Abstract
DNA vaccines can be greatly improved by polymer agents that simultaneously increase transgene expression and activate immunity. We describe here Pluronic P85 (P85), a triblock copolymer of ethylene oxide (EO) and propylene oxide (PO) EO26-PO40-EO26,. Using a mouse model we demonstrate that co-administration of a bacterial plasmid DNA with P85 in a skeletal muscle greatly increases gene expression in the injection site and distant organs, especially the draining lymph nodes and spleen. The reporter expression colocalizes with the specific markers of myocytes and keratinocytes in the muscle, as well as dendritic cells (DC) and macrophages in the muscle, lymph nodes and spleen. Furthermore, DNA/P85 and P85 alone increase the systemic expansion of CD11c+ (DC), and local expansion of CD11c+, CD14+ (macrophages) and CD49b+ (natural killer) cell populations. DNA/P85 (but not P85) also increases maturation of local DC (CD11c+CD86+, CD11c+CD80+, and CD11c+CD40+). We suggest that DNA/P85 promotes the activation and recruitment of the antigen-presenting cells, which further incorporate, express and carry the transgene to the immune system organs.
Keywords: Copolymer, gene expression, leukocyte, Pluronics
1. Introduction
Every year vaccination prevents three million children deaths due to infectious diseases. Furthermore, there are hopes that cancer vaccines can be useful in preventing or treating cancer [1-3]. Currently in the United States the Food and Drug Administration (FDA) approved two prophylactic vaccines against virus infections that lead to cancer. The first is the hepatitis B vaccine, which prevents infection with the hepatitis B virus associated with the liver cancer. The second is Gardasil™, a vaccine against the two types of human papillomavirus that cause 70% of cervical cancer cases worldwide. Despite these successes, the current vaccine technologies do not provide guaranteed protection against some of the major diseases such as HIV infection, mycobacterium tuberculosis infection, malaria parasites, cancer and others [4-7]. Therefore, there is a need to develop novel vaccines or enhance exiting vaccination approaches [8, 9].
One potential approach is DNA vaccination, which was shown to elicit potent cellular immune responses in small laboratory animals, but failed in human clinical trials [10]. The potency of DNA vaccines can be increased either by enhancing plasmid DNA expression or by directly stimulating the immune system using adjuvants. Examples of such adjuvants include alum salts [11], cytokines [12-15], liposomes [16-18], polymers and polymeric nanoparticles [19-26].
Recently, synthetic block copolymers of poly(ethylene oxide) (PEO) and poly(propylene oxide) (PPO) (called poloxamers or Pluronics) have attracted significant interest as chemical agents for non-viral gene therapy and vaccine therapy [27-31]. A number of studies has shown that poloxamers can facilitate cellular entry and nuclear transport of a plasmid DNA [32], and enhance transcription of the delivered DNA in cells [33] and altogether enhance transgene expression in vitro [34-37] and in vivo [38, 39]. It was also shown that certain poloxamers can activate NF-kB signaling pathway [39]. Furthermore, extensive studies have suggested that poloxamers can be used as adjuvants during vaccination [31, 40-49]. Based on this we posit that poloxamers may fulfill a dual function as DNA vaccine adjuvants, first, by increasing gene expression, and second, by activating pro-inflammatory signaling pathways and immune response. The present work reports for the first time that co-administration of a plasmid DNA with a poloxamer known as Pluronic P85 (P85) in a mouse skeletal muscle significantly increases gene expression not only in the muscle, but also in the draining lymph nodes and spleen. Furthermore, analysis of cellular distribution of reporter genes expression demonstrates that P85 enhances transgene expression in myocytes and keratinocytes in the muscle, as well as in dendritic cells (DC), and macrophages in the muscle, lymph nodes and spleen. We suggest that the intramuscular (i.m.) injection of a plasmid DNA with P85 induces pro-inflammatory response and leads to the activation and recruitment of the antigen-presenting cells (APC) at the injection site. The APC further incorporate, express and carry the transgene to the immune system organs. Altogether, the results suggest that P85 shows potential as a plasmid DNA adjuvant for the development of novel DNA vaccination modalities.
2. Materials and methods
2.1. Plasmids
The gWIZ™ high expression vectors encoding the reporter genes luciferase (gWIZ™Luc) and green fluorescent protein (GFP) (gWIZ™GFP) both under control of a optimized human cytomegalovirus (CMV) promoter followed by intron A from the CMV immediate-early (IE) gene were used throughout the study (Gene Therapy Systems, San Diego, CA). Both plasmids are expanded in DH5α E.coli and isolated using Qiagen endotoxin-free plasmid Giga-prep kits (Qiagen, Valencia, CA) according to the supplier's protocol.
2.2. Animals
Balb/c mice (6- to 8-weeks-old females, Charles River Laboratories, Wilmington, MA) were used throughout this study. The animals were kept in groups of five and fed ad libitum. In all studies except histological safety evaluation animals were euthanized by CO2.
2.3. Formulation of plasmid DNA with poloxamers
The poloxamers, Pluronic L61 (batch # WPNT-511B), P85 (batch # WPNT-511B), F127 (batch # WPNT-511B), F88 (batch # WPNT-511B) and F68 (batch # WPNT-511B) were purchased from BASF Co. (Mount Olive, NJ). A mixed composition of L61 (0.25%) and F127 (2%), SP1017 obtained from Supratek Pharma Inc. (Montreal, Canada) was also used in these studies. The 10% w/v stock solutions of block copolymers were prepared by dissolving the block copolymer in sterile Hank's Buffered Salt Solution (HBSS) and subsequently sterilized using 0.22 μm hydrophilic polysulfone membrane sterile filter unit (Nalgene, Rochester, NY). The plasmid DNA and block copolymer stock solutions were added to Gibco™ Hanks Balanced Salt Solution (HBSS) buffer (Invitrogen, Carlsbad, CA) and gently mixed to obtain required concentrations (in case of SP1017 the total block copolymer concentrations at various dilutions are presented). The formulations were used immediately for i.m. injections.
2.4.I.m. injection
Animals were anesthetized by intraperitoneal (i.p.) injection of 30 μL/mouse of a mixed solution of ketamine (80mg/ml) and xylazine (12mg/ml) (Sigma, St. Louis, MO). The 10 μg plasmid DNA in 50 μl of HBSS or 50 μl of the block copolymer solutions in HBSS was injected directly into right tibialis anterior muscles of the mice.
2.5. Histological safety evaluation
Animals were sacrificed via cervical dislocation; liver, spleen, kidney, salivary glands and muscle were then removed en bloc, inflated with 1 cc neutral buffered 10% formalin, fixed overnight in 10% formalin, embedded in paraffin, sectioned at 5 μm, and stained. Hematoxylin and eosin (H&E), periodic acid-Schiff with diastase (PAS) and Masson's trichrome (MT) stains were performed.
2.6. Luciferase expression
The mice were euthanized at the indicated time points. The muscle, kidney, spleen, liver and lymph nodes were dissected and weighted, and then rapidly homogenized with a Tissue Tearor in 400μL cell lysis buffer (Promega, Madison, WI). The extraction mixture was kept on ice for 30 min. and then centrifuged at 13,000 g for 2 min. The supernatants were kept for analysis of the luciferase activity. The assay was performed as follows: 10 μL of the supernatant was added to luminometric tubes and supplemented with 100 μL of luciferase substrate solution (Promega, Madison, WI). Light emission was measured with a luminometer (Promega, Madison, WI) for a period of 20 sec. The calibration curve was prepared using standard solution with known concentration of QuantiLum® recombinant luciferase (Promega, Madison, WI). The data were reported as pg luciferase per mg of tissue.
2.7.In vivo imaging of luciferase expression
Luciferase activity was measured in live animals using the Xenogen In Vivo Imaging System - IVIS 200 (Xenogen Corporation, Alameda, CA, USA). A charged-coupled device camera was used to detect bioluminescene emitted from D-luciferin (potassium salt; Xenogen Corporation, Alameda, CA, USA), which reacts with firefly luciferase in living animals. While under isoflurane anaesthesia, the mice received D-luciferin i.p. (30 mg/kg body weight). One minute after the infusion, the light emitted by luciferase was measured, with a 1 min integration time. The signal intensity was quantified as the sum of the photons detected and results were analyzed using Xenogen analysis software (Xenogen Corporation, Alameda, CA, USA).
2.8. GFP expression
The muscle, liver, kidney, spleen and inguinal lymph nodes were embedded in Tissue-Tek OCT (Bakura Finetec Inc. Torrance, CA), rapidly cooled to -80°C and sectioned with cryostat. The sections of 20 μm were atached to Superfrost® microscope slides (Fisher Scientific) dried for 1 hour at room temperature (r.t.) and kept in -80°C for subsequent use. The samples were analyzed for GFP fluorescence (excitation 488) using Zeiss 410 Confocal Laser Scanning Microscope equipped with an Argon-Krypton Laser or processed for immunohistochemistry as described below.
2.9. Immunohistochemistry
The muscle, spleen and inguinal lymph nodes specimens were harvested and processed as described above. Double staining immunofluorescence was performed in frozen muscle tissue sections cut on plain glass slides to determine which cell types express the GFP. The glass slides were sequentially treated with (a) polyclonal rabbit anti-GFP antibodies, anti-rabbit biotinylated secondary antibodies and Alexa 488 (green) strepatavidin conjugate to increase the green signal and then (b) with antibody to one of the markers defining the different cell types (myocytes, keratinocyte/epithelial cells, DC, macrophages/myeloid cells and T cells) and secondary anti-species (rat/mouse) antibodies conjugated to a Alexa 594 (red) fluorochrome. Specifically, the 20 μm frozen sections of tissues (five slides per each tissue specimen) were rinsed three times with Tris-buffered saline (100 mM Tris, 0.9% NaCl, pH=7.6), placed in the blocking/permeabilization solution (BPS) containing 0.5% saponin and 1% normal goat serum for 1 h at r.t., rinsed three times, and incubated overnight at 4°C with polyclonal rabbit anti-GFP primary antibody (Abcam Ltd, Cambridge, UK). After that the sections were rinsed again three times and incubated 2 h at r.t. with biotinylated goat anti-rabbit secondary antibody (1:200) and then with Alexa 488 labeled streptavidine (Dako, Carpinteria, CA USA). Sections processed for the GFP expression were rinsed twice with BPS and treated overnight at 4°C with one of the primary antibodies: 1) anti-CD11b/rat (BD Pharmingen, San Jose, CA USA) against macrophages/myeloid cell marker CD11b; 2) Clone NLDC-145/mouse (Cedarlane, Burlington, NC USA) against DC marker DEC-205; 3) Anti-CD3e rat (Dako, Carpinteria, CA USA) against T-cells marker CD3e; 4) Anti-Desmin(D33)/mouse (Dako, Carpinteria, CA USA) against myocytes marker desmin D33; or 5) Anti-Cytokeratin 5 and 8/mouse (RDI) against keratinocytes markers cytokeratin 5 and 8. Two negative control specimens (treated with isotype- and species-matched antibodies) were obtained from the same mouse (2 slides/each sample). The incubation was performed at 4°C overnight then slides were rinsed with PBS three times and stained for 45 min at r.t. with the secondary goat anti-mouse or goat anti-rat biotynalated antibodies conjugated to Alexa 594 (red) fluorochrome. Finally, the samples were counterstained with DAPI and analyzed by Zeiss 410 Confocal Laser Scanning Microscope equipped with an Argon-Krypton Laser (488 nm (GFP expression), 594 nm (cell marker) and 647 nm (nucleus)).
