Abstract
Terminal erythroid differentiation in vertebrates is characterized by progressive heterochromatin formation, chromatin condensation and, in mammals, culminates in nuclear extrusion. To date, although mechanisms regulating avian erythroid chromatin condensation have been identified, little is known regarding this process during mammalian erythropoiesis. To elucidate the molecular basis for mammalian erythroblast chromatin condensation, we used Friend virus-infected murine spleen erythroblasts that undergo terminal differentiation in vitro. Chromatin isolated from early and late stage erythroblasts had similar levels of linker and core histones, only a slight difference in nucleosome repeats, and no significant accumulation of known developmentally-regulated architectural chromatin proteins. However, histone H3(K9) dimethylation markedly increased while histone H4(K12) acetylation dramatically decreased and became segregated from the histone methylation as chromatin condensed. One histone deacetylase, HDAC5, was significantly upregulated during the terminal stages of Friend virus-infected erythroblast differentiation. Treatment with histone deacetylase inhibitor, trichostatin A, blocked both chromatin condensation and nuclear extrusion. Based on our data, we propose a model for a unique mechanism in which extensive histone deacetylation at pericentromeric heterochromatin mediates heterochromatin condensation in vertebrate erythroblasts that would otherwise be mediated by developmentally-regulated architectural proteins in nucleated blood cells.
Keywords: erythroid differentiation, enucleation, chromatin condensation, heterochromatin, histone deacetylation
Introduction
Mammalian terminal erythroid differentiation is characterized by extensive nuclear chromatin condensation and culminates in nuclear extrusion. Although chromatin condensation is also a feature of avian erythropoiesis, nuclei are retained in mature circulating erythrocytes. To date, mechanisms regulating avian erythroid chromatin condensation have been elucidated but little is known regarding this process in mammalian erythroblasts.
DNA in eukaryotic cells is configured into chromatin through its association with histones and nonhistone architectural proteins. The primary packing level consists of an array of repeating nucleosome units. Further compaction is achieved through a hierarchy of higher-order folding: first into the 30 nm chromatin fiber (secondary level) and then into more compact tertiary and quaternary higher order structures (Adkins et al., 2004; Woodcock and Dimitrov, 2001; Zlatanova and Leuba, 2003). One of the most intriguing phenomena related to chromatin higher order folding is the presence of two morphologically different types of chromatin within a single interphase nucleus: dispersed euchromatin and the condensed heterochromatin (Hennig, 1999; Richards and Elgin, 2002). Permanently repressed chromosomal loci such as pericentromeric or subtelomeric regions are associated with constitutive heterochromatin while facultative heterochromatin is associated with conditionally repressed portions of the genome. During eukaryotic differentiation, inactivation of many chromosomal loci is correlated with formation of abundant blocks of facultative heterochromatin (Francastel et al., 2000; Frenster, 1974). Recent genetic studies have revealed a complexity of protein factors, small RNA molecules, and histone modifications that mediate heterochromatin formation in multicellular eukaryotes (Bernstein and Allis, 2005; Huisinga et al., 2006; Jenuwein and Allis, 2001; Maison and Almouzni, 2004).
Studies in terminally differentiating cells show that chromatin undergoing extensive condensation acquires a special pattern of post-translational histone modifications typical for heterochromatin, such as histone H3(K9) methylation and histone H3 and H4 deacetylation and also accumulates specific developmentally-regulated protein factors that mediate chromatin condensation (Grigoryev et al., 2006). The spatial correlation of histone modifications with chromatin condensation has been well documented for chicken erythrocyte chromatin in general and dissected at a molecular level using the developmentally regulated β-globin gene (Bannister et al., 2005; Hebbes et al., 1994; Litt et al., 2001a; Litt et al., 2001b). During development, chicken erythrocytes show a marked change in the nucleosomal repeat (Weintraub, 1978) correlated with an increase of two developmentally-regulated chromatin-condensing factors which continue to be synthesized even when cell proliferation ceases: a special variant of linker histone, histone H5 (Sung et al., 1977) and a nonhistone protein MENT (Grigoryev et al., 1992). Histone H5 accumulates at repressed chromatin domains and decreases at active chromatin domains, such as the β-globin gene (Verreault and Thomas, 1993). MENT also accumulates at repressed heterochromatin and its interaction with chromatin is promoted by histone H3(K9) methylation, which is prevalent at heterochromatic loci and decreased at the active β-globin gene (Istomina et al., 2003). During chicken granulocyte differentiation, however, histone H5 is not expressed but MENT accumulates to a very high level: up to 2 molecules per nucleosome (Grigoryev and Woodcock, 1998). A closely-related nuclear serpin, MNEI, accumulates in chromatin of terminally differentiated human granulocytes (Popova et al., 2006). Thus various molecular pathways are involved in chromatin condensation during terminal differentiation in different types of blood cells.
To understand mechanisms governing chromatin condensation in mammalian erythroblasts, we examined chromatin organization in a well characterized model of terminal erythroid differentiation, Friend virus-infected murine spleen erythroblasts (FVA cells) undergoing differentiation and enucleation in vitro over 44–48 h (Koury et al., 1988; Koury et al., 1989; Krauss et al., 2005; Lee et al., 2003). Unlike the mouse erythroleukemia (MEL) cells derived from the leukemia phase of the Friend disease, the FVA cells that we use in our system do not undergo malignant transformation and are not expected to acquire global epigenetic changes associated with oncogenically-transformed genomes (Esteller, 2006; Feinberg et al., 2006). Here we report that chromatin from nuclei of early stage proerythroblasts and late stage orthochromatic erythroblasts contain similar levels of linker and core histones, similar nucleosome repeats, but surprisingly low levels of nonhistone proteins including nuclear serpins and most other architectural protein factors previously proposed to be associated with heterochromatin condensation such as HP1, MeCP2, MBD2, macroH2A, and H2A.Z (reviewed in (Grigoryev et al., 2006)). However, we did observe a significant increase in histone H3 dimethylated at Lysine 9 contrasting with a sharp decrease in the levels of histone H4 acetylated at Lysine 12 in late stage erythroblasts. The decrease in histone acetylation was concomitant with a marked increase in the level of one histone deacetylase, HDAC5. Treatment of differentiating erythroblasts with trichostatin A (TSA), a histone deacetylase (HDAC) inhibitor, maintained histone acetylation and strikingly inhibited chromatin condensation and nuclear extrusion. Our data suggest that the high degree of chromatin condensation in terminally differentiated mammalian erythroblasts, in contrast to other vertebrates, is mediated by post-translational histone modifications but does not require accumulation of known developmentally-regulated architectural proteins.
Materials and Methods
Reagents
All chemicals were purchased from Fisher Scientific (Pittsburgh, PA), unless otherwise noted. Antibodies against HP1α, HP1β, HP1γ, histones H3 trimethylated at Lysine 9 (H3me3K9), H3 dimethylated at Lysine 9 (H3me2K9), H3 acetylated at Lysines 9 and 14 (H3acK9,K14), H4 acetylated at Lysine 12 (H4acK12), macroH2A1.2 and H2A.Z were as described (Bulynko et al., 2006; Grigoryev et al., 2004) as were antibodies against lamins A/C and B (Chaudhary and Courvalin, 1993). Antibody against MBD2 (sheep, cat.# 07–198) and MeCP2 (rabbit, cat.#07–013) were from Upstate (Lake Placid, NY) and against HDAC5 (rabbit, cat.#2082) were from Cell Signalling Tech. (Beverly, MA). Secondary AlexaFluoro-conjugated antibodies (Molecular Probes, Eugene, OR) were used for immunofluorescence, and HRP-conjugated antibodies from Jackson ImmunoResearch (West Grove, PA) were used for Western blot analysis. Chicken blood was obtained from Bells & Evans/Farmers Pride Inc. (Fredericksburg, PA)
Isolation of Differentiating Murine Erythroblasts from Friend Virus-infected Spleens
Early erythroblasts were isolated using the system established by Koury et al (Koury et al., 1997; Koury et al., 1984; Krauss et al., 2005; Lee et al., 2003). In brief, CD2F1 mice were infected with the anemia-inducing strain of the Friend erythroleukemia virus (1 × 104 spleen focus-forming units injected per mouse). After eleven days, cells from two spleens were collected and separated by velocity sedimentation at unit gravity on a BSA (Bovine Serum Albumin) gradient. Cells sedimenting at approximately 6 mm/h or greater were pooled and yielded a population that was predominantly proerythroblasts (0% BFU-E, 10–20% CFU-E). Cells were cultured over 44 h with 1.0 units/ml recombinant human erythropoietin (epo). In culture, the cells proliferated and differentiated into late-stage erythroblasts, about one-third to one half of which underwent enucleation. At specific time points, cells were removed from culture for analysis. Cytospin slides were made using a Shandon Cytospin 4 (Thermo Electron Corp., Pittsburgh, PA). Slides were stained using 3,3′-dimethoxybenzidine and hematoxylin.
