Abstract
The tumor suppressor p53 is a master sensor of stress. Two human-specific polymorphisms, p53 codon 72 and MDM2 SNP309, influence the activities of p53. There is a tight association between cold winter temperature and p53 Arg72 and between low UV intensity and MDM2 SNP309 G/G in a cohort of 4029 individuals across Eastern Asia that suggests causative selection. Moreover, the two polymorphisms are not coselected. Haplotype-based selection analysis further suggests that this is a striking example of two functional polymorphisms being strongly selected for in human populations in response to environmental stresses.
Main Text
The tumor suppressor p53 (TP53 [MIM 191170]) is a master sensor of stress signals and is one of the most important tumor suppressors identified so far.1 As a nuclear protein, p53 is an evolutionarily conserved transcription factor and it can transactivate and transrepress a large number of target genes. Consequently, p53 is able to regulate many cellular functions including cell cycle progression, DNA repair, senescence, apoptosis, and cellular metabolism.2 The expression level of p53 is normally very low and this is due to its short half-life. However, p53 protein levels increase in response to various stress signals such as DNA damaging agents, oxidative stress, amino acid depletion, and temperature change.3
Among all cellular regulators of p53, the oncoprotein Mdm2 (MDM2 [MIM 164785]) is perhaps its most well known inhibitor.4,5 Interestingly, Mdm2 is also a transcriptional target of p53, and its expression level is tightly regulated by p53's transcriptional activity. This auto-regulatory loop ensures the precise regulation of the protein levels and activities of both p53 and Mdm2 and is vital for murine embryonic development and homeostasis.6
In humans, two single-nucleotide polymorphisms (SNPs) in the p53 pathway, p53 codon 72 and MDM2 SNP309, influence the activities of p53 and Mdm2, respectively.7,8 p53 codon 72 results in either a proline or an arginine as a result of a one nucleotide change, of the second base in the codon, from a C to G (rs1042522). The Arg allele is slightly more active in inducing apoptosis and exists only in humans.8 It therefore begs the question whether there is a positive selective pressure for the Arg allele in human evolution. If so, what factors influence such selection? Similar questions apply to the Mdm2 polymorphism MDM2 SNP309 (rs2279744), which is located in the MDM2 promoter sequence.9 A single-nucleotide change from T to G creates a binding site for the transcription factor SP1 (MIM 189906). As a result, homozygotes for the G allele express more Mdm2 mRNA and protein than do homozygotes for the T allele.9 Both polymorphisms affect the p53 stress response pathway in humans. It is, therefore, possible that certain alleles of either the p53 codon 72 or MDM2 SNP309 might be under evolutionary selection pressure in order to adapt to, or respond to, certain p53-activating environmental stress signals.
Previous studies have shown that the genotypes of both p53 codon 72 and MDM2 SNP309 are differentially distributed in different ethnic populations, originating from different parts of the world.10–12 The Arg and G alleles from p53 codon 72 and MDM2 SNP309, respectively, are more common in northern Europeans than in Africans or African-Americans, leading to the hypothesis that the frequency of these alleles is latitude dependent.10–12 It was suggested that the haplotype structure surrounding the MDM2 SNP309 polymorphism may indicate a recent selective sweep in European and Ashkenazi Jewish populations.10 Based on the observations made with p53 codon 72, it was hypothesized that varying UV radiation levels, a p53-activating stress, in different latitudes could underpin the latitude-dependent distribution of the genotypes.11 However, this study could not rule out the possibility that the differences in the allelic distributions of p53 codon 72 were a result only of the high level of divergence in the genetic makeup of the different ethnic populations studied.
In this report, we test the hypothesis that environmental stresses may play a role in selecting functional p53 polymorphisms in closely linked East Asian populations. In contrast to previous studies, our observations are made in either the same ethnic group (Han Chinese) or closely related ethnic groups in eastern Asia (mostly in China) that live in extremely different latitudes and are subsequently exposed to significantly different environmental stresses. The observations presented in this report suggest that environmental exposure is a selective force on both functional polymorphic loci in the p53 tumor suppressor pathway and that this selection is an on-going process in eastern Asian populations.