2.10. Immunostaining and flow cytometry
The spleen and lymph nodes were harvested and placed into a stomacher bag containing 10-15 ml complete RPMI (10% FCS). Cells were isolated from spleen and lymph nodes by disruption of tissue with a stomacher (Tekmar, Cincinnati, OH) and washed three times in PBS containing 2% FBS (PBS-FBS). Concentration of viable cells was determined using hemocytometer and 0.1% Trypan Blue solution and cell concentration was adjusted to 1×106 cells/ml. Cell were stained with various fluorescently labeled monoclonal antibodies against murine cell surface markers (eBioscience, San Diego, CA) (Table 1). Cell samples were incubated in the presence of optimized concentrations (Table 1) of each antibody for 30 min. and washed three times in cold PBS-FBS. Stained samples were fixed with stabilizing fixative (BD Biosciences, San Jose, CA), kept in dark and acquired with a FACSAria flow cytometer (Becton Dickinson, Boston, MA) at UNMC Cell Analysis Facility. Forward and side scatters were collected on a linear scale, while other signals were collected on a 4-decade log scale. Minimum 50,000 events were acquired and the frequency distribution of different cells was determined. Data analysis was performed using Attractors™ software packages (Becton Dickinson, San Jose, CA).
Table 1.
Monoclonal antibodies used in experiment to determine effects of treatments on immune cells populations.
| Monoclonal Antibodies | Excitation, nm | Emission, nm | Amount used, μg per 106 cells in 100 μl |
|---|---|---|---|
| Alexa Fluor® 700 anti-mouse CD11c (Integrin αx, p150/90) | 633 | 723 | 0.25 |
| Allophycocyanin anti-mouse CD11c (Integrin αx, p150/90) | 633 | 660 | 0.25 |
| Allophycocyanin anti-mouse CD14 | 633 | 660 | 0.5 |
| Allophycocyanin-Alexa Fluor® 750 anti-mouse CD4 (L3T4) | 633 | 779 | 0.25 |
| Fluorescein isothiocyanate anti-mouse pan-NK cells (CD49b, α2 integrin, very late antigen-2) | 488 | 518 | 0.5 |
| Phycoerythrin anti-mouse CD19 | 488 | 575 | 0.5 |
| Phycoerythrin anti-mouse CD40 | 488 | 575 | 0.5 |
| Phycoerythrin anti-mouse CD80 (B7-2) | 488 | 575 | 0.12 |
| Phycoerythrin anti-mouse CD86 (B7-2) | 488 | 575 | 0.125 |
| Phycoerythrin-Cy5 hamster anti-mouse CD3e (epsilon subunit; CD3e) | 488 | 670 | 0.25 |
| Phycoerythrin-Cy7 anti-mouse CD8a (alpha subunit, CD8a, Ly-2) | 488 | 760 | 0.5 |
2.11. Polymerase chain reaction (PCR)
The mice were sacrificed on 4th day after the i.m. injection of the gWIZ™Luc plasmid, each injected muscle, liver, kidney, spleen and lymph nodes were harvested and the total DNA was extracted. The PCRs were carried out for 35 cycles using Promega PCR Core System II kit (Promega, Madison WI). 5′TCAAAGAGGCGAACTGTGTG3′ and 5′GGTGTTGGAG-CAAGATGGAT3′ were used as the forward and reverse primers to amplify a 230 bp fragment of gWiz™Luc DNA. In addition a 474 bp fragment of glyceraldehydes-3-phosphate dehydrogenase (GAPDH) was amplified using 5′AAGGTGAAGGTCGGAG-TCAACG3′ and 5′AAGTTGTCATGGATGACCTTGG3′ as primers. The PCR products were visualized by electrophoresis in 2% agarose gels containing ethidium bromide (0.5 μg/mL). Images were acquired and analyzed as described above. The data were expressed as arbitrary units: density of luciferase/density of GAPDH band.
2.12. Reverse transcription polymerase chain reaction (RT-PCR)
The mice were sacrificed on 4th day after the i.m. injection of the gWiz™Luc and RNA were extracted from each injected muscle, liver, kidney, spleen and lymph nodes. 2μg of total DNase-treated RNA from each sample used for cDNA synthesis with M-MLV reverse transcriptase. The cDNA was further used for the RT-PCR. RT-PCR was performed using a Promega PCR Core System II kit (Promega). A typical reaction mixture contained 1.5 mM MgCI2, 0.2 mM dNTPs, 1.25 units of Taq DNA polymerase, and 0.5 μM of each primer. Amplifications were carried out for 30 cycles of 94°C for 1 min, 62°C for 30 sec, and 72°C for 30 sec followed by a final extension at 72°C for 5 min. PCR products were analyzed by electrophoresis in 2% agarose gels. Images were acquired using Gel Doc (Bio-Rad). For each sample, three pairs of primers were used to amplify gene fragment of luciferase (230 bp) and GAPDH. The primers were TCAAAGAGGCGAACTGTGTG and GGTGTTGGAGCAAGATGGAT (luciferase), and AAGGTGAAGGTCGGAGTCAACG and AAGTTGTCATGGATGACCTTGG (GAPDH). The PCR products were visualized by electrophoresis in 2% agarose gels containing EB (0.5 μg/mL). Images were acquired and analyzed as described above. The data were expressed as arbitrary units: density of luciferase/density of GAPDH band.
2.13. NFκB signaling pathway microarray
C2C12-CMVluc myocytes were treated with 1% P85 for 5 min, washed twice with PBS, and incubated for additional 3 h. Cells were collected and total RNA was isolated with TRIzol reagent, then quantified and qualified by UV spectrophotometry and Gel electrophoresis, respectively. Oligo GEArray® with 3 μg of total RNA of each sample was performed following the manufacturer's protocol (cDNA synthesis; cRNA synthesis, labeling and amplification; cRNA cleanup; Oligo GEArray hybridization and detection). Statistical analysis was performed using the GEArray® Expression Analysis Suite software.
3. Results
3.1. Effect of different poloxamers on luciferase gene expression in the muscle
Poloxamers (Pluronic block copolymers) consist of ethylene oxide (EO) and propylene oxide (PO) blocks arranged in a basic A-B-A structure: EOx/2-POy-EOx/2. This block arrangement results in an amphiphilic copolymer, in which the number of hydrophilic EO (x) and hydrophobic PO (y) units can be altered [50]. >Table 2 presents a list of copolymers used in this study and available from BASF Corp. Poloxamers with different number of EO and PO oxide units are characterized by different hydrophilic-lipophilic balance (HLB) [50]. In addition to individual block copolymers listed in the table we also used a mixture of L61 and F127 (1:8), SP1017, which was previously reported to be efficacious and safe in i.m. gene delivery [38, 51].
Table 2.
Physicochemical characteristics of selected poloxamer block copolymers.
| Copolymer | Molecular massa | Average no. EO units (x)b | Average no. PO units (y)c | Hydrophilic-lipophilic balance (HLB)c | Critical micelle concentration (CMC), % w/vd |
|---|---|---|---|---|---|
| Pluronic L61 | 2000 | 4.6 | 31.0 | 3 | 0.02 |
| Pluronic P85 | 4600 | 52.3 | 39.7 | 16 | 0.03 |
| Pluronic F127 | 12600 | 200 | 65.2 | 22 | 0.004 |
| Pluronic F68 | 8400 | 153 | 29.0 | 29 | 0.4 |
Average molecular weights provided by the manufacturer (BASF, Wyandotte, Michigan)
The average number of ethylene oxide (EO) and propylene oxide (PO) units
Hydrophilic-lipophilic balance (HLB) values of the copolymers and the cloud points were determined by manufacturer (BASF, Wyandotte, Michigan)
CMC values were determined previously using pyrene probe [48].
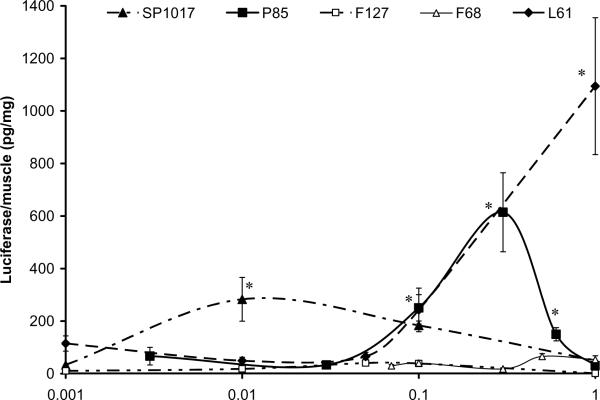
In order to rank the potency of various block copolymers, a plasmid containing a luciferase reporter gene under the control of CMV promoter (gWiz™Luc) was formulated with individual copolymers or SP1017 mixture and administered into tibialis anterior muscles of 6-8 week old Balb/c mice. The dose of the plasmid and the injection volume were kept constant throughout the study (10μg of plasmid in 50 μl HBSS solution), but the concentrations of the copolymers were varied. The reporter gene expression in the muscle was examined as described in the methods section [39]. The results suggest that P85 was among the most effective of the individual copolymers (Fig. 2). Specifically, the gene expression of a plasmid formulated with P85 reached a maximum at 0.3% w/v P85, and was increased ca. 20-fold compared to the naked DNA. At concentrations higher or lower than 0.3% w/v, the efficiency of the transgene expression declined. Interestingly, the optimal concentration was above the P85's critical micelle concentration (CMC) (Table 2).
Fig. 2.