Immunofluorescence Microscopy
For analysis of interphase nuclei, cells were resuspended in PBS (phosphate-buffered saline) buffer and 100 μl aliquots were either directly applied or cytospun on poly-L-lysine slides (Sigma). Cells were fixed in 2.7% paraformaldehyde (EMS, Hatfield, PA) in PBS, permeabilized in KCM buffer (120mM KCl, 20mM NaCl, 10mM Tris-HCl, pH7.7, 0.1% Triton X-100), blocked in 2% BSA, 10% non-fat dried milk in KCM buffer and incubated with primary and secondary antibodies diluted in the blocking solution. All samples were counterstained with Hoechst 33258 or DAPI DNA stains as described previously (Bulynko et al., 2006). Fluorescence microscopy was performed using a Nikon Eclipse E1000 automated microscope with 40x and 100x plan apo lens and Hamamatsu Orca-ER digital camera. Image capturing and analysis was performed using Image-Pro MC6.0 software. For deconvolution, 16 bit z-slices were captured at 0.4 μm steps with a cooled CCD camera and iteratively deconvolved (usually 20 cycles) using AutoDeblur software (AutoQuant Imaging, Watervliet, NY) as described (Irving et al., 2002). Deconvolved images of nuclei in Fig. 5 represent a single ‘z’ slice through the center of the nucleus.
Isolation of Murine and Chicken Nuclei
Fractionation of chicken blood cells and isolation of chicken erythrocytes was performed as described (Grigoryev and Woodcock, 1998). To isolate FVA cell or chicken erythrocyte nuclei, cell suspensions in PBS were centrifuged for 3 min at 1000×g and resuspended in 0.5% IGEPAL CA-630 (Sigma) in RSB (10mM NaCl, 3mM MgCl2, 10mM HEPES, pH 7.5) plus 1mM PMSF and protease inhibitors cocktail (Sigma). Cells were vortexed several times during 30 min incubation on ice and then centrifuged 10 min at 5000×g. The nuclei pellet was resuspended in RSB plus 1mM PMSF and protease inhibitors cocktail.
Nuclease Digestion
For micrococcal nuclease digestion, an aliquot of nuclear preparation containing 1 mg/ml DNA was resuspended in 5ml RSB, 0.5mM PMSF. CaCl2 was added to a final concentration of 1mM, and micrococcal nuclease (Nuclease S7, Roche Diagnostics, Indianapolis, IN) was added at 2.5 units/ml. The reaction was carried out at 37°C for various time intervals and terminated by adding EDTA to a final concentration of 10mM. Resulting soluble chromatin was treated with 50ug/ml Proteinase K plus SDS to 1% final concentration for 1 h at 55°C. DNA was purified by phenol/chloroform extraction and ethanol precipitation and subjected to electrophoresis in 1% agarose (UltraPure, Invitrogen, Carlsbad, CA) in Tris-Acetate-EDTA buffer (Grigoryev and Woodcock, 1998) with constant buffer recirculation.
Reverse-phase High-performance Liquid Chromatography (HPLC)
Analysis of major histone composition of chromatin from erythroblasts before (0 h) and 48 hafter erythropoietin induction was performed using reverse-phase HPLC on the 250×4.6 mm Vydac C18 column with 5μm pore size (Western Analytical Products, Marrieta, CA). Nuclei from erythroblasts (containing 0.3 mg of DNA) were acid-extracted with 0.2M H2SO4 for 30 min on ice, centrifuged in a tabletop microcentrifuge (Eppendorf North America, Westbury, NY) for 15 min at 14,000 rpm, and the supernatant (approximately 20 μg of total protein) was applied to the column equilibrated with 10% acetonitrile containing 0.1% trifluoroacetic acid (solution C). The column was washed with solution C for 8 min at 0.5ml/min flow rate, and histones were eluted using a two-step gradient: 0–60% solution B (100% acetonitrile, 0.1% TFA) for 120 min, then 60–100% solution B for 15 min at 0.5ml/min flow rate. 0.25 ml fractions were combined and analyzed by sodium dodecyl sulfate-containing polyacrylamide gel electrophoresis (SDS-PAGE).
SDS-PAGE Gel Electrophoresis and Western Blotting and
Isolated nuclei were dissolved in SDS-containing loading buffer and the electrophoresis was carried out in 15% polyacrylamide SDS-containing gels (Laemmli, 1970). Proteins were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore, Billerica, MA) and detected with primary and secondary HRP-conjugated antibodies as described (Bulynko et al., 2006). For semi-quantitative analysis of relative protein levels in nuclear samples, the Coomassie-stained gels or autoradiograph after ECL (Amersham, UK) detection were scanned and digitized and the intensity of protein bands was quantitated using the OptiQuant version 03.00 (Packard Instrument Co, Meriden, CT) software package.
Treatment of Cultured Erythroblasts with HDAC inhibitors
After 24 h of culture, 100 and 200 nM trichostatin A (Calbiochem) was added to culture medium and 0.2 % DMSO (vehicle) added to controls. At 48 h, control and trichostatin A treated cells were collected, total cells counted, cytospins prepared, and 500 cells were scored as erythroblasts, extruded nuclei, or reticulocytes. Brightfield images were acquired using a Nikon TE2000 microscope and a Plan Apo 60X/1.4NA oil objective (Nikon Instruments, Inc., Melville, NY) equipped with a Q-imaging RetigaEX CCD camera (Q IMAGING, Burnaby, BC Canada). For erythroblast populations, nuclear diameters were measured on randomly selected cells from cytospin slides using Image-Pro Plus software (MediaCybernetics, Silver Spring, MD).
Quantitative Real Time PCR
Friend virus infected erythroblasts were cultured and allowed to undergo terminal differentiation in vitro, as described above. RNA was extracted with RNAzol B (Tel-test, Friendswood, TX), every 8 h for 48 h. Total RNA (100 ng) was used to make cDNA, with random nonameric primers (100 ng), and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). Of the final reaction volume (20 μl), 1 μl of cDNA was used in the PCR reaction. The PCR reaction included Quantitech SYBR (Qiagen, Valencia, CA), sense and antisense PCR primers (0.25 μM), and cDNA. PCR primer sequences are listed in Supplemental Table 1. PCR reactions were loaded into a 96-well plate, and analyzed on a DNA Engine Opticon (MJ Research, Waltham, MA).
Results
Nucleosome Array Architecture and Histone Composition are Maintained in Condensed Chromatin of Late Stage Murine Erythroblasts
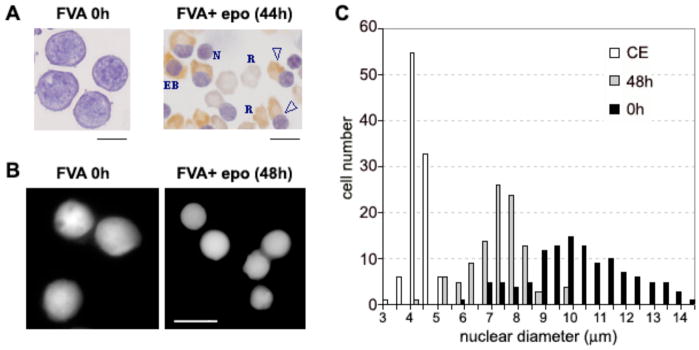
During chicken erythrocyte differentiation there is a substantial increase in nucleosomal repeats from 190 bp in 3–4 day erythroblasts to ~207 bp in 12–15 day embryo erythrocytes and 212 bp in adult erythrocytes, accompanying the global chromatin condensation (Weintraub, 1978). To determine if this is a feature of mammalian erythropoiesis, we utilized Friend virus-infected murine spleen erythroblasts (FVA cells), a well-characterized model of terminal erythroid differentiation. Employing this system, erythroblasts were observed to undergo differentiation and enucleation over a period of 44–48 h (Fig. 1A). Measurements of nuclear diameters of murine erythroblasts showed a significant decrease in average diameter from 9.6 to 6.8 μm during 48 h of terminal differentiation (Fig. 1B, C). Taking the diploid mouse genome size of 6.7 pg (Peterson et al., 1994), our results show a ca. 3-fold nuclear chromatin condensation: from 0.014 pg/μm3 at 0 h to 0.041 pg/μm3 at 48 h.
Fig. 1.
Nuclear condensation and extrusion during terminal differentiation of murine erythroblasts in the FVA model system. (A) Bright field micrograph of cytospin preparations at 0 h and 44 h. EB, late erythroblast; open triangle, enucleating erythroblast; R, reticulocyte; N, expelled nucleus. (B) Fluorescence microscopy of 0 and 48 h fixed cells stained with Hoechst 33258 for DNA. (C) Histogram showing nuclear diameter distribution in terminally differentiating murine erythroblasts at 0 h (black bars) and 48 h (gray bars) compared with chicken erythrocytes (open bars). Nuclear diameter measurements were performed by fluorescence microscopy on fixed cells stained with Hoechst 33258 for DNA. Scale bars, 10 μm.
