize with DNAs functionalized on AuNPs (Arm (5′) and Arm (3′)). After annealing substrate strand (39S-L), enzyme strand (39E), and AuNPs functionalized with Arm (5′) and Arm (3′) DNA strands, AuNP aggregate is formed. Heating the system above melting temperature will result in disassembly of gold nanoparticle due to dehybridization of the arm strands (Arm (5′) and Arm (3′)) from substrate strand (39S-L) which are 13 and 14 mer long, respectively (Figure 1A). In the presence of uranyl, however, substrate strand will be cleaved, which makes 9 base pairs between the cleaved RNA site and 3′ end of the enzyme strand (39E) the weakest linkage in the system. This difference in the melting temperature for samples with or without uranyl can be taken advantage for uranyl sensing.
Figure 1.
(A) The scheme of labeled colorimetric sensor based on AuNP disassembly in the absence and presence of UO22+. In the presence of UO22+, the length of the weakest complementary part in the aggregates becomes shorter due to UO22+ induced substrate cleavage. The substrate cleavage can decrease melting temperature of AuNP aggregates. An in the arm strands indicates 0A or 12A spacers (n=0 or 12). (B) As UO22+ is introduced into AuNP aggregates and temperature is controlled above the melting temperature of UO22+ treated aggregates, AuNP disassembles. (C) 32P assay result showing the cleavage kinetics in the presence of UO22+. (D) The melting curve of A12 aggregates with (blue curve) and without (red curve) UO22+. There is about 10 ° C decrease of melting temperature in the presence of UO22+. bps=base pairs.
The enzymatic cleavage activity of the DNAzyme-AuNP construct was first tested to compare its activity without AuNP attached to the DNAzyme. The activity assay was carried out by labeling 39S-L strands with 32P and running polyacrylamide gel electrophoresis (PAGE) (Figure 1C) with 1 μM uranyl at 300 mM NaCl and 50 mM MES (pH 5.5). The results show that the kinetics in DNAzyme-AuNP aggregates (blue curve) was ∼200 times slower than that in solution (black curve). We hypothesized that the significant slow down of the activity in the aggregates may be due to steric hindrance of AuNP attached immediately to the DNAzyme arm. Therefore we inserted a 12A spacer to relieve this steric hindrance (Figure 1A). Interestingly, insertion of a 12A spacer on 5′ end of Arm (5′) strand and 3′ end of Arm (3′) strand increased the initial cleavage rate by ∼71%.
We then measured the melting temperature of these DNAzyme-AuNP aggregates (called A12 aggregates) to investigate the temperature at which the colorimetric sensor can operate. The aggregates were prepared as described in the Materials and Methods, and were kept at room temperature overnight either in the absence or presence of 2.5 μM of UO22+. UV-vis spectrometer was used to monitor the extinction change of the samples at 260 nm. Increase of the extinction indicates the melting of the hybridized DNA. As shown in Figure 1D, in the presence of 300 mM NaCl, the sample with uranyl had melting temperature of ∼47 °C while one without uranyl had higher melting temperature of ∼57 °C. Since there is melting temperature difference of 10 °C between the two samples, heating the samples up to a temperature in between the two, 50 °C for example, will induce disassembly of the aggregates with uranyl but not for the one without uranyl. Even at 30 mM NaCl, which was the lowest salt concentration to allow stable AuNP aggregates, melting temperature was ∼40 °C in the absence of uranyl (Figure S1B). However, when uranyl is added to the aggregates in the presence of 30 mM NaCl at room temperature, only a small amount of uranyl induced disassembly could be observed (estimated ∼50% of disassembly happened after 30 minutes) (Figure S2). Since the disassembly was still too slow, it was necessary to optimize the system to facilitate the disassembly process close to room temperature.
Optimization of the system
Because a senor with fast response at room temperature is highly preferred for on-site and real-time operation, invasive DNAs56 of different lengths (Inva-0, Inva-2, Inva-4, and Inva-6) as well as arm strands with different poly A spacers (0A, 12A, and 24A) were investigated in order to induce disassembly at room temperature (see Figure 2).
Figure 2.
The schematic design of the labeled sensor including the sequences of the invasive DNAs (pink) and 0A, 12A, and 24A Arm strands (blue and red) used in this work.
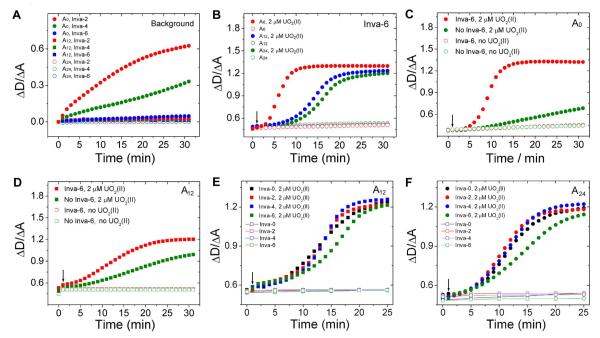
To demonstrate the effect of invasive DNAs on disassembly of AuNPs aggregates, 0A, 12A, 24A and 36A spacers were investigated in the presence of different invasive DNAs (Figure 3A). UV-vis spectrometer was used to record the plasmon peak shift of the AuNPs; integration ratio between 490 nm to 540 nm and 550 nm to 700 nm was chosen to monitor the color change (See Supporting Information). The integration between 490 nm to 540 nm and 550 nm to 700 nm represents dispersed (ΔD) and aggregated (ΔA) states of AuNPs, respectively.73 Lower ratio corresponds to aggregation of the AuNPs with blue color while higher ratio corresponds to disperse AuNPs with red color. Since the construct with 36A spacer did not show any color change after aggregation, probably due to the long distance between AuNPs controlled by the length of crosslinking DNA,46 it was not further investigated.
Figure 3.
The background increase of A0, A12, and A24 aggregates with invasive DNA strands in the absence of UO22+. (B) Disassembly kinetics difference between A0, A12, and A24 aggregates in the presence of Inva-6. The effect of Inva-6 on the UO22+ induced disassembly of A0 (C) and A12 (D) aggregates. The effect of longer invasive DNAs on A12 (E) and A24 (F) aggregates. UO22+ was added to samples one minute after UV-vis monitoring was started.