We genotyped the p53 codon 72 and the MDM2 SNP309 loci in 4029 eastern Asian individuals from 67 different populations (mostly from China [Figure S1 available online]). The protocol of this study was approved by the institutional review board of Kunming Insitute of Zoology, Chinese Academy of Sciences. The p53 codon 72 polymorphism was analyzed by PCR-RFLP method. The PCR primers used were: Forward primer 5′-GCT CTT TTC ACC CAT CTA CAG TCC-3′; Reverse 5′-AGG AGG CCC AGA CGG AAA CC-3′. The Arg allele can be recognized and cut by the restriction enzyme Bstu I. We also conducted sequencing (5 randomly selected samples from each 96-well sample plate) to double-check the typing result. The MDM2 SNP309 polymorphism was analyzed by sequencing method, via primers from Ma et al.13
The p53 codon 72 and MDM2 SNP309 polymorphism data was used to analyze frequency distribution across geographic regions by generating contour maps (Golden Software Surfer 7.0). Correlation analysis was conducted with SPSS13.0, and statistical significance was accessed by t test. The Hardy-Weinberg test was performed by Genepop 3.4, and the statistical significance was assessed by Fisher's exact test. The environmental data (UV radiation levels and temperature) were collected from the National Meteorological Information Centre of China. The UV data covered the years 1971 to 2000, whereas the temperature data covered the years 1951 to 2006. The average annual total of UV radiation (mega joules/year/m2) for each geographic site was calculated via the global solar radiation intensity over 30 years14 and was used in the correlation analysis. The monthly average temperature was calculated with the temperature records of the last 56 years. The average temperatures in July and January were taken as the highest and lowest temperatures, respectively, and used for the correlation analysis (Table S1).
The 67 populations analyzed in this study represent 41 ethnic nationalities living in China and other eastern Asian regions. Most of the samples were males and only five populations had both male and female samples (four Tibetan populations and one Yunnan Han population). Our statistical test showed that there was no significant allelic frequency difference between males and females of the same population; therefore, we analyzed all the samples without separating sexes. It is important to note that the genetic variation among populations living in eastern Asia is much smaller than that found between Africans and Northern Europeans.15 In China, although the majority of the population is Han Chinese (93.3%), there are 55 official minority nationalities (6.7%). Most of these have their own languages and are found predominantly in the peripheral regions of the country. Of the 67 populations in our study, 17 were Han Chinese. Almost all of the populations studied lived in different latitudinal regions of eastern Asia from latitude S11° to N65° (China N18° to N53°, Figure S1), and the results illustrate a clear latitude dependency for the occurrence of the Arg allele of p53 codon 72 (r = 0.64, p < 0.01, two-tailed t test; Table 1, Figure 1A and 1B; Table S2).
Table 1.