To analyze nucleosomal repeat length, we digested 0 and 48 h nuclei from murine erythroblast cultures with exogenous micrococcal nuclease and analyzed their digestion patterns. Both 0 and 48 h nuclei showed almost identical rates of digestion and produced very similar nuclease digestion patterns, and a similar width of the nuclease digestion bands indicating no increase in chromatin structural heterogeneity (Fig. 2A,B). By measuring the nuclease-digested DNA fragment length divided by the nucleosome number (Fig. 2C) we estimated that the size of the nucleosome repeat slightly decreased from 196 bp to 191 bp during murine erythroblast differentiation. This small decrease in size is strikingly different from the previously observed increase in the nucleosomal repeats in differentiating chicken erythroid cells (Weintraub, 1978). Thus our data suggest that nucleosomal array organization is minimally changed during murine erythroblast differentiation. Of note, as in earlier studies (Kelley et al., 1993; Krauss et al., 2005), we did not observe extensive DNA degradation in any samples, indicating no major activation of apoptotic pathways.
Fig. 2.
Nucleosome repeat length is not substantially changed during terminal differentiation of murine erythroblasts. (A, B) Agarose electrophoresis of DNA isolated from nuclei harvested from erythroblast cultures at 0 h (lanes 2–7, 16) and at 48 h (lanes 8–13, 17). Isolated nuclei were treated with 2.5 units/ml of micrococcal nuclease for 0 (lines 2, 8), 1 (3, 9), 3 (4, 10), 6 (5, 11), 12 (6, 12, 16, 17), and 30 (7, 13) minutes. Lanes 1, 14, and 15 show DNA molecular size markers. (C) Median sizes of the micrococcal nuclease digest bands for the erythroblasts before (0 h, solid line) and after erythropoietin induction (48 h, dotted line) were calculated from densitometry of the gel (A) and plotted against the nucleosome repeat number.
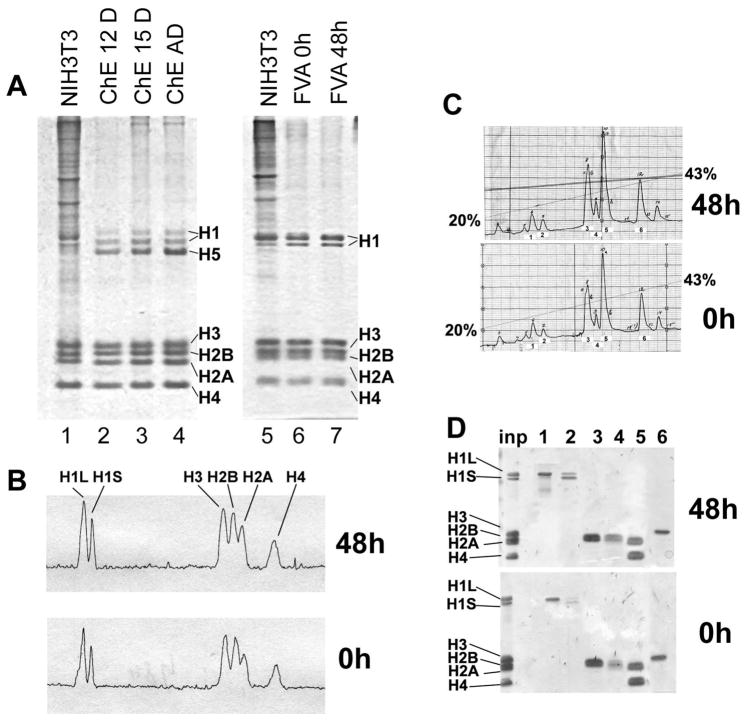
Developmentally-regulated transitions in chicken erythrocyte chromatin, in particular the increased base pairs of nucleosomal repeat, have been previously attributed to an increased level of linker histones (Woodcock et al., 2006) from 1 to 1.4 molecules per nucleosome due to expression of one developmentally-regulated linker histone variant, H5 (Bates and Thomas, 1981). To compare changes in chromatin protein composition during development of mouse and chicken erythrocytes, we isolated nuclei and soluble chromatin from the nuclease-treated 0 h and 48 h erythroblasts and examined their protein composition as previously described for chromatin from chicken erythrocytes and granulocytes (Grigoryev and Woodcock, 1998). Total nuclear protein analyzed by Laemmli PAGE, from both 0 h and 48 h samples showed a distinct chromatin protein pattern with the expected core histone (H2A, H2B, H3 and H4) and linker histone (H1) composition but, in contrast with chicken erythrocytes, no changes in the linker histone levels (Fig. 3A, lanes 2–4 and 6,7). Murine erythroblast nuclei also appeared to contain much less nonhistone protein than proliferating mouse NIH3T3 cell nuclei (Fig. 3A, lanes 1,5). Furthermore, densitometry of the histone region of the total nuclear protein separated by SDS-PAGE (Fig. 3B) and HPLC chromatography of acid-extracted histones (Fig. 3C,D) showed no significant changes between 0 and 48 h erythroblasts. Thus our data clearly show that no major developmentally-regulated architectural protein, predicted to act at a level stoichiometric with nucleosomes (McBryant et al., 2006), is expressed during the transition from proliferating to mature differentiated erythroblasts.
Fig. 3.
Histone composition in differentiating erythroblasts. (A) Total protein from nuclei analyzed by SDS-PAGE and stained with Coomassie R250. Mouse NIH3T3 cells, lanes 1 and 5; chicken erythroblasts from 12-day embryos, lane 2; 15-day chicken embryos, lane 3; adult chicken erythrocytes, lane 4; FVA erythroblasts at 0 h, lane 6; FVA erythroblasts at 48 h, lane 7. (B) Densitometry of lanes 6 and 7 showing that linker histone levels do not change during terminal murine erythroid differentiation. The peaks corresponding to the major linker histone subtypes: H1L (large H1 band) and H1S (small H1 band) and core histones: H2A, H2B, H3, and H4 are indicated. (C) HPLC chromatography of isolated core histones eluted with a linear gradient of 20–43% acetonitrile showing that histone variants do not change during terminal murine erythroid differentiation. Numbers 1–6 correspond to fractions analyzed in Panel D. (D) Pooled HPLC fractions from peaks marked by numbers 1–6 (Panel C) were separated on PAGE to identify by molecular weight the type of histone in each fraction. Inp: input of isolated core histone onto HPLC column.
Chromatin Condensation During Murine Erythropoiesis Does Not Involve Known Heterochromatin Architectural Proteins
In most eukaryotic cell types, constitutive heterochromatin is promoted by heterochromatin protein 1 (HP1) that is represented in vertebrate cells by 3 isoforms: α, β, and γ (Singh and Georgatos, 2002). However, the association of HP1 with facultative chromatin varies among different tissues. For example, in differentiating chicken erythrocytes where abundant facultative heterochromatin forms, Western blots probed with antibodies against the three known HP1 variants show absence of HP1γ, a marked decrease in HP1α, and a moderate decline in HP1β in adult erythrocytes relative to 12 day embryonic erythrocytes (Fig. 4A, lanes 4,5). Since the amount of cytologically-detectable heterochromatin is also dramatically increased during murine erythropoiesis (Koury et al., 1988), we compared the levels of HP1 isoforms in Western blots of early and late erythroblast nuclei. As a positive control, we used NIH/3T3 cells showing prominent bands with all three HP1 isoforms (Fig. 4A, lane 1). In 0 h murine erythroblasts levels of HP1α and β were high and HP1γ was low but detectable (Fig 4A, lane 2;Fig 4D). In contrast, in equal amounts of chromatin protein from 48 h cells, HP1α levels were slightly lower whereas HP1β and HP1γ were minimally detectable (Fig 4A, lane 3; Fig 4D). This suggests that the levels of HP1 proteins are not sufficient to physically condense chromatin during terminal murine erythroblast differentiation. These results are consistent with previous data showing a sharp decline of HP1 protein in other terminally differentiated blood cells (Gilbert et al., 2003; Istomina et al., 2003; Popova et al., 2006) and likely reflect the ability of HP1 to promote the secondary chromatin structure prevailing in constitutive heterochromatin (Fan et al., 2004) rather than the tertiary chromatin structure associated with facultative chromatin in terminally differentiated cells (see (Grigoryev et al., 2006) for review).
Fig. 4.