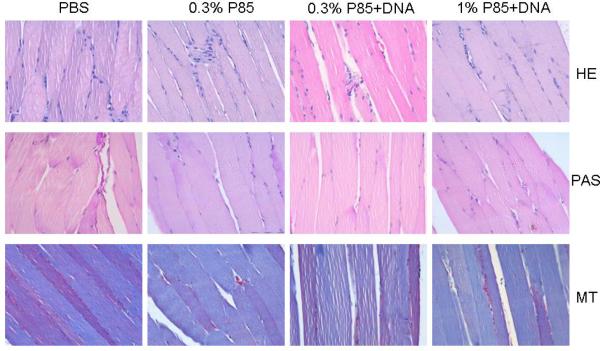
Tissue compatibility of P85 and DNA/P85 injections. Staining of muscle sections with H&E, PAS and MT were performed as described in Methods section.
A lipophilic copolymer L61 also appeared to show high efficacy at 0.1% and 1% w/v. However, at these doses L61 precipitated and was likely to be toxic [52]. Surprisingly, the same copolymer mixed with F127, SP1017, exhibited significant activity at a remarkably lower concentration, 0.01% w/v of the total copolymer. At this concentration SP1017 increased DNA expression in the muscle by ca. 15-fold compared to the DNA alone. It was also quite interesting that the second block copolymer in this formulation, F127 alone was practically inactive but appeared to amplify the activity of L61 at low concentrations. The optimal concentration of SP1017 was below the CMC of L61 and above the CMC of F127 (Table 2). Previous studies suggest that under these conditions L61 and F127 form mixed micelles [52]. Notably, SP1017 was previously clinically evaluated in combination with doxorubicin in cancer patients and was shown to be safe after intravenous (i.v.) administration at much higher doses [53, 54]. Similarly to F127 the hydrophilic copolymer F68 was practically inactive at least up to 1 % w/v (Table 2). For the subsequent studies, we used P85, which was characterized in our previous studies in vitro and in vivo as adjuvant for gene delivery [30, 33, 39, 55-57]
3.2. P85 safety evaluation
The safety of P85 was evaluated by injecting 50 μl of 1) P85 alone at 0.3% (optimal concentration) or 1% (maximal concentration), and 2) 0.3% P85 with 10 mg plasmid DNA (gWIZ™GFP). The 50 μl HBSS solution injections were used as a control. The injected muscles were harvested on day 4, fixed, dehydrated, and analyzed by H&E, PAS and MT staining. The representative images of the muscle samples in various groups are shown in Fig. 3. In summary, both P85 and P85/DNA injections induced low inflammatory responses with comparable number of monocytes and neutrophils at the injection site. There was no change in morphology associated with the increase in glycogen. No necrosis was observed for any of the groups. All together, P85 alone and P85 with DNA exhibited good tissue compatibility in mouse muscle.
Fig. 3.
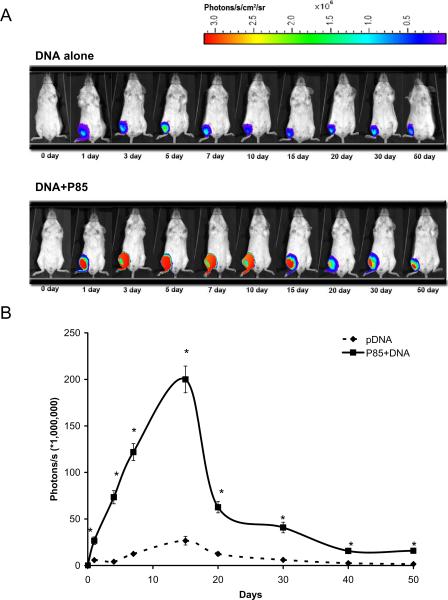
Effect of 0.3% P85 on luciferase gene expression in vivo after a single i.m. injection of 10 μg plasmid DNA in 50 μl HBSS. (A) A representative in vivo imaging in a mouse. (B) Quantitative data of in vivo imaging study. Data are mean ± SEM (n = 5). P values were obtained by the means of the Student's t test following logarithmic transformation of the data. Each P value corresponds to the comparison of naked DNA versus DNA formulated with P85: * p < 0.05.
3.3. P85 effect on the time course of plasmid DNA expression
In order to determine the time course of the luciferase expression in vivo the gWiz™Luc plasmid with/without 0.3 % P85 was injected i.m. in Balb/c mice (5 per group) and the animals were subjected to non-invasive bioluminescence detection on various days after injection (Fig. 4). The areas under a curve for period of 50 days were approx. 3.36 × 109 and 2.98 × 108 (photons/s) x days for DNA/P85 and naked DNA respectively as determined using trapezoidal rule (Fig. 4B). Therefore, the block copolymer enhanced the cumulative exposure of the animals to the transgene expression by 11-folds. Furthermore, the transgene expression was greatly prolonged and lasted for at least 50 days. The gene expression of the naked DNA peaked around the 5th day and appeared to be significantly less than that of the DNA/P85 at every time point.
Fig. 4.
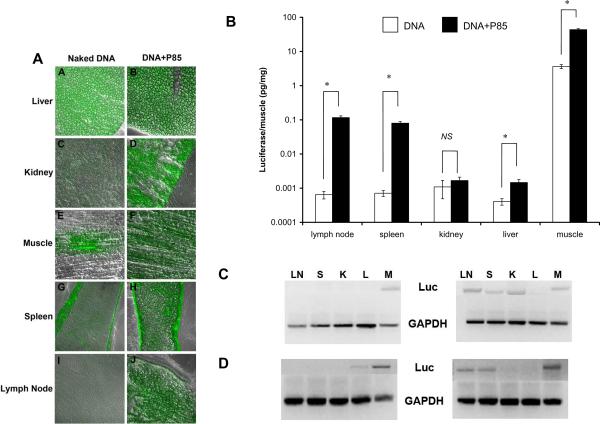
Effects of 0.3 % P85 on (A) GFP or (B) luciferase gene expression, and (C) DNA or (D) RNA levels in various organs. Tissue samples were collected on day 4 after single i.m. injections of 10 μg DNA in 50 μl HBSS solution with/without copolymer. (B) Data are mean ± SEM (n = 5). P values were obtained by the means of the Student's t test. Each P value corresponds to the comparison of naked DNA versus DNA formulated with P85: * p < 0.05, NS is not significant.
3.4. Effect of P85 on distribution of gene expression in the organs
The tissue distribution of gene expression was analyzed following single i.m. injections of gWiz™Luc and gWiz™GFP plasmids with/without 0.3% P85. Surprisingly, both GFP and luciferase data suggest that in addition to increased gene expression in the muscle there was a considerable increase in the gene expression in spleen, inguinal lymph nodes and liver (Fig. 5). For example, with the DNA/P85 the gene expression of luciferase in spleen and lymph nodes increased by over two-orders of magnitude and reached about 2 % and 3 % respectively of the levels observed in the muscle with the naked DNA. Furthermore, the RT-PCR and PCR experiments suggested that there were considerable increases in the luciferase mRNA and DNA levels in these organs (Fig. 5).
Fig. 5.
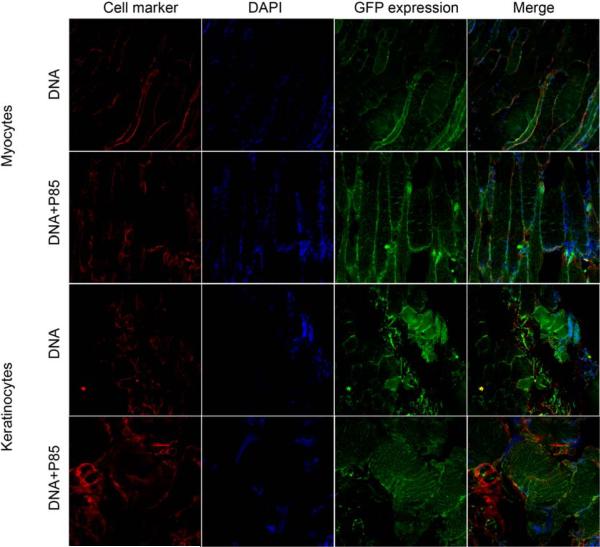
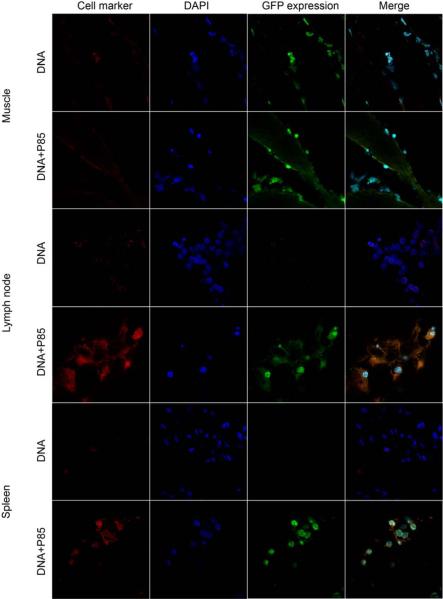
Confocal imaging of GFP expression in muscle tissue after single i.m. injection of naked DNA or DNA/P85. The color staining corresponds to myocytes or keratinocytes (red), nucleus (blue), and GFP (green). The last panels in each row present digitally superimposed images of three preceding panels to visualize the colocalization (yellow or white). Magnification 63×.
3.5. Co-localization of gene expression with cell-specific markers
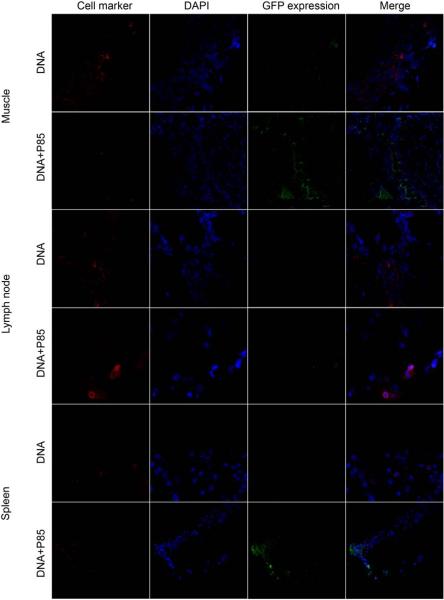
Co-localization studies were designed to determine which cell types express the transgene in the muscle, lymph nodes, and spleen. The gWiz™GFP plasmid with/without 0.3% w/v P85 was injected i.m. in the BALB/c mice, the tissues were harvested on day 4 after injection and processed for double staining immunohistochemistry and confocal microscopy. The images were digitally superimposed to determine which cells express the transgene. In the muscle tissues the reporter gene was expressed in myocytes and keratinocytes (Fig. 6) as well as DC (Fig. 7) and macrophages (Fig. 8), but not in T-cells (Fig. 9). Notably, in the muscle GFP expression co-localized with myocytes, keratinocytes, DC, and macrophages both after the naked DNA and DNA/P85 injections. In contrast, in the spleen and lymph nodes the GFP expression co-localized with DC (Fig. 7) and macrophages (Fig. 8) only in the case of DNA/P85. No colocalization with T-cells was observed (Fig. 9). It also worth mentioning that in the spleen and especially in lymph nodes the DNA/P85 appears to increase DC staining compare to the DNA. This may be caused by infiltration of DC or increased expression of the DC markers (DEC-205) as a result of activation of DC. Therfore, we further examined the effect of various treatments on the immune cells populations in spleen and lymph nodes.