In order to investigate the effects of different invasive DNAs on disassembly of AuNP aggregates, the integration ratio [ΔD/ΔA] of AuNP aggregates without poly A spacer [A0 aggregates] were monitored in the presence of Inva-2, Inva-4, and Inva-6 in the absence of uranyl (Figure 3A) at 40 mM NaCl, 50 mM MES (pH 5.5). Addition of Inva-2, Inva-4, and Inva-6 resulted in integration ratio increase of 0.62, 0.32, and 0.05 after 30 mins., respectively, suggesting that invasive DNA aided AuNP disassembly happens for both Inva-2 and Inva-4, but the extent of disassembly decreases as the length of invasive DNA becomes shorter and becomes negligible for Inva-6. Similar effects of invasive DNA has been observed previously.56 The same procedure was carried out with AuNP aggregates with 12A spacers [A12 aggregates] in the presence of same invasive DNAs. Surprisingly, the integration ratio increased by only 0.03, 0.02, and 0.01 for Inva-2, Inva-4, and Inva-6, respectively, indicating that little disassembly of AuNPs could be observed for invasive DNA of any length tested. The experiment was repeated with AuNP aggregates with 24A spacers [A24 aggregates] with all three invasive DNAs and again, integration ratio increase by only 0.02, 0.02, and 0.002 were observed for Inva-2, Inva-4, and Inva-6, respectively. This shows that invasive DNA induced disassembly of AuNPs did not happen regardless of the length of invasive DNAs for A24 aggregates. Since invasive DNA induced disassembly of AuNP aggregates increases background, it is preferred to use an invasive DNA causing smaller extent of disassembly. As Inva-6 shows negligible increase of disassembly not only for A12 and A24 aggregates but also for A0 aggregates, we concluded that Inva-6 is the invasive DNA that can be used for all three aggregates with negligible background increase.
Once Inva-6 is determined to be the most suitable invasive DNA, because of its negligible background increase over Inva-2 or Inva-4, we then investigated the effects of spacers with Inva-6 in the presence of μM uranyl (See Figure 3B) at 40 mM NaCl, 50 mM MES (pH 5.5). In the case of A0 aggregates, integration ratio started to increase in less than 5 minutes and saturated in about 10 minutes, suggesting that disassembly can be completed in less than 15 minutes. In contrary, both A12 and A24 aggregates showed much slower integration ratio increase and saturation at about 20-25 minutes, meaning that it takes about 25 minutes for A12 or A24 aggregates to be disassembled with Inva-6. Surprisingly, it shows that A0 aggregates disassemble the fastest while A12 and A24 aggregates were both significantly slower with Inva-6. What is observed here differs from the activity assay result shown in Figure 1C which showed that poly A spacers in the arm strands can help uranyl induced cleavage. This result made us wonder how the combination of the invasive DNA and spacer in arm strands affect the performance of the sensor, since these two factors were both shown to facilitate the disassembly of the aggregation individually. Is their combination constructive or destructive?
In order to answer the above question, we compared the performance of the sensor with and without invasive DNA in A0 aggregates and A12 aggregates in the presence of μM uranyl at 40 mM NaCl, 50 mM MES (pH 5.5). In the case of A0 aggregates, the integration ratio increased very slowly from 0.39 to 0.68 in 30 min. in the absence of Inva-6 (Figure 3B). In the presence of Inva-6, however, its integration ratio started to increase from 0.38 in about 4 min. after addition of uranyl and saturated in about 13 minutes up to the integration ratio of ∼1.32, which is about 224% increase based only on the integration ratio response in 30 min (Figure 3C). This result suggests that uranyl induced disassembly kinetics of A0 aggregates is significantly increased upon the addition of Inva-6. On the other hand, the integration ratio of A12 aggregates increased from ∼0.49 to 0.99 after 30 minutes of reaction which is approximately 2 times of the integration increase of A0 aggregates in the absence of Inva-6 (Figure 3D). However, even though Inva-6 is added to A12 aggregates, the integration ratio only increased from 0.53 to 1.2 and saturation only happened in about 25 minutes. This shows that even though Inva-6 helps to disassemble Au12 aggregates, it induces only 34% increase based on the integration ratio after 30 min. of reaction, which is much lower than A0 aggregates (224%). Therefore we conclude that Inva-6 works the most effectively with A0 aggregates and its contribution in A0 aggregates is more significant than that from A12 or A24 aggregates.
Considering the fact that Inva-6 was able to accelerate the disassembly more effectively in A0 aggregate than A12 and A24 aggregates, the possible reason might be lower activation energy necessary to replace arm strands. When AuNP are functionalized to arm strands without spacers, the stability of substrate strand and both arm strand duplexes (5′ and 3′) would be impaired and thus vulnerable to the attacking by invasive DNA due to the electrostatic repulsion between NPs, steric hindrance, and nonspecific adsorption of ssDNA strand on AuNP surface.46, 47, 74 For comparison, in the case A12 and A24 aggregates, it contains 12A and 24A spacers, respectively, which can minimize the destabilization of the duplex caused by AuNPs. Since substrate strand and complementary arm strands maintain their stability, invasive DNAs need sufficient energy to replace arm strands and in result it takes longer time for disassembly.
Since longer invasive DNAs are still available for A12 and A24 aggregates due to negligible increase in the background, they can still contribute to increase the disassembly kinetics of AuNP aggregates. So uranyl induced disassembly kinetics of A12 and A24 aggregates were monitored with Inva-0, 2, 4, and 6 to find whether there is any condition better than A0 aggregates with Inva-6 at 40 mM NaCl 50 mM MES (pH 5.5) (Figure 3E and F). The integration ratio increases from Inva-0, and Inva-4 were very similar and were slightly faster than Inva-6 for both A12 and A24 aggregates. However, most of the processes took about 20 minutes to saturate with the integration ratio increase from ranges 0.50-0.55 to 1.2-1.25, which is much slower than A0 with Inva-6. These data shows that increase of the length of invasive DNAs could not improve the kinetics significantly for A0 and A12 aggregates, and invasive DNA works the most effectively in A0 aggregates rather than A12 and A24 aggregates probably due to the lower activation energy.