Changes in heterochromatin proteins during erythroid terminal differentiation. (A) Total protein samples from the nuclei of mouse NIH3T3 cells (1), FVA cells before (2) and 48 h after erythropoietin induction (3), chicken erythroblasts from 12-day embryo (4), and adult chicken erythrocytes (5) were separated by SDS-PAGE and Western blots probed with antibodies against HP1α, β, and γ as indicated. The bottom panel showing control core histone (CH) loading was stained with Coomassie R250. (B) Nuclear protein samples were separated by SDS-PAGE and Western blots probed with antibodies against, MeCP2, histone H2AZ, lamins A/C and B1, and MBD2 as indicated. NIH3T3 cells, lane 6; FVA erythroblasts at 0 h, lane 7; and FVA erythroblasts at 48 h, lane 8. Controls stained for histones had equal loads (data not shown). (C) Nuclear protein samples were separated with SDS-PAGE and Western blots probed with antibodies against histones H3ac(K9, K14), H4acK12, H3me2K9 and H3me3K9, as indicated. Mouse NIH3T3 cells: lanes 9, 12; FVA erythroblasts at 0 h: lanes 10, 13, 14; 20 h: lane 15; 32 h: lane 16; 48 h: lanes 11, 17, 18. Lanes 13–14 and 17–18 represent two pairs of independent experiments. The bottom panel was stained with Coomassie R250 for histone loading controls. (D) Histogram showing densitometry of Western blots of NIH3T3 cells and FVA erythroblasts at 0 h and 48 h probed with antibodies to HP1α, β, and γ and antibodies to histones H3ac(K9, K14), H4acK12, H3me2K9 and H3me3K9. The signal intensities in NIH/3T3 were designated as equal to 100%. Error bars show Standard Deviation.
Other chromatin architectural proteins have been identified as having roles in chromatin compaction. In chicken and human granulocytes, chromatin condensation importantly involves accumulation of developmentally-regulated nuclear serpins MENT (Istomina et al., 2003) and MNEI (Popova et al., 2006). However, probing nuclear protein fractions with antibodies against MENT, as well as broad specificity panserpin antibodies recognizing all MENT- and MNEI-related serpins, did not detect any immunoreactive bands (data not shown). Additional chromatin architectural factors known to be involved in chromatin condensation in terminally-differentiated muscle cells include MeCP2 and MBD2 (Brero et al., 2005). Probing nuclear protein fractions with anti-MBD2 showed a decrease at 48 h relative to 0 h cells. While MeCP2 increased slightly at 48 h, it was markedly less than even the basal level present in proliferating NIH3T3 cells (Fig. 4B), a cell type with totally decondensed chromatin (Grigoryev et al., 2004). Additionally, the amount of MeCP2 expressed in FVA cells at 48 h is insufficient to cause chromatin condensation in other cell types such as muscle. Antibodies for the non-ubiquitinated form of H2A.Z, a histone variant associated with constitutive heterochromatin in differentiated trophoblasts (Rangasamy et al., 2003) and facultative XY-body chromatin in post-meiotic spermatids (Greaves et al., 2006), showed that its level was reduced in differentiated erythroblasts (Fig. 4B). Another histone variant, macroH2A1.2, associated with pericentromeric heterochromatin spreading in quiescent mammalian lymphocytes (Grigoryev et al., 2004) was similarly reduced during erythroblast murine differentiation (data not shown). We also analyzed expression of nuclear lamins that are associated with perinuclear heterochromatin formation. While lamin A (the top major band in NIH/3T3) was not detected in erythroblasts consistent with a sharp decrease of this lamin variant in differentiated mammalian erythroblasts (Martelli et al., 1992), neither the remaining lamin C (the middle major band in NIH/3T3) nor lamin B1 showed any notable change during erythroblast differentiation (Fig. 4B). Taken together these data substantiate the absence of known architectural chromatin proteins associated with heterochromatin spreading in mature mouse erythroblasts.
Histone Modification Levels Change During Murine Erythroid Differentiation
Methylation of histone H3 at lysine 9, in concert with loss of histone acetylation at this and several other histone amino acids, has been suggested to be a primary signal for establishing heterochromatin/euchromatin segregation (Jenuwein and Allis, 2001; Litt et al., 2001a). To detect histone H3 methylation, we used antibodies against an H3 peptide containing dimethylated Lysine-9 (anti-H3me2K9) and an H3 peptide containing trimethylated Lysine-9 (anti-H3me3K9). Anti- H3me3K9 is associated with pericentromeric constitutive heterochromatin while H3me2K9 is detected at facultative chromatin (Cowell et al., 2002). Western blotting (Fig. 4C, D) showed that the level of H3me3K9 did not appear different between proliferating and late stage erythroblasts. In comparison, the level of histone H3me2K9 was increased significantly approximately 6.5–fold during erythroid maturation (Fig. 4C, D). The specific increase in H3me2K9 is consistent with its association with facultative, developmentally-regulated heterochromatin while histone H3me3K9 is mostly associated with constitutive pericentric heterochromatin.
When we probed 0 and 48 h cells with antibodies against either acetylated histone H3acK9,K14 or histone H4acK12 we observed a very low level of H3acK9,K14 in both types of cells (Fig. 4C, lanes 9–11). With H4acK12, we observed that a significant level of this histone modification was maintained up to 32 h of cell differentiation, followed by a sharp decrease seen in 48 h erythroblasts (Fig. 4C, lanes 10–18) concomitantly with the overall chromatin condensation observed at this stage. As with the observed increase in H3me2K9, this transition is in agreement with the decrease of histone acetylation associated with heterochromatin spreading in other terminally differentiated cells (Grigoryev et al., 2004; Su et al., 2004a; Terranova et al., 2005). Thus, unlike architectural chromatin proteins, core histone modifications show global changes during murine erythroblast maturation and may play a significant role in condensation of facultative heterochromatin in terminally differentiated murine erythroblasts.
Histone Modification Transitions are Centered Around Constitutive Pericentric Heterochromatin
In mouse cells, the AT-rich pericentromeric heterochromatin from several chromosomes self-associates and forms a variable number of chromocenters that are readily detected by AT-rich DNA-specific fluorochromes such as Hoechst 33258 (Hilwig and Gropp, 1972). These Hoechst-positive pericentromeric heterochromatin foci such as seen on Fig. 5 (panels 1–4) provide cytological landmarks against which chromatin protein distribution could be monitored.
Fig. 5.
Spatial reorganization of histone modifications during erythroblast differentiation. Immunofluorescence microscopy of FVA cells before (0h) and after (48h) erythropoietin induction stained with Hoechst (blue, panels 1–4, 13, 16) for DNA or Hoechst plus antibodies against histones H3me2K9 (red, panels 5, 6), H4acK12 (green, panels 7, 8), and H3me3K9 (red, panels 14, 17) as indicated. Panels 1–4, 13, 16 show original images. Panels 5–8 and 14, 17 show overlayed images obtained after Autodeblur deconvolution (see Materials and Methods). Panels 9–12, 15 and 18 show line profiles (deconvolved images) illustrating the spatial fluorescence intensity changes of the specific antibody staining (red or green channel as indicated) and Hoechst (blue channel) plotted along the paths shown by the yellow lines on panels 5–8, 14 and 17. Scale bar, 10 μm. Panel 19 shows a histogram of average distances between Hoechst peaks and each of the three epigenetic histone modifications: H3me3K9, H3me2K9, and H4acK12. Distances were measured on multiple line profiles (such as those shown on panels 9 – 12, 15, 18) recorded after immunofluorescence deconvolution microscopy of FVA cells before (0h) and after (48h) erythropoietin induction. From 40 to 50 measurements were made for each category. Error bars show Standard Deviations. P-values shown over the brackets represent probability associated with a Student’s two-sample unequal variance t-Test with a two-tailed distribution.
In previous work with differentiated mouse lymphocytes we identified a distinct nuclear chromatin territory with a unique set of histone modifications (approximately 1 μm in thickness) adjacent to constitutive pericentromeric heterochromatin. We designated this territory as the apocentric nuclear zone (Grigoryev et al., 2004). It is within this zone that the most significant changes occur during the transition from proliferation to quiescence when facultative heterochromatin is formed. Specifically, in mouse lymphocytes, we found histone H3me2K9 accumulation and histone H4acK12 exclusion in apocentric zones around chromatin regions containing H3me3K9. In view of such nuclear distribution of histone modifications in quiescent lymphocytes, we analyzed the localization of these core histone modifications in proliferating and late stage non-proliferating murine erythroblasts.
Immunofluorescence microscopy of 0 h cells revealed that acetylated histone H4acK12 was distributed throughout the nucleus excluding only the pericentromeric heterochromatin marked by intensive Hoechst 33258 staining (Fig. 5, panels 3,7,11). Similar to quiescent lymphocytes (Grigoryev et al., 2004), H4acK12 showed a very distinct intranuclear distribution in the 48 h erythroblasts with acetylated histones being localized toward the periphery of the nucleus, and excluded from the intensely stained heterochromatic foci (Fig. 5, panels 4,8,12). Staining with antibodies against H3acK9,K14 showed a similar exclusion from heterochromatic foci (not shown). During differentiation, the average distance between histone H4acK12 peaks and pericentromeric Hoechst staining peaks was significantly (p<0.002) increased from 2.25 μm at 0 hr to 3.9 μm at 48 hr (panel 19). In contrast, immunofluorescence of 0 h cells with antibodies against histone H3Me2K9 tended to concentrate in the vicinity of the heterochromatin foci (Fig. 5, panels 1,5,9) from where H4acK12 was excluded. The fluorescence intensity profile (Fig. 5, panels 10 and 19) shows that in the 0 and 48 h erythroblasts the H3Me2K9 peaks cluster at a distance on average 1.3 μm from the heterochromatin foci respectively while the 0 hr (p>0.002) and even more so the 48 hr histone H4acK12 acetylation peaks (average 3.9 μm distance from Hoechst staining) retreat away from these areas. The heterochromatin foci were constitutively enriched with H3me3K9 that did not significantly change in the course of erythroid maturation (Fig. 5, panels 13–18, 19). These results show that during erythroblast differentiation the most significant changes in histone modifications take place in the close vicinity of pericentromeric chromatin leading to a dramatic increase in the ratio of histone H3me2K9 to H4acK12 at this nuclear territory contrasting with its sharp decrease at the nuclear periphery. Though H3me2K9 is distributed in patches rather than zones surrounding pericentromeric heterochromatin, the character of spatial distribution of the histone methylation and acetylation markers closely resembles the one previously observed in apocentric zones of differentiated mouse lymphocytes (Grigoryev et al., 2004).