Fig. 6.
Confocal imaging of GFP expression in DC in muscle, draining lymph nodes and spleen after single i.m. injection of naked DNA or DNA/P85. The color staining corresponds to DC (red), nucleus (blue), and GFP (green). The last panels in each row present digitally superimposed images of three preceding panels to visualize the colocalization (yellow or white). Magnification 63×.
Fig. 7.
Confocal imaging of GFP expression in macrophages in muscle, draining lymph nodes and spleen after single i.m. injection of naked DNA or DNA/P85. The color staining corresponds to macrophages (red), nucleus (blue), and GFP (green). The last panels in each row present digitally superimposed images of three preceding panels to visualize the colocalization (yellow or white). Magnification 63×.
Fig. 8.
Confocal imaging of GFP expression in T-cells in muscle, draining lymph nodes and spleen after single i.m. injection of naked DNA or DNA/P85. The color staining corresponds to T-cells (red), nucleus (blue), and GFP (green). The last panels in each row present digitally superimposed images of three preceding panels to visualize the colocalization (yellow or white). Magnification 63×.
Fig. 9.
CD49+, CD3e+, CD8a+, CD4+, CD14+, and CD11c+ cell frequency in spleen (A) and lymph nodes (B) of Balb/c mice injected with HBSS, DNA, P85 and DNA/P85. The cells frequency was determined by flow cytometry on day 4 after single i.m. injection. Data are mean ± SEM (n = 5). Statistical comparisons between the groups are made using ANOVA: * p < 0.05, NS is not significant.
3.6. Effect of treatments on immune cells populations
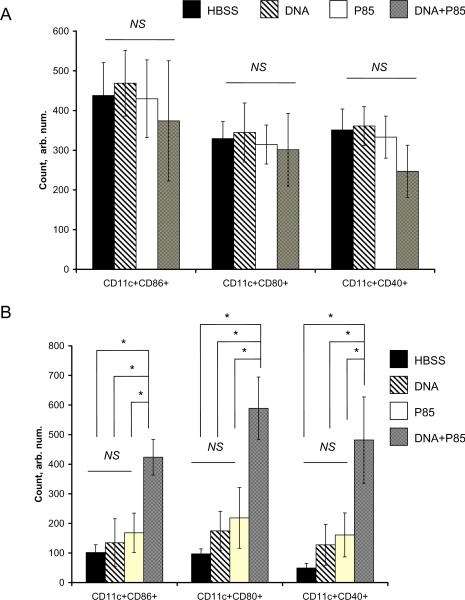
To evaluate effects of different treatments on expansion of immune cells in spleen and lymph nodes the BALB/c mice (5 per group) were injected i.m. with: 1) HBSS (control); 2) gWiz™GFP DNA, 3) P85 and 4) DNA/P85. The spleen and inguinal lymph nodes were harvested on the day 4 after injection and immune cells frequency was determined by flow cytometry. Injections of P85 alone and DNA/P85 increased systemic (spleen) expansion of CD11c+ (DC); and local (lymph nodes) expansion of CD11c+, CD14+ (macrophages) and CD49b+ (natural killer) cell populations (Fig. 10). No changes in systemic expansion of CD14+, CD49b+, CD3e+ (general T-cells), CD8a+ (cytotoxic T-cells), and CD4+ (helper T-cells) was observed with any treatment group (Fig. 10A). Similarly, no difference in local expansion of CD3e+, CD8a+, CD4+ populations was observed (Fig. 10B). Interestingly, naked DNA also significantly increased CD11c+ (but not CD14+ and CD49b+) cells in both spleen and lymph nodes. However, these increases (12 % spleen and 16% lymph nodes) were much less than the effects of P85 (59 % spleen, 97 % lymph nodes) and DNA/P85 (57 % spleen, 55% lymph nodes). (The differences between P85 and DNA/P85 treatment groups were not significant in spleen but significant in the lymph nodes.) Therefore, the naked DNA, the block copolymer and the DNA formulated with the block copolymer each increased the local and systemic population of DC, however, the effects of copolymer formulations were much greater than those of the naked DNA. We further examined the effects of different injections on the DC maturation (Fig. 11). No difference in splenic DC was observed with any treatment (Fig. 11A). However, in the lymphatic DC there were significant increases in each of CD11c+CD86+, CD11c+CD80+, and CD11c+CD40+ cells frequency after DNA/P85 treatment, but not after free DNA of P85 alone treatments (Fig. 11B). Therefore, the DNA/P85 combination promotes maturations of the DC.
Fig. 10.
CD11c+CD86+, CD11c+CD80+, and CD11c+CD40+ cell frequency in spleen (A) and lymph nodes (B) of Balb/c mice injected with HBSS, DNA, P85 and DNA/P85. The cells frequency was determined by flow cytometry on day 4 after single i.m. injection. Data are mean ± SEM (n = 5). Statistical comparisons between the groups are made using ANOVA: * p < 0.05, NS is not significant.
3.7. Activation of NFκB signaling pathway by P85
P85 was previously shown to activate the NFκB signaling pathway by inducing phosphorylation of IκB [33]. Here we examined the effect of P85 on NFκB-related genes using microarray (Table 3). The block copolymer significantly upregulated 39 out of 113 genes related to NFκB-mediated signal transduction.
Table 3.
Effect of P85 on expression of NFκB related genes in C2C12-CMVluc cells.a Total RNA was assayed using Oligo GEArray® Mouse NFκB Signaling Pathway Microarray and gene expression was analyzed by GEArray Expression Analysis Suite software (see http://www.superarray.com/gene_array_product/HTML/OMM-025.html for detailed list of genes and layout). Only genes exhibiting at least 2-fold increase in expression are presented. (i) NFκB pathway: (3, 4, 30, 72, 77, 78, 70, 84) transcription factors; (6, 10, 11, 108, 110, 112, 66, 74) other factors; (53, 54, 56, 60, 61, 64, 87, 88, 71, 80) kinases; (99, 103) ligands and receptors; (100, 113) inflammatory response, extracellular or membrane molecules. (ii) activation of NFκB pathway: (8, 105) ligands and receptors; (35, 5, 22) IκB kinase/NFkB cascade; (37) transcription factor; (32) extracellular or membrane molecules; (iii) NFκB responsive genes: (42, 48) ligands and receptors, or cytokines.
| Position | Symbol | Refseq | Description | Folds increased |
|---|---|---|---|---|
| 3 | Atfl | NM_007497 | Activating transcription factor 1 | 2.9 |
| 4 | Atf2 | NM_009715 | Activating transcription factor 2 | 4.6 |
| 30 | Hmgb1 | NM_010439 | High mobility group box 1 | 2.3 |
| 72 | Polr2a | NM_009089 | Polymerase (RNA) II (DNA directed) polypeptide A | 2.9 |
| 77 | Rel | NM_009044 | Reticuloendotheliosis oncogene | 4.5 |
| 78 | Rela | NM_009045 | V-rel reticuloendotheliosis viral oncogene homolog A | 2.4 |
| 70 | Pcaf | NM_020005 | P300/CBP-associated factor | 2.2 |
| 84 | Smad4 | NM_008540 | MAD homolog 4 (Drosophila) | 4.3 |
| 6 | Bcl3 | NM_033601 | B-cell leukemia/lymphoma 3 | 3.3 |
| 10 | Card14 | NM_130886 | Caspase recruitment domain family, member 14 | 2.2 |
| 11 | Card4 | NM_172729 | Caspase recruitment domain 4 | 2.2 |
| 108 | Tradd | NM_001033161 | TNFRSF1A-associated via death domain | 3.2 |
| 110 | Traf3 | NM_011632 | Tnf receptor-associated factor 3 | 2.1 |
| 112 | Traf6 | NM_009424 | Tnf receptor-associated factor 6 | 2.8 |
| 66 | Nalp12 | XM_355971 | NACHT, LRR and PYD containing protein 12 | 2.0 |
| 74 | Ppp5c | NM_011155 | Protein phosphatase 5, catalytic subunit | 3.6 |
| 53 | Map2k6 | NM_011943 | Mitogen activated protein kinase kinase 6 | 2.6 |
| 54 | Map3k1 | NM_011945 | Mitogen activated protein kinase kinase kinase 1 | 3.0 |
| 56 | Map3k3 | NM_011947 | Mitogen activated protein kinase kinase kinase 3 | 4.4 |
| 60 | Map4k2 | NM_009006 | Mitogen activated protein kinase kinase kinase kinase 2 | 4.4 |
| 61 | Mapk11 | NM_011161 | Mitogen-activated protein kinase 11 | 4.4 |
| 64 | Mapk8 | NM_016700 | Mitogen activated protein kinase 8 | 4.1 |
| 87 | Tgfbr1 | NM_009370 | Transforming growth factor, beta receptor I | 2.1 |
| 88 | Tgfbr2 | NM_009371 | Transforming growth factor, beta receptor II | 2.8 |
| 71 | Plk2 | NM_152804 | Polo-like kinase 2 (Drosophila) | 6.4 |
| 80 | Ripk1 | NM_009068 | Receptor (TNFRSF)-interacting serine-threonine kinase 1 | 4.0 |
| 99 | Tnfrsf10b | NM_020275 | Tumor necrosis factor receptor superfamily, member | 2.7 |
| 103 | Tnfrsf7 | NM_001033126 | 10b Tumor necrosis factor receptor superfamily, member 7 | 4.4 |
| 100 | Tnfrsf1a | NM_011609 | Tumor necrosis factor receptor superfamily, member 1a | 2.3 |
| 113 | Vapa | NM_013933 | Vesicle-associated membrane protein, associated protein A | 2.2 |
| 8 | Card1O | NM_130859 | Caspase recruitment domain family, member 10 | 5.1 |
| 105 | Tnfsf14 | NM_019418 | Tumor necrosis factor (ligand) superfamily, member 14 | 3.6 |
| 35 | Ikbkb | NM_010546 | Inhibitor of KappaB Kinase beta | 1.6 |
| 5 | Bcl10 | NM_009740 | B-cell leukemia/lymphoma 10 | 2.9 |
| 22 | Edaradd | NM_133643 | EDAR (ectodysplasin-A receptor)-associated death domain | 4.3 |
| 37 | Ikbkg | NM_010547 | Inhibitor of KappaB Kinase gamma | 1.5 |
| 32 | Icam1 | NM_010493 | Intercellular adhesion molecule | 3.9 |
| 42 | Il6 | NM_031168 | Interleukin 6 | 2.5 |
| 48 | Lta | NM_010735 | Lymphotoxin A | 4.7 |
Cells were exposed to 1% P85 for 5 min, washed by PBS and incubated for additional 3hxsx.