In the work described here, invasive DNA molecules are used to improve disassembly kinetics and thus sensing performance of the labeled sensing system. It relies on a detailed investigation of the role of invasive DNA in DNAzyme-AuNPs disassembly.56 To minimize variables in the system, introducing DNA mismatches, less stable base pairs, or alkylguanine to lower Tm are all good alternate strategies and will be pursued in the future.
Sensitivity and selectivity of labeled DNAzyme-AuNP colorimetric sensor
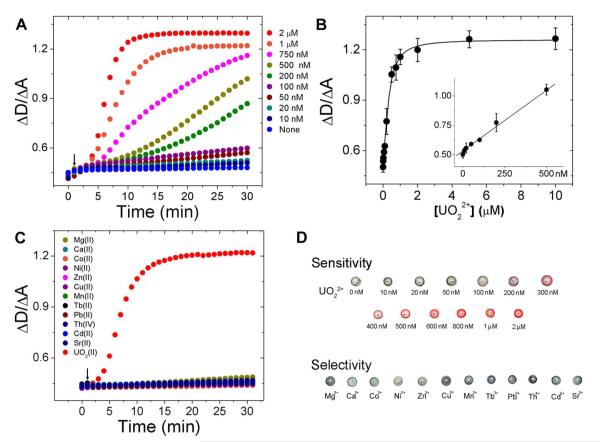
Through the optimization process, we determined that A0 aggregates have the best disassembly kinetics when Inva-6 was used as invasive DNA. This optimal construct was used to investigate the sensitivity of UO22+ dependent labeled DNAzyme-AuNP sensor, plasmon resonance peak shift of AuNPs was monitored by UV-vis for 30 minutes and the kinetics of the reaction is shown in Figure 4A based on the integration ratio between 490 nm to 540 nm and 550 nm to 700 nm at various UO22+ concentrations. The calibration curve based on the integration ratio of samples measured after 30 minutes of reaction is shown in Figure 4B. Based on the calibration curve, the detection limit of labeled method for uranyl sensor is 50 nM and the calibration curve saturated at 2 μM. The image of color change is shown in Figure 4D.
Figure 4.
(A) Disassembly of AuNP aggregates at various UO22+ concentrations and (B) calibration curve of labeled uranyl colorimetric sensor. (C) Disassembly of AuNPs in the presence of various metal ions including UO22+. (D) The color change of AuNP aggregates in the presence different concentrations of UO22+ and other metal ions. The concentration of all metal ions other than UO22+ is 2 μM. Metal ions were added to samples one minute after UV-vis monitoring was started.
To investigate the selectivity of the labeled DNAzyme-AuNP sensor, plasmon resonance peak shift of AuNPs was monitored by UV-vis for 30 minutes and the integration ratio change was compared for various metal ions including UO22+ (Figure 4C). Only the sample with UO22+ shows change in plasmon shift which means that the sensor only has response with uranyl and not with other metal ions. The colors of the sensor solution in the presence of several metal ions (2 μM) are shown in Figure 4D.
Label-free DNAzyme-AuNP colorimetric sensor
The scheme of the label-free method is illustrated in Figure 5. UO22+ cleavable Substrate-DNAzyme complex [complex] was first prepared separately and reacted with UO22+. In the presence of UO22+, substrate strand (39S) is cleaved and 10-mer ssDNA should be released, which can then be adsorbed onto AuNP to prevent the salt-induced aggregation. In the absence of UO22+, however, the complex should remain and will not interact with AuNPs, resulting in AuNP aggregation due to the screening effect from NaCl and thus inducing color change of AuNPs from red to blue.
Figure 5.
(A) The design and sequence of the label free sensor (complex). After UO22+ induced cleavage, 10 mer ssDNA is released which adsorbs on AuNP surface. (B) AuNP reaction in addition of UO22+ treated/untreated complex and additional NaCl. AuNPs aggregate in the absence of UO22+ but remain dispersed in its presence.
Stability of AuNPs upon addition of NaCl and ssDNA
In most of the reported label-free colorimetric sensors, ssDNA is adsorbed onto AuNPs surface first and salt is added afterwards to induce the color change. A 24-mer dsDNA is reported to be able to remain hybridized for about 10 minutes in the Au colloid without NaCl while introduction of a single mismatch will decrease the stability of dsDNA and thus cause dehybridization in 5 min.52 DNAzymes, however, contain a large number of mismatches between the enzyme strand and substrate strand (Figure 5A), which causes dehybridization of the complex in seconds in the absence of NaCl.75, 76 So unlike previously reported studies, DNA solution has to be added to AuNP solution together with sufficient amount of NaCl that can keep complex hybridized. In this case, since the stability of AuNPs is determined by the competition between the ssDNA adsorption on AuNP and electrostatic screening caused by NaCl which are introduced to AuNP solution at the same time, it is important to investigate whether DNA can still be adsorbed on AuNPs effectively and protect them in the presence of NaCl.
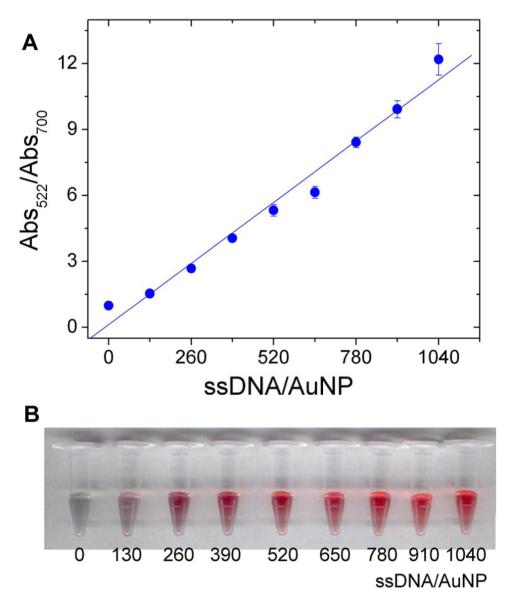
In order to investigate whether AuNPs can still be stabilized by ssDNA in the presence of NaCl, a 10 mer ssDNA was chosen as a model DNA strand to simulate the protection effect of the cleaved ssDNA from the substrate. Different amount of 10-mer ssDNA in 300 mM NaCl, 10 mM MES (pH 5.5) was added to AuNP solution and their color change was monitored based on the extinction ratio between 522 nm and 700 nm (see Figure 6A) (See Supporting Information).