HDAC5 Expression is Significantly Increased During Terminal Erythroid Differentiation
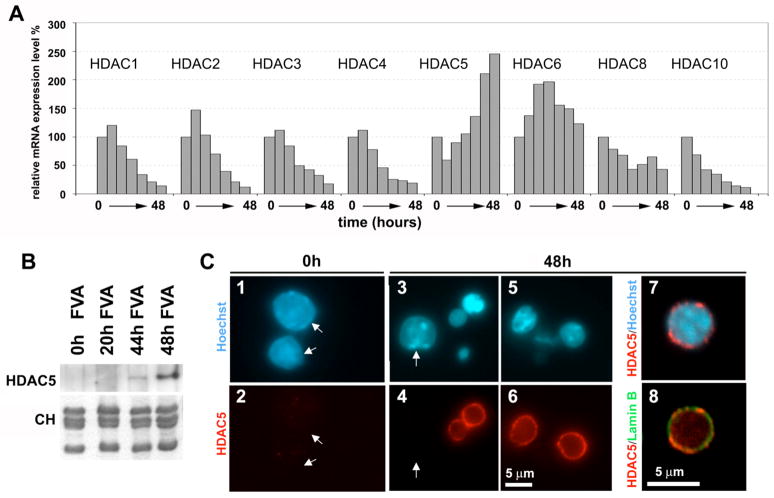
The abrupt decrease of histone acetylation in murine erythroblast nuclei with the most condensed chromatin as well as the recent finding that histone deacetylation may mediate chromatin condensation in terminal muscle differentiation (Terranova et al., 2005) suggested that histone deacetylases (HDACs) may play a role in chromatin condensation. Since a number of HDACs have been previously implicated in large-scale chromatin modification (Shahbazian and Grunstein, 2007), we used quantitative real-time PCR with cDNA isolated from the erythropoietin-induced FVA cells to examine the expression of a number of HDACs during terminal differentiation (Fig. 6A). Among all HDACs tested only HDAC5 showed an approximately 2.5 fold increase in expression level while the other HDACs either markedly declined (HDACs 1–4,10) or had levels that did not show a significant net change upon terminal differentiation (HDACs 6,8). To validate the increased expression of HDAC5 at the protein level, we probed nuclear protein samples for HDAC5 by Western blotting and observed a major increase of HDAC5 expression at the last stages of FVA erythroblast differentiation, reaching maximal expression after 48 h (Fig. 6B).
Fig. 6.
Increased mRNA expression of HDAC5 at the late stage of erythroblast differentiation. (A) Histograms showing relative mRNA expression levels (normalized to the maximal level shown as 100%) as determined by quantitative real-time PCR for HDACs 1, 2, 3, 4, 5, 6, 8, 10. For each HDAC, a set of 7 columns represents PCR experiments with cDNA obtained (left to right) from FVA cells incubated with erythropoietin for 0, 8, 16, 24, 32, 40, and 48 h.
(B) Total protein samples from the nuclei of mouse FVA cells before (0 h) and 20, 44, and 48 h after erythropoietin induction were separated by SDS-PAGE and Western blots probed with antibodies against HDAC5. The bottom panel shows histones from the same samples stained with Coomassie R250.
(C) Immunofluorescence microscopy of FVA cells before (0h) and after (48h) erythropoietin induction stained with Hoechst 33258 (panels 1, 3, 5, 7) for DNA, antibodies against HDAC5 (panels 2, 4, 6, 7) and antibodies against nuclear lamin B (panel 8). Arrows on panels 2, 4 indicate the positions of HDAC5-negative nuclei. Panels 7 and 8 show superimpositions of confocal images (at higher magnification) stained with Hoechst 33258 and anti-HDAC5 (7), and anti-HDAC5 and anti-lamin B (8).
The population of 48 h FVA cells has some heterogeneity in nuclear diameters (Fig. 1B) where the smallest nuclei with the most condensed chromatin are associated with the lowest levels of histone acetylation (Fig. 5 panel 8). To examine if HDAC5 expression is indeed associated with condensed nuclei, we used immunofluorescence microscopy to detect HDAC5 in FVA cells. Consistent with Western blotting results, the 0 h FVA cells showed no detectable signal of HDAC5 (Fig. 6C, panels 1, 2). In contrast, the 48 h cells showed strong HDAC5 signals, predominantly associated with the periphery of small nuclei with condensed chromatin. The majority of larger nuclei either did not express detectable HDAC5 or showed lesser levels at the nuclear periphery (Fig. 6C, panels 3–7). Thus, most of HDAC5 protein expressed at 48 h of FVA erythroblast differentiation is detected specifically in cells with condensed chromatin. The peripheral HDAC5 was proximal to lamin B (Fig. 6C panel 8) but formed a distinct pattern of local intensities.
Inhibition of Histone Deacetylation Blocks Chromatin Condensation and Erythroblast Enucleation
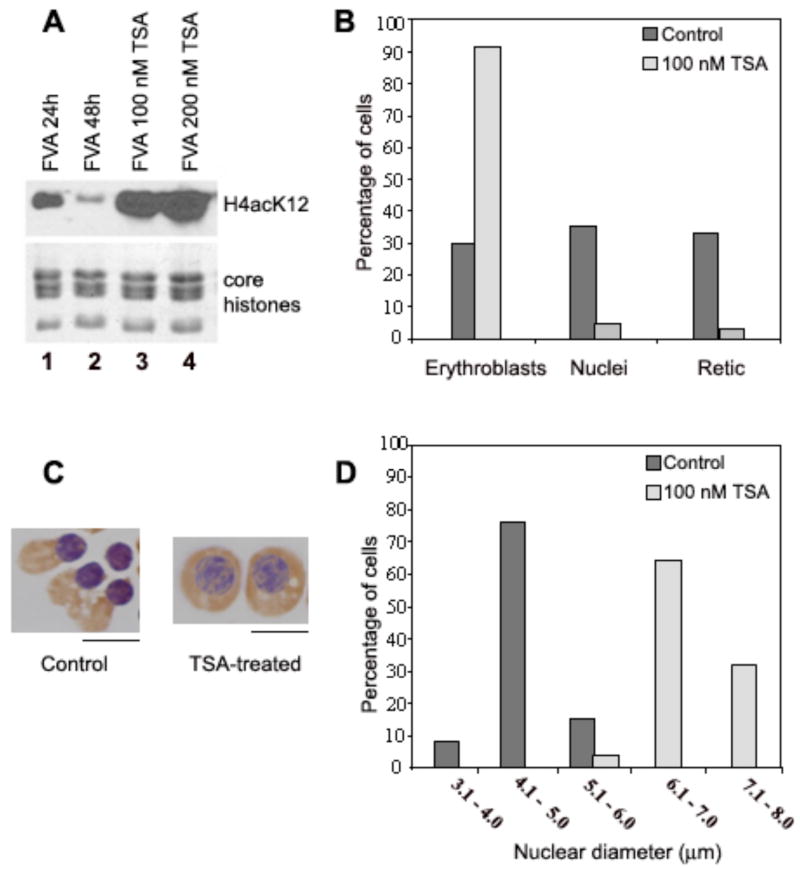
We next asked if HDAC activity could be instrumental in mechanisms for chromatin condensation in differentiating FVA cells. To test this, we treated mouse erythroblasts after 24 h in culture with the HDAC-specific inhibitor trichostatin A (TSA). We then compared specific parameters of differentiation in the treated cells with untreated control cells 20 h later. Western blotting of H4K12Ac showed that 100 or 200 nM TSA-treated erythroblasts had the predicted dramatic increase in histone H4(K12) acetylation (Fig. 7A, lanes 3,4) in contrast to the significant decrease seen at 48 h in control cells (Fig 7A, lane 2). We also scored the categories of cells present after exposure to 100 nM TSA. The percentage of erythroblasts was increased ~3 fold in the 44 h cultures treated with 100 nM TSA compared to untreated controls (Fig. 7B). Furthermore, correlated with an increased percentage of erythroblasts, there was a marked reduction in extruded nuclei and reticulocytes (Fig. 7B) after TSA treatment. A very similar level of acetylation and inhibition of chromatin condensation was observed after treatment of cells with 1 mM or 2 mM sodium butyrate, another HDAC inhibitor (data not shown).