4. Discussion
Previous studies have demonstrated that certain poloxamers such as SP1017 mixture, F68 (poloxamer 188), P85, and L64 (PE6400) can increase transgene expression of the naked DNA administered to muscle tissue [32, 38, 58-61]. It has also been long known that poloxamers display immuno-adjuvant activity with respect to polypeptides [23, 25, 62-66]. Pro-inflammatory effects of certain poloxamers such as SP1017 and P85 have been also reported [39, 47, 48, 67]. In this study we report for the first time, that P85 co-administered with plasmid DNA a) significantly enhances plasmid expression in the spleen and lymph nodes; b) facilitates plasmid DNA uptake and expression in myocytes, keratinocytes, DC and macrophages; c) stimulates local and systemic expansion of APC and local maturation of DC.
The DNA or DNA/poloxamer compositions were administered as a single i.m. injection, which is a generally accepted route for DNA vaccination [68, 69]. A successful DNA vaccination requires efficient transgene expression in the host organism [70-72]. Therefore, it is important that select poloxamer compositions have greatly increased DNA expression in the muscle. Interestingly, the hydrophilic copolymers, F68 and F127 were practically ineffective, while more lipophilic copolymer, P85 or a mixture of lipophilic L61 and hydrophilic F127 (SP1017 mixture) exhibited high activity. Furthermore, this study reinforced our previous reports that these active copolymers greatly prolong the transgene expression in the muscle. This alone may be a significant advantage for future DNA vaccination approach compared to a naked DNA, which may not enable sufficient duration of expression to elicit strong immune response [38, 39, 73, 74].
For the first time we report that DNA/P85 administration results in a drastic increase in the transgene expression not only in the immediate site of injection in the muscle but also in the distant sites, most noticeably, the spleen and draining lymph nodes. A significant increase in the systemic levels of transgene in the plasma following DNA/SP1017 administration was previously reported by us [38]. In the present study, following DNA/P85 injection in the muscle the transgene expression in the spleen and lymph nodes increased by over two orders of magnitude and reached ca. 2 to 3 % of the levels observed in the muscle with the naked DNA. This may enhance the transgene presentation to the immune system because APC in the central immune organs are abundant, as well as phenotypically and functionally immature [75]. Moreover, the transgene expression in the muscle, spleen and lymph nodes co-localized with APC such as macrophages and DC. Based on this, it is reasonable to suggest that in response to administration of the DNA/P85 the APC incorporated, expressed and transported the transgene to the central immune organs. This is reinforced by the fact that the spleen and lymph nodes in addition to the protein product also exhibited increased levels of transgene mRNA and DNA. Notably, in the muscle the transgene expression was also abundant in myocytes and keranotinocytes. Therefore, we cannot exclude that along with the incorporated plasmid DNA the APC also incorporated the protein product expressed by the muscle tissue and then carried this product to the immune organs. Altogether, this should enhance representation of the expressed transgene to the immune system after i.m. administration of the DNA/poloxamer compositions.
The increase in the immune system engagement in response to the copolymer-based compositions is critical because the DNA-encoded antigens alone may not be sufficiently immunogenic to initiate APC-mediated adaptive immune responses [76]. The ability of poloxamers to activate immune system was previously described by Hunter and others [23, 25, 26, 62-65]. In particular, a lipophilic copolymer, Pluronic L121 injected in an oil-in-water emulsion, was found to be a powerful adjuvant for increasing antibody formation to polypeptides [23, 25, 63]. Another, lipohphilic copolymer, Pluronic L101, was less effective in antibody formation, but induced granulomatous inflammation. In addition, such poloxamers promoted histamine release in mediator containing cells [22, 25, 26, 63, 77-79]. Notably, these studies concluded that strong adjuvant activities of poloxamers were most profound in the case of most lipophilic copolymers with HLB values of 2 or less [23, 25, 63]. At the same time, it is known that such poloxamers induce profound muscle toxicity, which is proportional to the copolymer lipohilicity - the more lipophilic the copolymer, the more severe the lesions [80]. Therefore, it is significant that our studies have focused on adjuvant effects of less lipophilic or hydrophilic copolymers such P85 (HLB = 16) and SP1017 (HLB = 20 for L61 and F127 blend). The latter, SP1017 was shown to be safe in two clinical trials following i.v. injections (in combination with Doxorubicin) [54, 81]. Furthermore, based on the present study P85 did not induce toxicity in the muscle after single injection. It is remarkable, therefore, that this copolymer also exhibited immunoajuvant properties, both alone and in combination with the plasmid DNA. The infiltration of DC to the site of injection and local DC population expansion after i.m. injections of adjuvant Fms-like tyrosine kinase (Flt3L) plasmid formulated with SP1017 was also observed previously [47, 48]. Interestingly, the present study provides evidence of both local and systemic expansion of APC in response to administration of DNA/P85 or P85 alone in the absence of a strong DNA adjuvant such as Flt3L or granulocyte macrophage-colony stimulating factor (GMCSF). Furthermore, locally, the maturation of DC was observed when DNA was administered with P85 (but not the copolymer alone). It appears, therefore, that P85 enhances intrinsic immunogenicity of a plasmid DNA. This immunogenicity is related to the presence of unmethylated CpG rich sites or traces of bacterial lipopolysaccharide (LPS), which stimulates the innate immune response through recognition by toll-like receptor 4 [82]. Notably, Ratanamart et al. reported that SP1017 can increase pro-inflammatory response to a CpG-rich bacterial plasmid. However, no increases in gene expression in APC were reported in that study. Furthermore, contrary to the plasmids used in Ratanamart et al., in our case the plasmids were endotoxin-free and did not induce toxicity or strong inflammatory responses in the muscle [67]. Despite increased transgene expression and activation of APC, injections of the P85/DNA formulations resulted in sustained gene expression for at least 50 days, which further reinforces the lack of cytotoxic immune response in the muscle. Altogether, it appears that in addition to enhancing gene expression, P85 can stimulate the innate immune system by attracting and activating APC, which can result in development of a long lasting, antigen specific immune response [83].
The adjuvant effects of poloxamers in 1) increasing transgene expression and 2) activating the immune system may be interrelated. We have shown that P85 can activate the NFκB signaling pathway (by inducing phosphorylation of IκB) [33] and upregulate many genes related to NFκB-mediated signal transduction in the muscle cells (this study). Activation of NFκB was related to the ability of poloxamers, in particular P85, to activate transcription of transgenes driven by the NFκB-responsive promoter (such as CMV) in stably transfected cells [33]. A more recent study demonstrated that poloxamers also enhance DNA delivery into cells, including the nuclear translocation of NFκB-responsive plasmids [84]. Based on this it appears that following DNA/P85 injections in the muscle the copolymer plays a critical role by increasing the cellular delivery and subsequent transcription of the plasmid DNA. This is further reinforced by the fact that the increase in the gene expression in the muscle by P85 (and SP1017) was promoter-selective for NFκB-responsive plasmids [39]. Altogther, the enhancement in the transgene expression observed with DNA/P85 appears to be due to the activation of the NFκB signaling pathway by the copolymer. The same signaling pathway is implicated in the activation of APC and the overall immunity [85]. This may explain the simultaneous increases in gene expression in the muscle tissue and antigen presenting cells following administration of DNA/P85.
It needs to be highlighted that the mechanisms of the poloxamer effects may not include their interactions with the DNA proper. It has been specifically pointed out before that in the range of the concentrations used in this study P85 did not bind with and condense the plasmid DNA, In addition, no retardation in the movement of the DNA upon mixing with Pluronic P85 and any protection offered by Pluronic P85 upon treatment of the mixture with DNase [39]. Therefore, it cannot protect DNA from degradation or promote DNA transport into the cell like cationic molecules do. Most likely, following the injections of DNA/P85 the plasmid is internalized into cells through the regular pathways that are also employed by the naked DNA. However, the cellular transport through these pathways and overall transgene expression are enhanced by the copolymer as described above. We posit that the critical role in exhibition of the adjuvant effects of poloxamers belongs to their interactions with the lipophilic domains of the cellular membranes. In this respect it is important that hydrophilic poloxamers, such as F68 or F127, that do not penetrate well into the cellular membranes, also have little if any effect on the gene expression [86]. In contrast, more lipophilic copolymers including P85 were shown to bind with membranes, presumably by inserting their hydrophobic PPO chains into the lipid environment [86]. Most recently it was shown that below the critical micelle concentration (CMC) P85 co-localizes with caveolae markers and is transported into cells via caveolae-mediated endocytosis [57]. As the concentration of the P85 increases above the CMC the copolymer appears to disrupt the caveolae and inhibit the caveolae-mediated endocytosis. Notably, caveolae are known to cluster multiple receptors, such as EGF, CCK, Endothelin, Tyrosine Kinases etc., which enable signal integration and amplification [87, 88]. They also host signal transducers, for example, PKC, MAPK, eNOS and calmodulin, transporters (e.g. IP3 receptor and Porin) as well as structural molecules (e.g. actin and myosin). Therefore, activation of cell signaling by P85 may be related to disruption of caveolae by the PPO chains. We cannot exclude, however, that the PEO chains of poloxamers may additionally contribute to their adjuvant activities. It is noteworthy that maximal transgene expression of DNA formulated with P85 and SP1017 is observed in the conditions when the micelles are formed. In particular, P85 micelles are spherical core-shell structures of approximately 14.6 nm in diameter [89]. They are composed of approximately 60 P85 molecules and have a PPO core of 5.4 nm in diameter surrounded by 3.7 nm PEO shell. Despite of the fact that PEO is often considered inert some interactions of the copolymers with high PEO content with the surface of cell membranes have been suggested [86]. This may include one or combination of hydrophobic and hydrogen binding interactions, for example, with membrane proteins. Interestingly, several studies suggested that either PEO alone or PEO-covered nanoparticles may induce complement activation [31]. The structural similarity between the terminal region of the PEO chain and carbohydrates has also been noted, which may result in PEO binding with lectins [90] as well as carbohydrate receptors [91] of APC. Therefore, detailed characterization of interactions of poloxamers with cell membranes may provide foundation for future understanding of the diverse biological response-modifying activities of these copolymers, including activation of cell signaling, gene expression and immunity.