Figure 6.
(A) Extinction ratio dependence on the number of 10 mer ssDNA per 13 nm AuNP. The stability of AuNP increases as more ssDNA exist per one AuNP. (B) The color change of AuNPs with different ratio of DNA per AuNP.
The concentration of NaCl in the final solution was 0.1 M. The extinction ratio (Abs522nm/Abs700nm) of AuNPs was linearly dependent on the amount of DNA at 0.1 M NaCl, suggesting that ssDNA can still stabilize AuNPs even though it is introduced to AuNP solution with NaCl at the same time. The extinction ratio reached 11 when ∼ 1000 equiv. of ssDNA was used per one AuNP. However, since extinction ratio change from 1 to 5 is sufficient for detection, 500 equiv. of ssDNA per one AuNP was used in the following experiments because it could stabilize the Au NPs and make the color change from blue to red.
Quenching UO22+ dependent cleavage reaction by shifting pH
Hybridization of substrate and enzyme strand was carried out at pH 5.5, where UO22+ dependent cleavage reaction occurs most efficiently.39 Since the UO22+ dependent cleavage reaction happens very quickly in several minutes, if the reaction cannot be effectively stopped during the measurements, significant error could occur, which makes the sensor impractical. Since biochemical investigation of this uranyl specific DNAzyme showed that its activity is highly pH dependent, having the activity peak occurring around pH 5.5 with dramatic decrease of activity at either higher or lower pH,70 we hypothesized that the DNAzyme might not be active at pH 8. So to quench the reaction, small amount of concentrated TRIS (2-Amino-2-(hydroxymethyl)propane-1,3-diol) base solution was added to the solution containing complex to shift pH from 5.5 to around 8.
UV-vis spectroscopy was used to monitor the quenching effect of uranyl induced cleavage reaction using TRIS base solution (see Figure 7A). When the complex was added to AuNP solution without addition of uranyl, an extinction ratio of ∼1.4 was observed, suggesting AuNP aggregation. On the other hand, when the complex was treated with 500 nM uranyl for 6 minutes, an extinction ratio of ∼4 was observed, indicating AuNP dispersion. When the reaction was quenched by adding TRIS base solution after 1 minute of uranyl induced reaction followed by addition to AuNP solution, an extinction ratio of ∼2.2 was observed, suggesting partial AuNPs dispersion. This means that the uranyl induced cleavage ration has been stopped by quenching reaction. To make sure that quenching reaction was complete and no further uranyl induced reaction happened afterwards, control experiment was performed which has 5 minutes interval between uranyl induced cleavage reaction (1 minute) and mixture in AuNP solution. It is shown that even though there is 5 minutes of interval between quenching and mixture with AuNP, there is no further uranyl induced cleavage reaction. This result indicates the TRIS base solution could quench the reaction very effectively and in time. Furthermore, it turned out that TRIS also helps to aggregate AuNPs more effectively and thus helps to lower the background signal (Figure 7B).
Figure 7.
(A) The quenching efficiency of label free sensor by shifting the pH of the solution from 5.5 to 8. AuNPs are aggregated in the absence of UO22+ (blue curve) but remains dispersed after 6 minutes reaction with 2 μM UO22+ (red curve). AuNPs show less amount of disassembly after 1 minute of reaction with 2 μM UO22+ and quenching (purple curve). No further disassembly of AuNP aggregates after 5 minutes of holding in between quenching and AuNP addition (green curve) shows that quenching efficiency is very high and quick. The color change of each sample is also shown in the inset. (B) The color change difference before (upper) and after (lower) addition of TRIS base solution. Concentration of uranyl is 2 μM.
Sensitivity and selectivity of label-free colorimetric sensor for UO22+
In order to check the sensitivity of UO22+ dependent label-free colorimetric sensor, plasmon resonance peak shift of AuNPs was monitored by UV-vis and the extinction ratio between 522 nm and 700 nm was compared at various UO22+ concentrations (Figure 8A). The detection limit was determined to be 1 nM and the linear fitting range was from 1 nM to 100 nM. The calibration curve saturated at 700 nM, which is similar to fluorescence based uranyl sensor.39 Since the UO22+ dependent cleavage reaction was made in concentrated DNAzyme solution in optimized conditions, followed by the addition of AuNP, UO22+ dependent cleavage reaction can happen very efficiently, which helps to keep high sensitivity. Furthermore, as reacted DNA solution containing NaCl is added to AuNP solution after quenching, the color of AuNP solution change occurs immediately and does not change much afterwards.
Figure 8.
(A) Calibration curve of label free sensor. Sensor has detection limit of 1 nM. (B) The color change of AuNP solution with different concentration of UO22+ in the solution. (C) The color change of AuNP solution with various metal ions including UO22+ (2 μM).
In the low concentration range (Figure 8A), the extinction ratio increases with increasing concentration of uranyl until it saturates ∼700 nM, beyond which the ratio decreases with increasing uranyl concentrations. This ‘bell-shape” metal-dependent curve appears to be originated from uranyl dependent DNAzyme activities,39 and similar curves have been reported in other DNAzymes.32-34 The image of color change is shown in Figure 8B. Since UO22+ dependent cleavage reaction can easily be quenched by shifting pH from 5.5 to 8, it might be possible to tune the dynamic range simply by changing the reaction time.
To investigate the selectivity of label-free sensor, several metal ions including uranyl were added to sensor solution and their color changes are shown in Figure 8C. The result clearly shows that the sensor only have response in the existence of uranyl, showing that the sensor has excellent selectivity.