Fig. 7.
Trichostatin A inhibits histone deacetylation, nuclear condensation and nuclear extrusion. (A) Total nuclear proteins were separated on SDS-PAGE and detected by Western blotting with antibodies against acetylated histone H4(K12). FVA erythroblasts were cultured for 24 or 48 h with erythropoietin (lanes 1 and 2) or additionally treated for the final 24 h with 100 (3) or 200 (4) nM TSA and then nuclei were isolated. The bottom panel shows histone loading controls stained with Coomassie R250. (B) Histogram of percent erythroblasts, reticulocytes and expelled nuclei in untreated cultures and in cells cultured for 24 h and then treated for the final 24 h with 100 nM TSA. (C) Examples of cytospin preparations at 44 h of untreated, control erythroblasts and erythroblasts treated with 100 nM TSA. Scale bar, 5 μm. (D) Histogram showing nuclear diameter distribution in cytospin preparations of control and TSA-treated cells at 44 h.
Evaluation of hemoglobinization of erythroblasts by benzidine/hemotoxylin staining showed that hemoglobin synthesis continued after TSA, indicating the cells remained metabolically active (Fig. 7C). We also observed that erythroblast nuclei did not decrease in size and their chromatin did not condense in TSA-treated cells relative to 44 h controls (Fig. 7C, D). Treatment with 2mM butyrate resulted in a very similar cell phenotype (not shown) strengthening the argument that the effect was due to HDAC inhibition. Taken together, it appears that HDAC inhibitors significantly inhibit erythroblast nuclear condensation and extrusion. Thus our data support a model in which histone deacetylase activity, in the absence of other known heterochromatin-promoting factors, plays a significant mechanistic role in the global chromatin condensation occurring in differentiating murine erythroblasts.
Discussion
Chromatin in mature terminally-differentiated vertebrate tissues (e.g. nucleated erythrocytes, white blood cells, brain, or liver) is highly condensed and shows a propensity for selective self-association of repressed genes generating significant changes in chromatin higher-order folding (reviewed in (Grigoryev, 2001)). This progressive increase in chromatin condensation and selective self-association during terminal differentiation raises the question of which developmentally-regulated factors are responsible for changes in chromatin higher order structure. Previous studies of constitutive and facultative heterochromatin revealed several different architectural factors that promote heterochromatin formation. These include three subtypes of heterochromatin protein 1, a key factor in promoting chromatin-mediated repression and heterochromatin spreading (Eissenberg and Elgin, 2000; Li et al., 2003; Verschure et al., 2005); methyl CpG-binding proteins MeCP2 and MBD2 promoting heterochromatin clustering in muscle cells (Brero et al., 2005); erythroid tissue-specific linker histone H5 that promotes chromatin condensation by gluing together nucleosome arrays (Weintraub, 1984), along with closely related erythrocyte-specific linker histone H1 variants also found in other vertebrates such as fish (Wright et al., 1987) and amphibians (Koutzamani et al., 2002); and nuclear serpin MENT expressed in chicken blood cells, including nucleated erythrocytes and, in synergy with linker histone H5, promoting chromatin condensation (Springhetti et al., 2003). Surprisingly, none of these factors have been found to significantly accumulate in mature murine erythroblasts. Furthermore, even among unidentified protein bands in murine erythrocyte nuclei we observed no single one with an expression level comparable to histones (Fig. 3A). Accordingly, no single protein in the mouse erythroblast nucleus appears to be present in sufficient quantity to cause global chromatin transitions by physically altering the chromatin fiber folding as the known chromatin architectural proteins do (McBryant et al., 2006).
In differentiating erythroid and lymphoid mammalian cells, spreading of heterochromatin structure is associated with juxtaposition of silenced euchromatic genes with constitutive heterochromatin (Brown et al., 1997; Schubeler et al., 2000). In differentiated mouse lymphocytes histone H3 methylation is increased and acetylation is decreased (Su et al., 2004b). In these cells, we have previously observed a special apocentric zone enriched in histone H3me2K9 and depleted in histone acetylation (Grigoryev et al., 2004). In the current study, we found a similar enrichment in histone H3 methylation and spatial segregation from histone acetylation at the heterochromatin periphery in terminally differentiated erythroblasts.
Furthermore, inhibition of histone deacetylase activity by TSA or sodium butyrate treatment blocked characteristics of terminal differentiation including chromatin condensation and nuclear extrusion, although in some other systems they have been reported to promote differentiation (Cho et al., 2005). Interestingly, the processes we observed in differentiating mouse erythroblasts resemble those of terminally differentiating muscle tissue where heterochromatin clustering was accompanied by enhanced histone H3(K9) methylation and prevented by HDAC inhibition (Terranova et al., 2005). However, in contrast to erythroblasts, chromatin condensation in myogenic cells involved major contributions from two architectural heterochromatin factors, MeCP2 and MBD2 (Brero et al., 2005).
The results reported here suggest a strikingly different organization of condensed chromatin in mammalian erythroblasts compared to other vertebrates, whose erythrocytes remain nucleated. The apparent absence of stage-specific heterochromatin architectural proteins in terminally-differentiated murine erythroblasts suggests a new model for chromatin condensation (Fig. 8) where histone deacetylation per se is directly responsible for chromatin condensation. Indeed, extensive acetylation of histone N-terminal domains are well known to inhibit chromatin folding and self-association (Tse et al., 1998) and acetylation of only one amino acid (Lys 16) in histone H4 is particularly efficient in inhibiting chromatin compaction (Shogren-Knaak et al., 2006). In addition to histone deacetylation, other histone modifications, such as linker histone phosphorylation (Yellajoshyula and Brown, 2006) may contribute to chromatin condensation in mammalian erythroblasts.
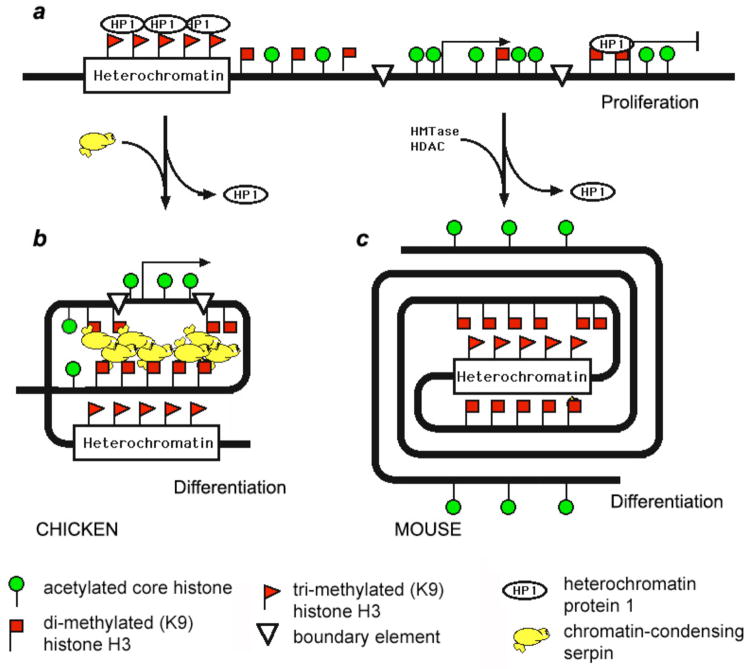
Fig. 8.
Model showing different molecular mechanisms proposed to mediate chromatin condensation in avian (nucleated) erythrocytes vs. mammalian erythroblasts before enucleation. (a) In proliferating cells histone H3me2K9 (square flags) is interspersed with histone acetylation (circle signs) in the euchromatin. Histone H3me3K9 (triangle flags) and HP1 are associated with pericentromeric heterochromatin. (b) During terminal differentiation in avian erythrocytes, the chromatin condensing factors (H5 and MENT) replace HP1 and cause chromatin condensation at multiple and dispersed foci of facultative heterochromatin marked by H3me2K9. Active chromosomal domains are structurally insulated by boundary elements (open triangles) that inhibit spreading of chromatin condensing factors onto active genes. (c) In differentiating mammalian erythroblasts, HP1 is also removed from the chromatin but is not replaced by another chromatin-condensing architectural factor. Instead, the increased histone methyl transferase and HDAC activities lead to a sharp decrease in histone acetylation in the apocentric zone formed around the constitutive heterochromatin. The apocentric chromatin condenses as a result of loss of histone acetylation and, by the end of the differentiation process, forms a facultative heterochromatin territory spatially segregated from the residual active chromatin territory remaining at the nuclear periphery.