Conclusion
We have investigated the effect of non-ionic block copolymer Pluronic P85 administered i.m. on plasmid DNA expression in mouse tissues, organs and immune cells. In summary, the results of the present study demonstrate that co-administration of a plasmid DNA with P85 in a skeletal muscle greatly increases gene expression in the injection tissue site and distant organs, especially the draining lymph nodes and spleen. Reporter expression colocalizes with the specific markers of myocytes and keratinocytes in the muscle, as well as DC and macrophages in the muscle, lymph nodes and spleen. In addition, combination of DNA with P85 and P85 alone increase the systemic expansion of DC, and local expansion of DC, macrophages and natural killer cell populations; but only combination of DNA with P85 increases maturation of local DC. The results presented provide a strong rationale for further development of poloxamer-based compositions for DNA vaccination.
Fig. 1.
Dose dependent effect of various poloxamers on luciferase gene expression in skeletal (tibialis anterior) muscle of BALB/c mice after single i.m. injection of 10 μg plasmid DNA in 50 μl HBSS solution with or without block copolymers (5 mice/group). Muscles tissue was harvested on day 4 after i.m. injection and luciferase activity was measured using a standard procedure described in the Methods section. Data are mean ± SEM (n = 5).The P values were obtained by the means of the Student's t test following logarithmic transformation of the data. Each P value corresponds to the comparison of naked DNA versus DNA formulated with P85: * p < 0.05
Acknowledgments
This study was supported by the National Institutes of Health grant R01 CA116591 (AVK) and the American Heart Association fellowship 0610065Z (ZG). Michael Karas (Supratek Pharma Inc.) provided valuable advice to flow cytometry and immunostaining experiments. The assistance of the following individuals at the University of Nebraska Medical Center (UNMC) is also gratefully acknowledged: Janice A. Taylor and James R. Talaska (confocal microscopy); Charles A. Kuszynski, Linda M. Wilkie, and Victoria B. Smith (flow cytometry); Margaret “Anita” Jennings (histology and immunohistochemistry); Donald R. Johnson and Barbara Switzer (cell isolation); and Terence A. Lawson (in vivo imaging). These studies were carried out using the following UNMC facilities: Confocal Laser Scanning Microscope Core, Cell Analysis Facility, Flow Cytometry Research Core, Molecular Phenotyping Core, Monoclonal Antibody Core, and IVIS 200 In vivo Imaging Core. These facilities are supported by the Nebraska Research Initiative, the UNMC-Eppley Cancer Center and the Nebraska Center for Virology.
References
- [1].Odunsi K, Sabbatini P. Harnessing the immune system for ovarian cancer therapy. Am J Reprod Immunol. 2008 Jan;59(1):62–74. doi: 10.1111/j.1600-0897.2007.00560.x. [DOI] [PubMed] [Google Scholar]
- [2].Lesterhuis WJ, Aarntzen EH, De Vries IJ, Schuurhuis DH, Figdor CG, Adema GJ, et al. Dendritic cell vaccines in melanoma: from promise to proof? Crit Rev Oncol Hematol. 2008 May;66(2):118–34. doi: 10.1016/j.critrevonc.2007.12.007. [DOI] [PubMed] [Google Scholar]
- [3].Brichard VG, Lejeune D. GSK's antigen-specific cancer immunotherapy programme: pilot results leading to Phase III clinical development. Vaccine. 2007 Sep 27;25(Suppl 2):B61–71. doi: 10.1016/j.vaccine.2007.06.038. [DOI] [PubMed] [Google Scholar]
- [4].Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000 Feb;6(2):207–10. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- [5].Flynn JL, Goldstein MM, Triebold KJ, Koller B, Bloom BR. Major histocompatibility complex class I-restricted T cells are required for resistance to Mycobacterium tuberculosis infection. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):12013–7. doi: 10.1073/pnas.89.24.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bojang KA, Milligan PJ, Pinder M, Vigneron L, Alloueche A, Kester KE, et al. Efficacy of RTS,S/AS02 malaria vaccine against Plasmodium falciparum infection in semi-immune adult men in The Gambia: a randomised trial. Lancet. 2001 Dec 8;358(9297):1927–34. doi: 10.1016/S0140-6736(01)06957-4. [DOI] [PubMed] [Google Scholar]
- [7].Terando AM, Faries MB, Morton DL. Vaccine therapy for melanoma: Current status and future directions. Vaccine. 2007 Jul 5; doi: 10.1016/j.vaccine.2007.06.033. [DOI] [PubMed] [Google Scholar]
- [8].Markel H. The search for effective HIV vaccines. N Engl J Med. 2005 Aug 25;353(8):753–7. doi: 10.1056/NEJMp058146. [DOI] [PubMed] [Google Scholar]
- [9].Sharpe S, Hanke T, Tinsley-Bown A, Dennis M, Dowall S, McMichael A, et al. Mucosal immunization with PLGA-microencapsulated DNA primes a SIV-specific CTL response revealed by boosting with cognate recombinant modified vaccinia virus Ankara. Virology. 2003 Aug 15;313(1):13–21. doi: 10.1016/s0042-6822(03)00282-4. [DOI] [PubMed] [Google Scholar]
- [10].Forde GM. Rapid-response vaccines--does DNA offer a solution? Nat Biotechnol. 2005 Sep;23(9):1059–62. doi: 10.1038/nbt0905-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ulmer JB, DeWitt CM, Chastain M, Friedman A, Donnelly JJ, McClements WL, et al. Enhancement of DNA vaccine potency using conventional aluminum adjuvants. Vaccine. 1999 Aug 20;18(1-2):18–28. doi: 10.1016/s0264-410x(99)00151-6. [DOI] [PubMed] [Google Scholar]
- [12].Barouch DH, Santra S, Steenbeke TD, Zheng XX, Perry HC, Davies ME, et al. Augmentation and suppression of immune responses to an HIV-1 DNA vaccine by plasmid cytokine/Ig administration. J Immunol. 1998 Aug 15;161(4):1875–82. [PubMed] [Google Scholar]
- [13].Kim JJ, Trivedi NN, Nottingham LK, Morrison L, Tsai A, Hu Y, et al. Modulation of amplitude and direction of in vivo immune responses by co-administration of cytokine gene expression cassettes with DNA immunogens. Eur J Immunol. 1998 Mar;28(3):1089–103. doi: 10.1002/(SICI)1521-4141(199803)28:03<1089::AID-IMMU1089>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- [14].Xin KQ, Hamajima K, Sasaki S, Tsuji T, Watabe S, Okada E, et al. IL-15 expression plasmid enhances cell-mediated immunity induced by an HIV-1 DNA vaccine. Vaccine. 1999 Feb 26;17(7-8):858–66. doi: 10.1016/s0264-410x(98)00271-0. [DOI] [PubMed] [Google Scholar]
- [15].Seaman MS, Peyerl FW, Jackson SS, Lifton MA, Gorgone DA, Schmitz JE, et al. Subsets of memory cytotoxic T lymphocytes elicited by vaccination influence the efficiency of secondary expansion in vivo. J Virol. 2004 Jan;78(1):206–15. doi: 10.1128/JVI.78.1.206-215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nabel GJ, Nabel EG, Yang ZY, Fox BA, Plautz GE, Gao X, et al. Direct gene transfer with DNA-liposome complexes in melanoma: expression, biologic activity, and lack of toxicity in humans. Proc Natl Acad Sci U S A. 1993 Dec 1;90(23):11307–11. doi: 10.1073/pnas.90.23.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Perrie Y, Frederik PM, Gregoriadis G. Liposome-mediated DNA vaccination: the effect of vesicle composition. Vaccine. 2001 Apr 30;19(23-24):3301–10. doi: 10.1016/s0264-410x(00)00432-1. [DOI] [PubMed] [Google Scholar]
- [18].Mannino RJ, Canki M, Feketeova E, Scolpino AJ, Wang Z, Zhang F, et al. Targeting immune response induction with cochleate and liposome-based vaccines. Adv Drug Deliv Rev. 1998 Jul 6;32(3):273–87. doi: 10.1016/s0169-409x(98)00014-3. [DOI] [PubMed] [Google Scholar]
- [19].Goula D, Benoist C, Mantero S, Merlo G, Levi G, Demeneix BA. Polyethylenimine-based intravenous delivery of transgenes to mouse lung. Gene Ther. 1998 Sep;5(9):1291–5. doi: 10.1038/sj.gt.3300717. [DOI] [PubMed] [Google Scholar]
- [20].Bos GW, Kanellos T, Crommelin DJ, Hennink WE, Howard CR. Cationic polymers that enhance the performance of HbsAg DNA in vivo. Vaccine. 2004 Dec 9;23(4):460–9. doi: 10.1016/j.vaccine.2004.06.020. [DOI] [PubMed] [Google Scholar]
- [21].Funhoff AM, van Nostrum CF, Lok MC, Kruijtzer JA, Crommelin DJ, Hennink WE. Cationic polymethacrylates with covalently linked membrane destabilizing peptides as gene delivery vectors. J Control Release. 2005 Jan 3;101(1-3):233–46. doi: 10.1016/j.jconrel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- [22].Bennett B, Check IJ, Olsen MR, Hunter RL. A comparison of commercially available adjuvants for use in research. J Immunol Methods. 1992 Aug 30;153(1-2):31–40. doi: 10.1016/0022-1759(92)90302-a. [DOI] [PubMed] [Google Scholar]
- [23].Hunter R, Strickland F, Kezdy F. The adjuvant activity of nonionic block polymer surfactants. I. The role of hydrophile-lipophile balance. J Immunol. 1981 Sep;127(3):1244–50. [PubMed] [Google Scholar]
- [24].Hunter RL. Overview of vaccine adjuvants: present and future. Vaccine. 2002 May 31;20(Suppl 3):S7–12. doi: 10.1016/s0264-410x(02)00164-0. [DOI] [PubMed] [Google Scholar]