Comparison between labeled and label-free based colorimetric sensors
Since both colorimetric methods have been demonstrated to successfully detect uranyl, it gave us the opportunity to compare the properties of both sensors using the same DNAzyme and AuNPs.
In terms of performance of sensors, as labeled sensing method depends on UO22+ induced cleavage of substrate strands in aggregated state and the release of AuNP aggregates, certain amount of UO22+ and time is necessary for the cleavage to complete, which could explain the relatively higher detection limit (50 nM) and slower kinetics (30 minutes). On the other hand, as label-free sensor allows UO22+ based cleavage reaction to occur separately without AuNPs and AuNPs solution is added afterwards for signaling, the cleavage could be completed with less amount of UO22+ and less time, resulting relatively lower detection limit (1 nM) and shorter sensing time (6 minutes). Despite this difference, both sensors have detection limits that are much lower than the maximum contamination level defined by the US EPA (130 nM)71 and have excellent selectivity over other metal ions. Furthermore, both sensors operate at room temperature.
For preparation and handling of sensors, it takes more time and effort to prepare the labeled sensor, but the labeled sensor is more convenient to handle once the sensor is prepared, as sensing can be carried out through one step of addition of uranyl. On the other hand, the label-free sensor can be easily prepared, but requires three processing steps, uranyl addition, quench, and AuNP addition, for detection. In doing so, the label-free method contains many more variables that are uncertain for on-site and real-time detection. Therefore, for practical applications, it is desirable for professionals to spend more time and efforts to prepare the sensors in a laboratory in order to minimize steps for on-site operation; the more processing steps for on-site operation, the more likely errors could occur, especially when used by non-professional operators.
In addition, since the AuNPs in labeled sensor are modified with thiolate oligonucleotides, they are much more stable than those in the label-free sensor and thus can be stored for a long time and used under a variety of conditions. In contrast, the AuNPs used in label-free sensors are much less stable and requires much narrow range of conditions such as ionic strength to operate. The stability of AuNP is very important for practical applications because it is the key ingredient for the sensor. In addition, labeled system could be easily incorporated into lateral flow devices under a variety of conditions,59 while a much more carefully controlled condition may be required if label-free AuNPs were to be used. Finally, the labeled method resulted in a turn-on sensor, going from purple AuNP aggregates into red disassembled AuNPs upon addition of uranyl, while the label-free DNAzyme-AuNP sensor is a turn-off sensor; the red AuNPs remains red in the presence of uranyl but changes to blue in the absence of uranyl. Turn-on sensors are preferred as they are much less vulnerable to false positive signals due to interfering metal ions, other species or conditions. Furthermore, the labeled methods can be designed as turn-off sensors 23, 55 demonstrating versatility in sensor design for the labeled system.
In conclusion, uranyl specific colorimetric sensors using gold nanoparticles and uranyl selective DNAzyme were prepared by both labeled and label-free methods and the general properties of the two colorimetric sensors were compared in various aspects. The labeled sensor is simpler to use as it requires one step process once the sensor is prepared and is more versatile as DNA functionalized on AuNPs provides stability and turn-on sensing. On the other hand, the label-free sensor has better sensitivity, shorter operation time and lower costs. Despite the difference, both sensors have detection limits lower than the maximum contamination level defined by the US EPA (130 nM), have excellent selectivity over other metal ions, and operate at room temperature. If operation requires very low detection limit, the label-free should be a better choice. When detection limit is adequate, such as in the case of uranyl sensing, the labeled method should be used due to turn-on sensing, and high stability of the system that allows operation under a variety of conditions.
Experimental section
Oligonucleotide and reagents
All Oligonucleotides were purchased from Integrated DNA Technologies Inc. (Coralville, IA). DNAzyme strand (39E) and both substrate strands (39S-L for DNA-disassembly sensor and 39S for NaCl-aggregation sensor) were purified by HPLC by the company while the arm strands with thiolate modifications and invasive DNA strands were standard desalted. HAuCl4 (99.999%), sodium citrate dehydrate (99+%) were purchased from Aldrich and used without further purification.
Labeled sensor
Preparation and functionalization of AuNPs
Gold nanoparticles (13 nm diameter) were synthesized by reduction of HAuCl4 by sodium citrate, and the AuNP-DNA conjugate were prepared following the published protocol.44 In order to activate thiol modification on Arm (5′) strand, 9 μL of 1 mM Arm (5′) strand, 1 μL of 500 mM pH 5.5 MES buffer, and 1.5 μL of 10 mM TCEP solution were mixed in a microcentrifuge tube and kept for an hour. In a separate tube, a parallel experiment was done with Arm (3′) strand to activate thiol modification on Arm (3′) strand.
At the same time, two scintillation vials were incubated in fresh 10 M NaOH solution for an hour and then rinsed with distilled water for several times and then with Millipore water (18 MΩ ) for copious times to prevent AuNP sticking on the surface of glass vials.
In order to attach AuNP with Arm (5′) DNA strand, 3 mL of 13 nm AuNP solution was placed in one scintillation vial and activated 9 μL Arm (5′) strand was added and left overnight in a dark place for reaction after gentle shaking. In the other scintillation vial, another 3 mL of 13 nm AuNP solution was mixed with activated 9 μL Arm (3′) strand and then was treated in the same way.
300 μL of 1 M NaCl and 15 μL of 500 mM TRIS-acetate buffer (pH 7.6) was added on the next day and again kept in a dark place overnight after gentle shaking.