In our experimental system, the low levels of histone acetylation in late erythroblasts may result from two parallel developmentally-regulated processes. One mechanism entails increased expression of histone deacetylases while the second mechanism involves accumulation of histone H3 methylation in the vicinity of heterochromatin (Fig. 8). This methylation may act to further inhibit histone acetylation since histone methylation and acetylation involve similar amino acids according to the “histone code” in which a precise set of histone modifications determines chromatin structure and transcriptional activity (Jenuwein and Allis, 2001). Furthermore, decreased acetylation may result in detachment of chromatin from existing nuclear structures (e.g. those containing bromodomain proteins) and thus contribute to chromatin condensation without any additional chromatin architectural proteins.
In contrast, chicken erythrocyte differentiation does not involve either increased histone H3 methylation nor spatial segregation between histone methylation and acetylation (Istomina et al., 2003) consistent with the primary roles of developmentally-regulated structural proteins H5 and MENT in the condensation process (Fig. 8). Interestingly, chicken β-globin gene contains a unique boundary element ensuring structural segregation between the active chromosomal loci and the nearby heterochromatin (Felsenfeld et al., 2004). The reason that such a well-defined boundary element is absent from mammalian β-globin genes may be related to the spatial rather than structural segregation between active and repressed chromatin in the mammalian blood cells.
Our study identified HDAC5 as a histone deacetylase with an increased expression level in differentiating murine erythroblasts. Remarkably, HDAC5 has been recently shown to be associated with histone H3 N-terminal peptide requiring lysine 9 for its association (Heo et al., 2007). HDAC5 enrichment at the nuclear periphery (Fig. 6C), where the residual histone acetylation is also localized (Fig. 5, panel 8), suggests that HDAC5 is tethered to the areas of its action through direct association with histone H3 N-tail. HDAC5 has been previously implied in regulating cell growth and differentiation (Zhang et al., 2002) and, in particular, its accumulation in the nucleus of erythroleukemia (MEL) cells has been shown to interfere with activity of the key factor of erythroid differentiation, GATA-1 (Watamoto et al., 2003). Our present work shows that HDAC5 may play a dual role in mammalian erythropoiesis: first by acting locally to inhibit a differentiation inducer at the early stage of erythroid differentiation, and secondly to condense chromatin by promoting global histone deacetylation at the later stages of erythroblast maturation. In view of increasing use of deacetylase inhibitors for chemotherapy of hematologic malignancies (Claus and Lubbert, 2003), it is extremely important to elucidate the role of histone modifications in terminal erythroid differentiation and minimize their possible adverse effects on erythropoiesis.
Supplementary Material
Acknowledgments
We are thankful to Drs. P. Singh (Borstel, Germany), D. Tremethick (Canberra, Australia), and N. Chaudhary (Woodlands, TX) for their kind gifts of antibodies against HP1, H2A.Z, and lamins A/C and B. Supported in part by National Institutes of Health Grants DK32094, DK56267, DK59079, and CA084214; National Science Foundation grant MCB-0615536, and by the Director, Office of Health and Environment Research Division, US Department of Energy, under Contract DE-AC03-76SF00098; and by Merit Review Award from the Dept. of Veteran Affairs.
List of abbreviations
- FVA cells
Friend virus-infected murine spleen erythroblasts
- H3acK9
K14, histone H3 acetylated at Lysines 9 and 14
- H3me2K9
histone H3 dimethylated at Lysine 9
- H3me3K9
histone H3 trimethylated at Lysine 9
- H4acK12
histone H4 acetylated at Lysine 12
- HDAC
histone deacetylase
- HP1
heterochromatin protein 1
- HPLC
High-performance Liquid Chromatography
- KCM buffer
120mM KCl, 20mM NaCl, 10mM Tris-HCl, pH7.7, 0.1% Triton X-100
- MEL
mouse erythroleukemia cells
- PBS
phosphate-buffered saline
- RSB
Reticulocyte Standard Buffer (10mM NaCl, 3mM MgCl2, 10mM HEPES, pH7.5)
- SDS-PAGE
sodium dodecyl sulfate-containing polyacrylamide gel electrophoresis
- TSA
trichostatin A
References
- Adkins NL, Watts M, Georgel PT. To the 30-nm chromatin fiber and beyond. Biochim Biophys Acta. 2004;1677:12–23. doi: 10.1016/j.bbaexp.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of Di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Bates DL, Thomas JO. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 1981;9:5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Brero A, Easwaran HP, Nowak D, Grunewald I, Cremer T, Leonhardt H, Cardoso MC. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J Cell Biol. 2005;169:733–743. doi: 10.1083/jcb.200502062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Bulynko YA, Hsing LC, Mason RW, Tremethick DJ, Grigoryev SA. Cathepsin L stabilizes the histone modification landscape on the Y chromosome and pericentromeric heterochromatin. Mol Cell Biol. 2006;26:4172–4184. doi: 10.1128/MCB.00135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary N, Courvalin JC. Stepwise reassembly of the nuclear envelope at the end of mitosis. J Cell Biol. 1993;122:295–306. doi: 10.1083/jcb.122.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HH, Park HT, Kim YJ, Bae YC, Suh KT, Jung JS. Induction of osteogenic differentiation of human mesenchymal stem cells by histone deacetylase inhibitors. J Cell Biochem. 2005;96:533–542. doi: 10.1002/jcb.20544. [DOI] [PubMed] [Google Scholar]
- Claus R, Lubbert M. Epigenetic targets in hematopoietic malignancies. Oncogene. 2003;22:6489–6496. doi: 10.1038/sj.onc.1206814. [DOI] [PubMed] [Google Scholar]
- Cowell IG, Aucott R, Mahadevaiah SK, Burgoyne PS, Huskisson N, Bongiorni S, Pantera G, Fanti L, Pimpinelli S, Wu R, Gilbert DM, Shi W, Fundele R, Morrisin H, Jeppesen P, Singh PB. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma. 2002;111:22–36. doi: 10.1007/s00412-002-0182-8. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Elgin SC. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer. 2006;94:179–183. doi: 10.1038/sj.bjc.6602918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Burgess-Beusse B, Farrell C, Gaszner M, Ghirlando R, Huang S, Jin C, Litt M, Magdinier F, Mutskov V, Nakatani Y, Tagami H, West A, Yusufzai T. Chromatin boundaries and chromatin domains. Cold Spring Harb Symp Quant Biol. 2004;69:245–250. doi: 10.1101/sqb.2004.69.245. [DOI] [PubMed] [Google Scholar]
- Francastel C, Schubeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1:137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- Frenster JH. Ultrastructure and function of heterochromatin and euchromatin. In: Busch H, editor. The Cell Nucleus. Vol. 1. Academic Press; New York: 1974. pp. 565–581. [Google Scholar]
- Gilbert N, Boyle S, Sutherland H, de Las Heras J, Allan J, Jenuwein T, Bickmore WA. Formation of facultative heterochromatin in the absence of HP1. Embo J. 2003;22:5540–5550. doi: 10.1093/emboj/cdg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Rangasamy D, Devoy M, Marshall Graves JA, Tremethick DJ. The X and Y Chromosomes Assemble into H2A.Z, Containing Facultative Heterochromatin, following Meiosis. Mol Cell Biol. 2006;26:5394–5405. doi: 10.1128/MCB.00519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryev SA. Higher-order folding of heterochromatin: protein bridges span the nucleosome arrays. Biochem Cell Biol. 2001;79:227–241. [PubMed] [Google Scholar]
- Grigoryev SA, Bulynko YA, Popova EY. The end adjusts the means: heterochromatin remodelling during terminal cell differentiation. Chromosome Res. 2006;14:53–69. doi: 10.1007/s10577-005-1021-6. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Nikitina T, Pehrson JR, Singh PB, Woodcock CL. Dynamic relocation of epigenetic chromatin markers reveals an active role of constitutive heterochromatin in the transition from proliferation to quiescence. J Cell Sci. 2004;117:6153–6162. doi: 10.1242/jcs.01537. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Solovieva VO, Spirin KS, Krasheninnikov IA. A novel nonhistone protein (MENT) promotes nuclear collapse at the terminal stage of avian erythropoiesis. Exp Cell Res. 1992;198:268–275. doi: 10.1016/0014-4827(92)90379-m. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Woodcock CL. Chromatin structure in granulocytes. A link between tight compaction and accumulation of a heterochromatin-associated protein (MENT) J Biol Chem. 1998;273:3082–3089. doi: 10.1074/jbc.273.5.3082. [DOI] [PubMed] [Google Scholar]
- Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. Embo J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig W. Heterochromatin. Chromosoma. 1999;108:1–9. doi: 10.1007/s004120050346. [DOI] [PubMed] [Google Scholar]
- Heo K, Kim B, Kim K, Choi J, Kim H, Zhan Y, Ranish JA, An W. Isolation and characterization of proteins associated with histone H3 tails in vivo. J Biol Chem. 2007;282:15476–15483. doi: 10.1074/jbc.M610270200. [DOI] [PubMed] [Google Scholar]
- Hilwig I, Gropp A. Staining of constitutive heterochromatin in mammalian chromosomes with a new fluorochrome. Exp Cell Res. 1972;75:122–126. doi: 10.1016/0014-4827(72)90527-7. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Brower-Toland B, Elgin SC. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 2006;115:110–122. doi: 10.1007/s00412-006-0052-x. [DOI] [PubMed] [Google Scholar]
- Irving JA, Shushanov SS, Pike RN, Popova EY, Bromme D, Coetzer TH, Bottomley SP, Boulynko IA, Grigoryev SA, Whisstock JC. Inhibitory activity of a heterochromatin-associated serpin (MENT) against papain-like cysteine proteinases affects chromatin structure and blocks cell proliferation. J Biol Chem. 2002;277:13192–13201. doi: 10.1074/jbc.M108460200. [DOI] [PubMed] [Google Scholar]
- Istomina NE, Shushanov SS, Springhetti EM, Karpov VL, Krasheninnikov IA, Stevens K, Zaret KS, Singh PB, Grigoryev SA. Insulation of the Chicken β–globin Chromosomal Domain from a Chromatin-Condensing Protein, MENT. Mol Cell Biol. 2003;23:6455–6468. doi: 10.1128/MCB.23.18.6455-6468.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kelley LL, Koury MJ, Bondurant MC, Koury ST, Sawyer ST, Wickrema A. Survival or death of individual proerythroblasts results from differing erythropoietin sensitivities: a mechanism for controlled rates of erythrocyte production. Blood. 1993;82:2340–2352. [PubMed] [Google Scholar]
- Koury MJ, Park DJ, Martincic D, Horne DW, Kravtsov V, Whitlock JA, del Pilar Aguinaga M, Kopsombut P. Folate deficiency delays the onset but increases the incidence of leukemia in Friend virus-infected mice. Blood. 1997;90:4054–4061. [PubMed] [Google Scholar]
- Koury MJ, Sawyer ST, Bondurant MC. Splenic erythroblasts in anemia-inducing Friend disease: a source of cells for studies of erythropoietin-mediated differentiation. J Cell Physiol. 1984;121:526–532. doi: 10.1002/jcp.1041210311. [DOI] [PubMed] [Google Scholar]
- Koury ST, Koury MJ, Bondurant MC. Morphological changes in erythroblasts during erythropoietin-induced terminal differentiation in vitro. Exp Hematol. 1988;16:758–763. [PubMed] [Google Scholar]
- Koury ST, Koury MJ, Bondurant MC. Cytoskeletal distribution and function during the maturation and enucleation of mammalian erythroblasts. J Cell Biol. 1989;109:3005–3013. doi: 10.1083/jcb.109.6.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutzamani E, Loborg H, Sarg B, Lindner HH, Rundquist I. Linker histone subtype composition and affinity for chromatin in situ in nucleated mature erythrocytes. J Biol Chem. 2002;277:44688–44694. doi: 10.1074/jbc.M203533200. [DOI] [PubMed] [Google Scholar]
- Krauss SW, Lo AJ, Short SA, Koury MJ, Mohandas N, Chasis JA. Nuclear substructure reorganization during late-stage erythropoiesis is selective and does not involve caspase cleavage of major nuclear substructural proteins. Blood. 2005;106:2200–2205. doi: 10.1182/blood-2005-04-1357. Epub 2005 Jun 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee G, Spring FA, Parsons SF, Mankelow TJ, Peters LL, Koury MJ, Mohandas N, Anstee DJ, Chasis JA. Novel secreted isoform of adhesion molecule ICAM-4: potential regulator of membrane-associated ICAM-4 interactions. Blood. 2003;101:1790–1797. doi: 10.1182/blood-2002-08-2529. [DOI] [PubMed] [Google Scholar]
- Li Y, Danzer JR, Alvarez P, Belmont AS, Wallrath LL. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development. 2003;130:1817–1824. doi: 10.1242/dev.00405. [DOI] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001a;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Recillas-Targa F, Prioleau MN, Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. Embo J. 2001b;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Billi AM, Gilmour RS, Manzoli L, Di Primio R, Cocco L. Mouse and human hemopoietic cell lines of erythroid lineage express lamins A,B and C. Biochem Biophys Res Commun. 1992;185:271–276. doi: 10.1016/s0006-291x(05)80986-6. [DOI] [PubMed] [Google Scholar]
- McBryant SJ, Adams VH, Hansen JC. Chromatin architectural proteins. Chromosome Res. 2006;14:39–51. doi: 10.1007/s10577-006-1025-x. [DOI] [PubMed] [Google Scholar]
- Peterson DG, Stack SM, Healy JL, Donohoe BS, Anderson LK. The relationship between synaptonemal complex length and genome size in four vertebrate classes (Osteicthyes, Reptilia, Aves, Mammalia) Chromosome Res. 1994;2:153–162. doi: 10.1007/BF01553494. [DOI] [PubMed] [Google Scholar]
- Popova EY, Claxton DF, Lukasova E, Bird PI, Grigoryev SA. Epigenetic heterochromatin markers distinguish terminally differentiated leukocytes from incompletely differentiated leukemia cells in human blood. Exp Hematol. 2006;34:453–462. doi: 10.1016/j.exphem.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Rangasamy D, Berven L, Ridgway P, Tremethick DJ. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. Embo J. 2003;22:1599–1607. doi: 10.1093/emboj/cdg160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Schubeler D, Francastel C, Cimbora DM, Reik A, Martin DI, Groudine M. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Singh PB, Georgatos SD. HP1: facts, open questions, and speculation. Journal of Structural Biology. 2002;140:10–16. doi: 10.1016/s1047-8477(02)00536-1. [DOI] [PubMed] [Google Scholar]
- Springhetti EM, Istomina NE, Whisstock JC, Nikitina TV, Woodcock CL, Grigoryev SA. Role of the M-loop and Reactive Center Loop Domains in the Folding and Bridging of Nucleosome Arrays by MENT. J Biol Chem. 2003;278:43384–43393. doi: 10.1074/jbc.M307635200. [DOI] [PubMed] [Google Scholar]
- Su RC, Brown KE, Saaber S, Fisher AG, Merkenschlager M, Smale ST. Dynamic assembly of silent chromatin during thymocyte maturation. Nat Genet. 2004a;36:502–506. doi: 10.1038/ng1351. [DOI] [PubMed] [Google Scholar]
- Su RC, Brown KE, Saaber S, Fisher AG, Merkenschlager M, Smale ST. Dynamic assembly of silent chromatin during thymocyte maturation. Nat Genet. 2004b;36:502–506. doi: 10.1038/ng1351. Epub 2004 Apr 2018. [DOI] [PubMed] [Google Scholar]
- Sung MT, Harford J, Bundman M, Vidalakas G. Metabolism of histones in avian erythroid cells. Biochemistry. 1977;16:279–285. doi: 10.1021/bi00621a019. [DOI] [PubMed] [Google Scholar]
- Terranova R, Sauer S, Merkenschlager M, Fisher AG. The reorganisation of constitutive heterochromatin in differentiating muscle requires HDAC activity. Exp Cell Res. 2005;20:20. doi: 10.1016/j.yexcr.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A, Thomas JO. Chromatin structure of the beta-globin chromosomal domain in adult chicken erythrocytes. Cold Spring Harb Symp Quant Biol. 1993;58:15–24. doi: 10.1101/sqb.1993.058.01.005. [DOI] [PubMed] [Google Scholar]
- Verschure PJ, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter AE, Belmont AS, van Driel R. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watamoto K, Towatari M, Ozawa Y, Miyata Y, Okamoto M, Abe A, Naoe T, Saito H. Altered interaction of HDAC5 with GATA-1 during MEL cell differentiation. Oncogene. 2003;22:9176–9184. doi: 10.1038/sj.onc.1206902. [DOI] [PubMed] [Google Scholar]
- Weintraub H. The nucleosome repeat length increases during erythropoiesis in the chick. Nucleic Acids Res. 1978;5:1179–1188. doi: 10.1093/nar/5.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. Histone-H1-dependent chromatin superstructures and the suppression of gene activity. Cell. 1984;38:17–27. doi: 10.1016/0092-8674(84)90522-1. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Dimitrov S. Higher order structure of chromatin and chromosomes. Curr Opin in Gen Dev. 2001;11:130–135. doi: 10.1016/s0959-437x(00)00169-6. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- Wright JM, Wiersma PA, Dixon GH. Use of protein blotting to study the DNA-binding properties of histone H1 and H1 variants. Eur J Biochem. 1987;168:281–285. doi: 10.1111/j.1432-1033.1987.tb13418.x. [DOI] [PubMed] [Google Scholar]
- Yellajoshyula D, Brown DT. Global modulation of chromatin dynamics mediated by dephosphorylation of linker histone H1 is necessary for erythroid differentiation. Proc Natl Acad Sci U S A. 2006;103:18568–18573. doi: 10.1073/pnas.0606478103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Leuba SH. Chromatin Fibers, One-at-a-time. J Mol Biol. 2003;331:1–19. doi: 10.1016/s0022-2836(03)00691-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.