- [25].Hunter RL, Bennett B. The adjuvant activity of nonionic block polymer surfactants. II. Antibody formation and inflammation related to the structure of triblock and octablock copolymers. J Immunol. 1984 Dec;133(6):3167–75. [PubMed] [Google Scholar]
- [26].Snippe H, De Reuver MJ, Strickland F, Willers JM, Hunter RL. Adjuvant effect of nonionic block polymer surfactants in humoral and cellular immunity. Int Arch Allergy Appl Immunol. 1981;65(4):390–8. doi: 10.1159/000232780. [DOI] [PubMed] [Google Scholar]
- [27].Newman MJ, Actor JK, Balusubramanian M, Jagannath C. Use of nonionic block copolymers in vaccines and therapeutics. Crit Rev Ther Drug Carrier Syst. 1998;15(2):89–142. doi: 10.1615/critrevtherdrugcarriersyst.v15.i2.10. [DOI] [PubMed] [Google Scholar]
- [28].Newman MJ, Todd CW, Balusubramanian M. Design and development of adjuvant-active nonionic block copolymers. J Pharm Sci. 1998 Nov;87(11):1357–62. doi: 10.1021/js980072c. [DOI] [PubMed] [Google Scholar]
- [29].Todd CW, Balusubramanian M, Newman MJ. Development of adjuvant-active nonionic block copolymers. Adv Drug Deliv Rev. 1998 Jul 6;32(3):199–223. doi: 10.1016/s0169-409x(98)00011-8. [DOI] [PubMed] [Google Scholar]
- [30].Kabanov A, Zhu J, Alakhov V. Pluronic Block Copolymers for Gene Delivery. Adv Genet. 2005;53PA:231–61. doi: 10.1016/S0065-2660(05)53009-8. [DOI] [PubMed] [Google Scholar]
- [31].Reddy ST, van der Vlies AJ, Simeoni E, Angeli V, Randolph GJ, O'Neil CP, et al. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat Biotechnol. 2007 Oct;25(10):1159–64. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- [32].Pitard B, Pollard H, Agbulut O, Lambert O, Vilquin JT, Cherel Y, et al. A nonionic amphiphile agent promotes gene delivery in vivo to skeletal and cardiac muscles. Hum Gene Ther. 2002 Sep 20;13(14):1767–75. doi: 10.1089/104303402760293592. [DOI] [PubMed] [Google Scholar]
- [33].Sriadibhatla S, Yang Z, Gebhart C, Alakhov VY, Kabanov A. Transcriptional activation of gene expression by pluronic block copolymers in stably and transiently transfected cells. Mol Ther. 2006 Apr;13(4):804–13. doi: 10.1016/j.ymthe.2005.07.701. [DOI] [PubMed] [Google Scholar]
- [34].Astafieva I, Maksimova I, Lukanidin E, Alakhov V, Kabanov A. Enhancement of the polycation-mediated DNA uptake and cell transfection with Pluronic P85 block copolymer. FEBS Lett. 1996 Jul 8;389(3):278–80. doi: 10.1016/0014-5793(96)00607-2. [DOI] [PubMed] [Google Scholar]
- [35].Liu F, Yang J, Huang L, Liu D. Effect of non-ionic surfactants on the formation of DNA/emulsion complexes and emulsion-mediated gene transfer. Pharm Res. 1996 Nov;13(11):1642–6. doi: 10.1023/a:1016480421204. [DOI] [PubMed] [Google Scholar]
- [36].Cho CW, Cho YS, Kang BT, Hwang JS, Park SN, Yoon DY. Improvement of gene transfer to cervical cancer cell lines using non-viral agents. Cancer Lett. 2001 Jan 10;162(1):75–85. doi: 10.1016/s0304-3835(00)00629-7. [DOI] [PubMed] [Google Scholar]
- [37].Cho CW, Cho YS, Lee HK, Yeom YI, Park SN, Yoon DY. Improvement of receptor-mediated gene delivery to HepG2 cells using an amphiphilic gelling agent. Biotechnol Appl Biochem. 2000 Aug;32(Pt 1):21–6. doi: 10.1042/ba20000022. [DOI] [PubMed] [Google Scholar]
- [38].Lemieux P, Guerin N, Paradis G, Proulx R, Chistyakova L, Kabanov A, et al. A combination of poloxamers increases gene expression of plasmid DNA in skeletal muscle. Gene Ther. 2000 Jun;7(11):986–91. doi: 10.1038/sj.gt.3301189. [DOI] [PubMed] [Google Scholar]
- [39].Yang Z, Zhu J, Sriadibhatla S, Gebhart C, Alakhov V, Kabanov A. Promoter- and strain-selective enhancement of gene expression in a mouse skeletal muscle by a polymer excipient Pluronic P85. J Control Release. 2005 Nov 28;108(2-3):496–512. doi: 10.1016/j.jconrel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- [40].Kang ML, Jiang HL, Kang SG, Guo DD, Lee DY, Cho CS, et al. Pluronic F127 enhances the effect as an adjuvant of chitosan microspheres in the intranasal delivery of Bordetella bronchiseptica antigens containing dermonecrotoxin. Vaccine. 2007 Jun 6;25(23):4602–10. doi: 10.1016/j.vaccine.2007.03.038. [DOI] [PubMed] [Google Scholar]
- [41].Selinsky C, Luke C, Wloch M, Geall A, Hermanson G, Kaslow D, et al. A DNA-based vaccine for the prevention of human cytomegalovirus-associated diseases. Hum Vaccin. 2005 Jan-Feb;1(1):16–23. doi: 10.4161/hv.1.1.1335. [DOI] [PubMed] [Google Scholar]
- [42].Vilalta A, Mahajan RK, Hartikka J, Rusalov D, Martin T, Bozoukova V, et al. I. Poloxamer-formulated plasmid DNA-based human cytomegalovirus vaccine: evaluation of plasmid DNA biodistribution/persistence and integration. Hum Gene Ther. 2005 Oct;16(10):1143–50. doi: 10.1089/hum.2005.16.1143. [DOI] [PubMed] [Google Scholar]
- [43].Coeshott CM, Smithson SL, Verderber E, Samaniego A, Blonder JM, Rosenthal GJ, et al. Pluronic F127-based systemic vaccine delivery systems. Vaccine. 2004 Jun 23;22(19):2396–405. doi: 10.1016/j.vaccine.2003.11.064. [DOI] [PubMed] [Google Scholar]
- [44].Westerink MA, Smithson SL, Srivastava N, Blonder J, Coeshott C, Rosenthal GJ. ProJuvant (Pluronic F127/chitosan) enhances the immune response to intranasally administered tetanus toxoid. Vaccine. 2001 Dec 12;20(5-6):711–23. doi: 10.1016/s0264-410x(01)00423-6. [DOI] [PubMed] [Google Scholar]
- [45].Spitzer N, Jardim A, Lippert D, Olafson RW. Long-term protection of mice against Leishmania major with a synthetic peptide vaccine. Vaccine. 1999 Mar 17;17(11-12):1298–300. doi: 10.1016/s0264-410x(98)00363-6. [DOI] [PubMed] [Google Scholar]
- [46].Ashour AE, Turnquist HR, Burns N, Wang X, Lin X, Tremayne J, et al. Flt3 ligand delivered in a pluronic formulation prolongs the survival of mice with orthotopic pancreatic adenocarcinoma. Cancer Biother Radiopharm. 2007 Apr;22(2):235–8. doi: 10.1089/cbr.2007.336. [DOI] [PubMed] [Google Scholar]
- [47].Robinson SN, Chavez JM, Pisarev VM, Mosley RL, Rosenthal GJ, Blonder JM, et al. Delivery of Flt3 ligand (Flt3L) using a poloxamer-based formulation increases biological activity in mice. Bone Marrow Transplant. 2003 Mar;31(5):361–9. doi: 10.1038/sj.bmt.1703816. [DOI] [PubMed] [Google Scholar]
- [48].Sang H, Pisarev VM, Munger C, Robinson S, Chavez J, Hatcher L, et al. Regional, but not systemic recruitment/expansion of dendritic cells by a pluronic-formulated Flt3-ligand plasmid with vaccine adjuvant activity. Vaccine. 2003 Jun 20;21(21-22):3019–29. doi: 10.1016/s0264-410x(03)00143-9. [DOI] [PubMed] [Google Scholar]
- [49].Reddy ST, Swartz MA, Hubbell JA. Targeting dendritic cells with biomaterials: developing the next generation of vaccines. Trends Immunol. 2006 Dec;27(12):573–9. doi: 10.1016/j.it.2006.10.005. [DOI] [PubMed] [Google Scholar]
- [50].Kabanov AV, Alakhov VY. Pluronic block copolymers in drug delivery: from micellar nanocontainers to biological response modifiers. Crit Rev Ther Drug Carrier Syst. 2002;19(1):1–72. doi: 10.1615/critrevtherdrugcarriersyst.v19.i1.10. [DOI] [PubMed] [Google Scholar]
- [51].Lemieux P, Vinogradov SV, Gebhart CL, Guerin N, Paradis G, Nguyen HK, et al. Block and graft copolymers and NanoGel copolymer networks for DNA delivery into cell. J Drug Target. 2000;8(2):91–105. doi: 10.3109/10611860008996855. [DOI] [PubMed] [Google Scholar]
- [52].Alakhov V, Klinski E, Li S, Pietrzynski G, Venne A, Batrakova E, et al. Block copolymer-based formulation of doxorubicin. From cell screen to clinical trials Colloids Surf B Biointerfaces. 1999;16(1):113–34. [Google Scholar]
- [53].Armstrong JK, Meiselman HJ, Wenby RB, Fisher TC. Modulation of red blood cell aggregation and blood viscosity by the covalent attachment of Pluronic copolymers. Biorheology. 2001;38(2-3):239–47. [PubMed] [Google Scholar]
- [54].Danson S, Ferry D, Alakhov V, Margison J, Kerr D, Jowle D, et al. Phase I dose escalation and pharmacokinetic study of pluronic polymer-bound doxorubicin (SP1049C) in patients with advanced cancer. Br J Cancer. 2004 Jun 1;90(11):2085–91. doi: 10.1038/sj.bjc.6601856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Batrakova EV, Miller DW, Li S, Alakhov VY, Kabanov AV, Elmquist WF. Pluronic P85 enhances the delivery of digoxin to the brain: in vitro and in vivo studies. J Pharmacol Exp Ther. 2001 Feb;296(2):551–7. [PubMed] [Google Scholar]
- [56].Batrakova EV, Li S, Alakhov VY, Elmquist WF, Miller DW, Kabanov AV. Sensitization of cells overexpressing multidrug-resistant proteins by pluronic P85. Pharm Res. 2003 Oct;20(10):1581–90. doi: 10.1023/a:1026179132599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sahay G, Batrakova E, Kabanov A. Different Internalization Pathways of Polymeric Micelles and Unimers and Their Effects on Vesicular Transport. Bioconjug Chem. 2008 doi: 10.1021/bc8002315. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Riera M, Chillon M, Aran JM, Cruzado JM, Torras J, Grinyo JM, et al. Intramuscular SP1017-formulated DNA electrotransfer enhances transgene expression and distributes hHGF to different rat tissues. J Gene Med. 2004 Jan;6(1):111–8. doi: 10.1002/jgm.463. [DOI] [PubMed] [Google Scholar]