In order to prepare labeled sensor, DNA functionalized AuNPs were first purified to remove free DNA in AuNP solution. 500 μL of AuNP functionalized with Arm (5′) strand and same amount of AuNP functionalized with Arm (3′) strand were placed in two 1.5 mL micro-centrifuge tubes, respectively, and were centrifuged at 16,110g for 15 minutes. After that, supernatant in both solutions was replaced with fresh 500 μL 100 mM NaCl, 50 mM MES (pH 5.5) solution. After purification process was repeated, the supernatant was replaced with 250 μL 300 mM NaCl, 50 mM MES (pH 5.5) buffer. After mixing two AuNP solutions, 10 μL of 10 μM elongated substrate strand (39S-L), and 20 μL of 10 μM enzyme strand (39E) were added and annealed from 55 °C to room temperature for about 1 hour. The color of AuNP solution changed from red to purple which shows that DNA directed AuNPs assembly happened. The AuNPs aggregates were centrifuged by a micro-centrifuge for about a minute and the supernatant was replaced with 120 μL of 300 mM NaCl, 50 mM MES (pH 5.5) solution to remove free DNA (39S-L and 39E) in the sensor solution.
Activity assays
To prepare aggregates containing 32P-labeled substrate, ∼0.1% of 32P-labeled substrates (in respect to the total substrate amount) were added, while keeping other conditions the same. 32P-labeled aggregates were added to a solution with 1 μM UO22+, 300 mM NaCl and 50 mM MES, pH 5.5. Aliquots were taken out at designated time points and quenched in a solution containing 8 M urea and 200 mM EDTA. The quenched aliquots were heated at 60 °C to fully release substrate strands from aggregates and then loaded to 20% denaturing polyacrylamide gel. 32P-labeling and procedures for single-turnover solution phase activity assays were the same as reported elsewhere.72
Uranyl detection
In order to detect uranyl using labeled sensor, 381 μL of 50 mM MES buffer (pH 5.5), 4.5 μL of 0.2 mM Inva-6 (5′), 4.5 μL of 0.2 mM Inva-6 (3′) were mixed and 60 μL of sensor solution in 300 mM NaCl, 50 mM MES (pH 5.5) was added just before UV-vis measurement. The concentration of invasive DNA is μM in the final solution. Uranyl was added 1 minute after the measurement has started and the whole reaction was made for 30 minutes. Calibration curve was made based on the data collected on the 31st minutes after 30 minutes of reaction.
Label-free sensor
AuNPs stabilized by ssDNA in the presence of NaCl
Different amount (from 0 μL to 8 μL) of 10 mer DNA (5′-CAT GCT ACT G-3′, 100 μM) was added into 70 μL 300 mM NaCl, 10 mM MES buffer solution (pH 5.5) in 0.6 mL microcentrifuge tube. A mixture of 1.19 μL of 500 mM TRIS base solution and appropriate amount of Millipore water was added to make the total volume as 134 μL. After vortex, 76 μL 10 nM Au nanoparticles (13 nm) were added and the surface plasma absorption was collected by UV-vis spectra.
Sensor preparation and uranyl detection
UV-Vis was used to check the exact concentration of 39E and 39S strands. This process is very important because very small number of unhybridized ssDNA can still stabilize AuNP and increase the background. Based on the measured concentration, 4 μL of 100 μM 39S strand and equal amount of 39E strand were mixed in 70 μL 300 mM NaCl, 10 mM MES buffer solution (pH 5.5) in 0.6 mL microcentrifuge tube. After vortex, sample was heated up to 80 °C and cooled down to room temperature in one hour and a half. Hybridization solution can be multiplied by preparing in large scale in volume. After that, 77 μL of solution containing hybridized DNAzyme and enzyme strand was transferred into a new tube and cleaved by uranyl for 6 minutes. In order to quench UO22+ dependent cleavage reaction, a mixture of 1.19 μL of 500 mM TRIS base solution and 56 μL Millipore water was added to the same tube and tube is vortexed quickly which in result shifts the pH from 5.5 to 8. 76 μL 10 nM Au nanoparticles (13 nm) was transferred to the tube containing DNA. The solution will show color change corresponding to the concentration of uranyl in the solution. The color change can be monitored by eye or by plasmon peak shift in UV-vis spectra.
Supplementary Material
Table 1.
Comparison between labeled and label-free colorimetric UO22+ sensors
| Sensors | Labeled Sensor | Label-Free Sensor |
|---|---|---|
| Detection range | 50 nM-2 μM | 1 nM-700 nM |
| Detection limit | 50 nM | 1 nM |
| Linear range | 50-500 nM | 1-100 nM |
| Saturation point | 2 μM | 700 nM |
| Working time | 30 minutes | 6 minutes |
| Working temperature | Room temperature | Room temperature |
| Operation step | 1 step | 3 steps |
| Quenching | No | Possible (by shifting pH) |
| Std. Dev. | ∼10 % of the signal change when saturated |
∼10 % of the signal change when saturated |
| Color Change | Purple to red (in the presence of analyte) |
Red to blue (in the absence of analyte) |
| Type | Turn on | Turn off |
| Stability of AuNPs | Stable | Less stable |
ACKNOWLEDGMENT
This work has been supported by the Office of Science (BER), U. S. Department of Energy, Grant No. DE-FG02-08ER64568, and the U. S. National Science Foundation (CTS-0120978, DMR-0117792, and DMI-0328162).
Footnotes
Supporting Information Available: Data on melting curves of A0 and A12 aggregates at 30 mM and 100 mM NaCl and uranyl induced disassembly of A12 aggregates in the presence of 30 mM NaCl at room temperature. The reason of using integration ratio method for labeled and extinction ratio method for label free systems. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Konstantin B. Gongalsky. Environ. Monit. Assess. 2003;89:197–219. doi: 10.1023/a:1026031224658. [DOI] [PubMed] [Google Scholar]
- (2).Craft E, Abu-Qare A, Flaherty M, Garofolo M, Rincavage H, Abou-Donia M. J. Toxicol. Environ. Health B Crit. Rev. 2004;7:297–317. doi: 10.1080/10937400490452714. [DOI] [PubMed] [Google Scholar]
- (3).Zhou P, Gu B. Environ. Sci. Technol. 2005;39:4435–4440. doi: 10.1021/es0483443. [DOI] [PubMed] [Google Scholar]
- (4).Abbasi SA. Int. J. Environ. Anal. Chem. 1989;36:163–172. doi: 10.1080/03067318108071554. [DOI] [PubMed] [Google Scholar]
- (5).Brina R, Miller AG. Anal. Chem. 1992;64:1413–1418. [Google Scholar]
- (6).Boomer DW, Powell MJ. Anal. Chem. 1987;59:2810–2813. doi: 10.1021/ac00150a019. [DOI] [PubMed] [Google Scholar]
- (7).Mlakar M, Branica M. Anal. Chim. Acta. 1989;221:279–287. [Google Scholar]
- (8).Li J, Lu Y. J. Am. Chem. Soc. 2000;122:10466–10467. [Google Scholar]
- (9).Burdette SC, Walkup GK, Spingler B, Tsien RY, Lippard SJ. J. Am. Chem. Soc. 2001;123:7831–7841. doi: 10.1021/ja010059l. [DOI] [PubMed] [Google Scholar]
- (10).Chen P, He C. J. Am. Chem. Soc. 2004;126:728–729. doi: 10.1021/ja0383975. [DOI] [PubMed] [Google Scholar]
- (11).Yang L, McRae R, Henary MM, Patel R, Lai B, Vogt S, Fahrni CJ. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11179. doi: 10.1073/pnas.0406547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yoon S, Albers AE, Wong AP, Chang CJ. J. Am. Chem. Soc. 2005;127:16030–16031. doi: 10.1021/ja0557987. [DOI] [PubMed] [Google Scholar]
- (13).Nolan EM, Lippard SJ. J. Am. Chem. Soc. 2007;129:5910. doi: 10.1021/ja068879r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wegner SV, Okesli A, Chen P, He C. J. Am. Chem. Soc. 2007;129:3474. doi: 10.1021/ja068342d. [DOI] [PubMed] [Google Scholar]
- (15).Tsien RY. Fluorescent Chemosensors for Ion and Molecule Recognition. Washington, DC: 1993. pp. 130–146. ed. Vol. [Google Scholar]
- (16).Jiang P, Guo Z. Coord. Chem. Rev. 2004;248:205–229. [Google Scholar]
- (17).Lim MH, Lippard SJ. Acc. Chem. Res. 2007;40:41. doi: 10.1021/ar950149t. [DOI] [PubMed] [Google Scholar]
- (18).Homola J, Piliarik M. Springer Series on Chemical Sensors and Biosensors. 2006;4:45–67. [Google Scholar]
- (19).Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Angew. Chem. Int. Ed. 2005;44:5456. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- (20).Li D, Yan Y, Wieckowska A, Willner I. Chem. Comm. 2007:3544–3546. doi: 10.1039/b704731b. [DOI] [PubMed] [Google Scholar]
- (21).Willner I, Zayats M. Angew. Chem. Int. Ed. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- (22).Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078–1080. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- (23).Liu J, Lu Y. J. Am. Chem. Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- (24).Hazarika P, Ceyhan B, Niemeyer CM. Angew. Chem. Int. Ed. 2004;43:6469–6471. doi: 10.1002/anie.200461887. [DOI] [PubMed] [Google Scholar]
- (25).Liu J, Lu Y. Angew. Chem. Int. Ed. 2006;45:90–94. [Google Scholar]
- (26).Rohwer H, Rheeder N, Hosten E. Anal. Chim. Acta. 1997;341:263–268. [Google Scholar]
- (27).Sessler JL, Melfi PJ, Seidel D, Gorden AEV, Ford DK, Palmer PD, Tait CD. Tetrahedron. 2004;60:11089–11097. [Google Scholar]
- (28).Blake RC, II, Pavlov AR, Khosraviani M, Ensley HE, Kiefer GE, Yu H, Li X, Blake DA. Bioconjug. Chem. 2004;15:1125–1136. doi: 10.1021/bc049889p. [DOI] [PubMed] [Google Scholar]
- (29).Safavi A, Bagheri M. Anal. Chim. Acta. 2005;530:55–60. [Google Scholar]
- (30).Greene PA, Copper CL, Berv DE, Ramsey JD, Collins GE. Talanta. 2005;66:961–966. doi: 10.1016/j.talanta.2004.12.061. [DOI] [PubMed] [Google Scholar]
- (31).Breaker RR, Joyce GF. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- (32).Cuenoud B, Szostak JW. Nature. 1995;375:611–614. doi: 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]
- (33).Carmi N, Shultz LA, Breaker RR. Chem. Biol. 1996;3:1039–1046. doi: 10.1016/s1074-5521(96)90170-2. [DOI] [PubMed] [Google Scholar]
- (34).Wang W, Billen LP, Li Y. Chem. Biol. 2002;9:507–517. doi: 10.1016/s1074-5521(02)00127-8. [DOI] [PubMed] [Google Scholar]
- (35).Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF., III J. Am. Chem. Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- (36).Mei SHJ, Liu Z, Brennan JD, Li Y. J. Am. Chem. Soc. 2003;125:412–420. doi: 10.1021/ja0281232. [DOI] [PubMed] [Google Scholar]
- (37).Bruesehoff P,J, Li J, Augustine AJ, Lu Y. Comb. Chem. High T. Scr. 2002;5:327–335. doi: 10.2174/1386207023330264. [DOI] [PubMed] [Google Scholar]