- [59].Hartikka J, Sukhu L, Buchner C, Hazard D, Bozoukova V, Margalith M, et al. Electroporation-facilitated delivery of plasmid DNA in skeletal muscle: plasmid dependence of muscle damage and effect of poloxamer 188. Mol Ther. 2001 Nov;4(5):407–15. doi: 10.1006/mthe.2001.0483. [DOI] [PubMed] [Google Scholar]
- [60].Perales MA, Fantuzzi G, Goldberg SM, Turk MJ, Mortazavi F, Busam K, et al. GM-CSF DNA induces specific patterns of cytokines and chemokines in the skin: implications for DNA vaccines. Cytokines Cell Mol Ther. 2002;7(3):125–33. doi: 10.1080/13684730310000923. [DOI] [PubMed] [Google Scholar]
- [61].Dona M, Sandri M, Rossini K, Dell'Aica I, Podhorska-Okolow M, Carraro U. Functional in vivo gene transfer into the myofibers of adult skeletal muscle. Biochem Biophys Res Commun. 2003 Dec 26;312(4):1132–8. doi: 10.1016/j.bbrc.2003.11.032. [DOI] [PubMed] [Google Scholar]
- [62].Hunter R, Olsen M, Buynitzky S. Adjuvant activity of non-ionic block copolymers. IV. Effect of molecular weight and formulation on titre and isotype of antibody. Vaccine. 1991 Apr;9(4):250–6. doi: 10.1016/0264-410x(91)90108-i. [DOI] [PubMed] [Google Scholar]
- [63].Hunter RL, Bennett B. The adjuvant activity of nonionic block polymer surfactants. III. Characterization of selected biologically active surfaces. Scand J Immunol. 1986 Mar;23(3):287–300. doi: 10.1111/j.1365-3083.1986.tb01970.x. [DOI] [PubMed] [Google Scholar]
- [64].Hunter RL, McNicholl J, Lal AA. Mechanisms of action of nonionic block copolymer adjuvants. AIDS Res Hum Retroviruses. 1994;10(Suppl 2):S95–8. [PubMed] [Google Scholar]
- [65].Allison AC, Byars NE. Adjuvant formulations and their mode of action. Semin Immunol. 1990 Sep;2(5):369–74. [PubMed] [Google Scholar]
- [66].Allison AC, Byars NE. An adjuvant formulation that selectively elicits the formation of antibodies of protective isotypes and of cell-mediated immunity. J Immunol Methods. 1986 Dec 24;95(2):157–68. doi: 10.1016/0022-1759(86)90402-3. [DOI] [PubMed] [Google Scholar]
- [67].Ratanamart J, Huggins CG, Kirby JA, Shaw JA. In vitro and in vivo evaluation of intrinsic immunogenicity of reporter and insulin gene therapy plasmids. J Gene Med. 2007 Aug;9(8):703–14. doi: 10.1002/jgm.1066. [DOI] [PubMed] [Google Scholar]
- [68].Davis HL, Millan CL, Watkins SC. Immune-mediated destruction of transfected muscle fibers after direct gene transfer with antigen-expressing plasmid DNA. Gene Ther. 1997 Mar;4(3):181–8. doi: 10.1038/sj.gt.3300380. [DOI] [PubMed] [Google Scholar]
- [69].Davis HL, Mancini M, Michel ML, Whalen RG. DNA-mediated immunization to hepatitis B surface antigen: longevity of primary response and effect of boost. Vaccine. 1996 Jun;14(9):910–5. doi: 10.1016/0264-410x(95)00255-y. [DOI] [PubMed] [Google Scholar]
- [70].Babiuk LA, Pontarollo R, Babiuk S, Loehr B, van Drunen Littel-van den Hurk S. Induction of immune responses by DNA vaccines in large animals. Vaccine. 2003 Jan 30;21(7-8):649–58. doi: 10.1016/s0264-410x(02)00574-1. [DOI] [PubMed] [Google Scholar]
- [71].Babiuk S, Baca-Estrada ME, Foldvari M, Storms M, Rabussay D, Widera G, et al. Electroporation improves the efficacy of DNA vaccines in large animals. Vaccine. 2002 Sep 10;20(27-28):3399–408. doi: 10.1016/s0264-410x(02)00269-4. [DOI] [PubMed] [Google Scholar]
- [72].Babiuk S, Baca-Estrada M, Babiuk LA, Ewen C, Foldvari M. Cutaneous vaccination: the skin as an immunologically active tissue and the challenge of antigen delivery. J Control Release. 2000 May 15;66(2-3):199–214. doi: 10.1016/s0168-3659(99)00274-6. [DOI] [PubMed] [Google Scholar]
- [73].Calarota S, Bratt G, Nordlund S, Hinkula J, Leandersson AC, Sandstrom E, et al. Cellular cytotoxic response induced by DNA vaccination in HIV-1-infected patients. Lancet. 1998 May 2;351(9112):1320–5. doi: 10.1016/S0140-6736(97)09440-3. [DOI] [PubMed] [Google Scholar]
- [74].Wang R, Doolan DL, Le TP, Hedstrom RC, Coonan KM, Charoenvit Y, et al. Induction of antigen-specific cytotoxic T lymphocytes in humans by a malaria DNA vaccine. Science. 1998 Oct 16;282(5388):476–80. doi: 10.1126/science.282.5388.476. [DOI] [PubMed] [Google Scholar]
- [75].Wilson NS, El-Sukkari D, Belz GT, Smith CM, Steptoe RJ, Heath WR, et al. Most lymphoid organ dendritic cell types are phenotypically and functionally immature. Blood. 2003 Sep 15;102(6):2187–94. doi: 10.1182/blood-2003-02-0513. [DOI] [PubMed] [Google Scholar]
- [76].Singh M, O'Hagan D. Advances in vaccine adjuvants. Nat Biotechnol. 1999 Nov;17(11):1075–81. doi: 10.1038/15058. [DOI] [PubMed] [Google Scholar]
- [77].Atkinson TP, Bullock JO, Smith TF, Mullins RE, Hunter RL. Ion transport mediated by copolymers composed of polyoxyethylene and polyoxypropylene. Am J Physiol. 1988 Jan;254(1 Pt 1):C20–6. doi: 10.1152/ajpcell.1988.254.1.C20. [DOI] [PubMed] [Google Scholar]
- [78].Atkinson TP, Smith TF, Hunter RL. Histamine release from human basophils by synthetic block co-polymers composed of polyoxyethylene and polyoxypropylene and synergy with immunologic and non-immunologic stimuli. J Immunol. 1988 Aug 15;141(4):1307–10. [PubMed] [Google Scholar]
- [79].Atkinson TP, Smith TF, Hunter RL. In vitro release of histamine from murine mast cells by block co-polymers composed of polyoxyethylene and polyoxypropylene. J Immunol. 1988 Aug 15;141(4):1302–6. [PubMed] [Google Scholar]
- [80].Johnston TP, Miller SC. Toxicological evaluation of poloxamer vehicles for intramuscular use. J Parenter Sci Technol. 1985 Mar-Apr;39(2):83–9. [PubMed] [Google Scholar]
- [81].Armstrong A, Brewer J, Newman C, Alakhov V, Pietrzynski G, Campbell S, et al. SP1049C as first-line therapy in advanced (inoperable or metastatic) adenocarcinoma of the oesophagus: A phase II window study. J Clin Oncology. 2006;2006(24):4080. [Google Scholar]
- [82].Ratanamart J, Shaw JA. Plasmid-mediated muscle-targeted gene therapy for circulating therapeutic protein replacement: a tale of the tortoise and the hare? Curr Gene Ther. 2006 Feb;6(1):93–110. doi: 10.2174/156652306775515583. [DOI] [PubMed] [Google Scholar]
- [83].Sasaki S, Takeshita F, Xin KQ, Ishii N, Okuda K. Adjuvant formulations and delivery systems for DNA vaccines. Methods. 2003 Nov;31(3):243–54. doi: 10.1016/s1046-2023(03)00140-3. [DOI] [PubMed] [Google Scholar]
- [84].Yang Z, Sahay G, Sriadibhatla S, Kabanov A. Amphiphilic Block Copolymers Enhance Cellular Uptake and Nuclear Entry of Polyplex-Delivered DNA. Bioconjug Chem. 2008 doi: 10.1021/bc800144a. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Nicchitta CV. Re-evaluating the role of heat-shock protein-peptide interactions in tumour immunity. Nat Rev Immunol. 2003 May;3(5):427–32. doi: 10.1038/nri1089. [DOI] [PubMed] [Google Scholar]
- [86].Batrakova EV, Li S, Alakhov VY, Miller DW, Kabanov AV. Optimal structure requirements for pluronic block copolymers in modifying P-glycoprotein drug efflux transporter activity in bovine brain microvessel endothelial cells. J Pharmacol Exp Ther. 2003 Feb;304(2):845–54. doi: 10.1124/jpet.102.043307. [DOI] [PubMed] [Google Scholar]
- [87].Kabanov AV. Polymer genomics: an insight into pharmacology and toxicology of nanomedicines. Adv Drug Deliv Rev. 2006 Dec 30;58(15):1597–621. doi: 10.1016/j.addr.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Anderson RG. The caveolae membrane system. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- [89].Kabanov AV, Nazarova IR, Astafieva IV, Batrakova EV. Micelle Formation and Solubilization of Fluorescent Probes in Poly(oxyethylene-b-oxypropylene-boxyethylene) Solutions. Macromolecules. 1995;28(7):2303. [Google Scholar]
- [90].Moghimi SM, Hunter AC, Dadswell CM, Savay S, Alving CR, Szebeni J. Causative factors behind poloxamer 188 (Pluronic F68, Flocor)-induced complement activation in human sera. A protective role against poloxamer-mediated complement activation by elevated serum lipoprotein levels. Biochim Biophys Acta. 2004 Jun 28;1689(2):103–13. doi: 10.1016/j.bbadis.2004.02.005. [DOI] [PubMed] [Google Scholar]
- [91].Klink D, Yu QC, Glick MC, Scanlin T. Lactosylated poly-L-lysine targets a potential lactose receptor in cystic fibrosis and non-cystic fibrosis airway epithelial cells. Mol Ther. 2003 Jan;7(1):73–80. doi: 10.1016/s1525-0016(02)00016-3. [DOI] [PubMed] [Google Scholar]
- [92].Kozlov MY, Melik-Hubarov NS, Batrakova E, Kabanov A. Relationship between pluronic block copolymer structure, critical micellization concentration and partitioning coefficients of low molecular mass solutes. Macromolecules. 2000;33(9):3305–13. [Google Scholar]