- (38).Wang Y, Silverman SK. J. Am. Chem. Soc. 2003;125:6880–6881. doi: 10.1021/ja035150z. [DOI] [PubMed] [Google Scholar]
- (39).Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2056. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Niazov T, Pavlov V, Xiao Y, Gill R, Willner I. Nano Lett. 2004;4:1683. [Google Scholar]
- (41).Rupcich N, Chiuman W, Nutiu R, Mei S, Flora KK, Li Y, Brennan JD. J. Am. Chem. Soc. 2006;128:780–790. doi: 10.1021/ja053336n. [DOI] [PubMed] [Google Scholar]
- (42).Liu J, Lu Y. J. Am. Chem. Soc. 2007;129:9838–9839. doi: 10.1021/ja0717358. [DOI] [PubMed] [Google Scholar]
- (43).Liu J, Lu Y. Angew. Chem. Int. Ed. 2007;46:7587–7590. doi: 10.1002/anie.200702006. [DOI] [PubMed] [Google Scholar]
- (44).Liu J, Lu Y. Nat. Protoc. 2006;1:246–252. doi: 10.1038/nprot.2006.38. [DOI] [PubMed] [Google Scholar]
- (45).Zhao W, Gonzaga F, Li Y, Brook MA. Adv. Mater. 2007;19:1766–1771. [Google Scholar]
- (46).Storhoff JJ, Lazarides AA, Mucic RC, Mirkin CA, Letsinger RL, Schatz GC. J. Am. Chem. Soc. 2000;122:4640–4650. [Google Scholar]
- (47).Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- (48).Sato K, Hosokawa K, Maeda M. J. Am. Chem. Soc. 2003;125:8102–8103. doi: 10.1021/ja034876s. [DOI] [PubMed] [Google Scholar]
- (49).Lim II, Ip W, Crew E, Njoki PN, Mott D, Zhong CJ, Pan Y, Zhou S. Langmuir. 2007;23:826–833. doi: 10.1021/la062334t. [DOI] [PubMed] [Google Scholar]
- (50).Lee JH, Wernette DP, Yigit MV, Liu J, Wang Z, Lu Y. Angew. Chem. Int. Ed. 2007;46:9006–9010. doi: 10.1002/anie.200702569. [DOI] [PubMed] [Google Scholar]
- (51).Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- (52).Li H, Rothberg L. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Pavlov V, Xiao Y, Shlyahovsky B, Willner I. J. Am. Chem. Soc. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- (54).Huang C-C, Huang Y-F, Cao Z, Tan W, Chang H-T. Anal. Chem. 2005;77:5735–5741. doi: 10.1021/ac050957q. [DOI] [PubMed] [Google Scholar]
- (55).Liu J, Lu Y. J. Am. Chem. Soc. 2004;126:12298–12305. doi: 10.1021/ja046628h. [DOI] [PubMed] [Google Scholar]
- (56).Liu J, Lu Y. J. Am. Chem. Soc. 2005;127:12677–12683. doi: 10.1021/ja053567u. [DOI] [PubMed] [Google Scholar]
- (57).Lee J-S, Han MS, Mirkin CA. Angew. Chem. Int. Ed. 2007;46:4093–4096. doi: 10.1002/anie.200700269. [DOI] [PubMed] [Google Scholar]
- (58).Liu J, Lu Y. Chem. Comm. 2007:4872–4874. doi: 10.1039/b712421j. [DOI] [PubMed] [Google Scholar]
- (59).Liu J, Mazumdar D, Lu Y. Angew. Chem. Int. Ed. 2006;45:7955–7959. doi: 10.1002/anie.200603106. [DOI] [PubMed] [Google Scholar]
- (60).Li HX, Rothberg LJ. J. Am. Chem. Soc. 2004;126:10958–10961. doi: 10.1021/ja048749n. [DOI] [PubMed] [Google Scholar]
- (61).Bloomfield VA, Crothers DM, Tinoco I., Jr . University Science Books; Sausalito,CA: 2000. p. 800. 1999. [Google Scholar]
- (62).Li H, Rothberg L. Anal. Chem. 2005;77:6229–6233. doi: 10.1021/ac050921y. [DOI] [PubMed] [Google Scholar]
- (63).Wang L, Liu X, Hu X, Song S, Fan C. Chem. Comm. 2006:3780–3782. doi: 10.1039/b607448k. [DOI] [PubMed] [Google Scholar]
- (64).Liu CW, Hsieh YT, Huang CC, Lin ZH, Chang HT. Chem. Comm. 2008;19:2242–2244. doi: 10.1039/b719856f. [DOI] [PubMed] [Google Scholar]
- (65).Li D, Wieckowska A, Willner I. Angew. Chem. Int. Ed. 2008;47:3927–3931. doi: 10.1002/anie.200705991. [DOI] [PubMed] [Google Scholar]
- (66).Wang L, Zhang J, Wang X, Huang Q, Pan D, Song S, Fan C. Gold Bulletin. 2008;41:37–41. [Google Scholar]
- (67).Zhao W, Chiuman W, Brook MA, Li Y. Chembiochem. 2007;8:727–731. doi: 10.1002/cbic.200700014. [DOI] [PubMed] [Google Scholar]
- (68).Zhang J, Wang L, Pan D, Song S, Boey FY, Zhang H, Fan C. Small. 2008;4:1196–1200. doi: 10.1002/smll.200800057. [DOI] [PubMed] [Google Scholar]
- (69).Wei H, Li B, Li J, Wang E, Dong S. Chem. Comm. 2007;36:3735–3737. doi: 10.1039/b707642h. [DOI] [PubMed] [Google Scholar]
- (70).Wang Z, Lee JH, Lu Y. Adv. Mater. 2008;20:3263–3267. [Google Scholar]
- (71).Wei H, Li B, Li J, Dong S, Wang E. Nanotechnology. 2008;19:095501/1–095501/5. doi: 10.1088/0957-4484/19/9/095501. [DOI] [PubMed] [Google Scholar]
- (72).Zhao W, Chiuman W, Lam JCF, McManus SA, Chen W, Cui Y, Pelton R, Brook MA, Li Y. J. Am. Chem. Soc. 2008;130:3610–3618. doi: 10.1021/ja710241b. [DOI] [PubMed] [Google Scholar]
- (73).Laromaine A, Koh L, Murugesan M, Ulijn RV, Stevens MM. J. Am. Chem. Soc. 2007;129:4156–4157. doi: 10.1021/ja0706504. [DOI] [PubMed] [Google Scholar]
- (74).Demers LM, Oestblom M, Zhang H, Jang N-H, Liedberg B, Mirkin CA. J. Am. Chem. Soc. 2002;124:11248–11249. doi: 10.1021/ja0265355. [DOI] [PubMed] [Google Scholar]
- (75).Liu J, Lu Y. Anal. Chem. 2003;75:6666–6672. doi: 10.1021/ac034924r. [DOI] [PubMed] [Google Scholar]
- (76).Wernette DP, Mead C, Bohn PW, Lu Y. Langmuir. 2007;23:9513–9521. doi: 10.1021/la701303k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.