The Latitude, UV Radiation, and Winter Temperature Correlation Analysis on p53 Codon 72 and MDM2 SNP309
| Environmental Factors | Populations |
p53 Codon 72 |
MDM2 SNP309 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| PP | RP | RR | R | TT | TG | GG | G | ||
| Latitude | all populations (67) | −0.478∗∗ | −0.221 | 0.611∗∗ | 0.640∗∗ | 0.008 | 0.195 | −0.193 | −0.153 |
| Han populations (17) | −0.378 | −0.372 | 0.699∗∗ | 0.672∗∗ | −0.140 | 0.212 | −0.132 | −0.039 | |
| UV radiation | all populations (67) | −0.211 | −0.013 | 0.189 | 0.232 | −0.044 | 0.455∗∗ | −0.426∗∗ | −0.319∗∗ |
| Han populations (17) | −0.027 | −0.144 | 0.173 | 0.130 | −0.112 | 0.569∗ | −0.509∗ | −0.344 | |
| Winter temperature | all populations (67) | 0.512∗∗ | 0.213 | −0.631∗∗ | −0.670∗∗ | 0.036 | −0.177 | 0.153 | 0.104 |
| Han populations (17) | 0.420 | 0.377 | −0.739∗∗ | −0.772∗∗ | 0.043 | −0.124 | 0.100 | 0.058 | |
| Summer temperature | all populations (67) | 0.197 | 0.046 | −0.209 | −0.237 | 0.026 | −0.183 | 0.169 | 0.125 |
| Han populations (17) | 0.028 | 0.270 | −0.305 | −0.219 | −0.117 | −0.194 | 0.265 | 0.258 | |
Note: the numbers are the correlation coefficient values. ∗p < 0.05, ∗∗p < 0.01
Figure 1.
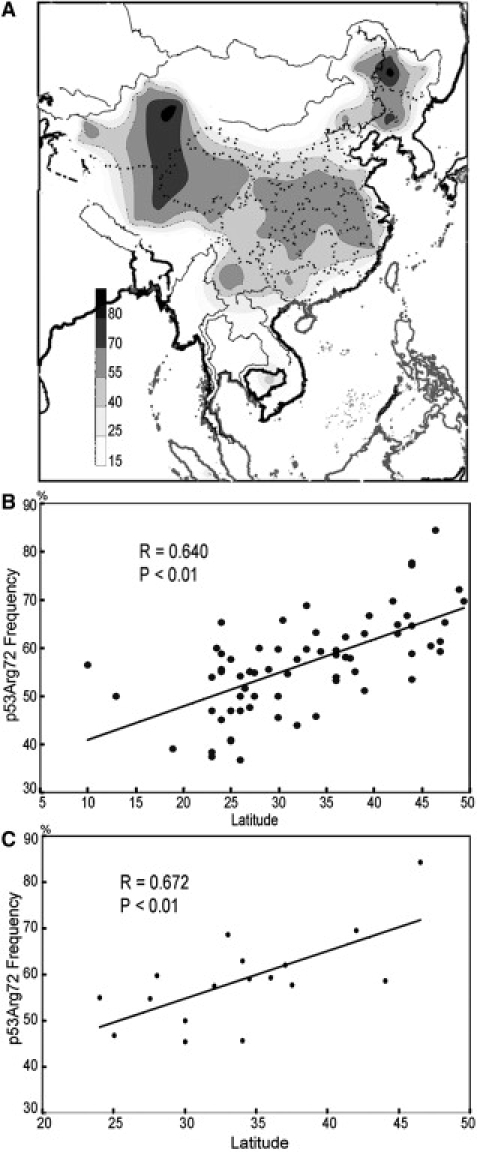
The Distribution of the p53 Arg72 Allele Frequencies and Their Correlation with the Latitudes
(A) A contour map of p53 Arg72 allele frequency distribution of the 67 populations in eastern Asia showing strong latitude dependency. Black shading indicates a high frequency of p53 Arg72.
(B) The correlation of p53 Arg72 allele frequencies with latitudes for the 67 populations studied.
(C) The correlation of p53 Arg72 allele frequencies with latitudes for the 17 Han Chinese populations.
Importantly, we made similar observations when comparing populations of the same ethnicity. Of the 67 populations in our study group, the 17 Han Chinese populations had been residing at different latitudinal regions of China for several thousand years, namely from latitude N24° to N46.5° (Figure S1). Interestingly, the same correlation with high latitude was observed for high frequency of p53 Arg72 (r = 0.67, p < 0.01, two-tailed t test) (Figure 1C; Tables S1 and S2). However, this correlation did not exist for MDM2 SNP309 G allele (r = −0.153, p = 0.216; Table 1). Together, these observations suggest a strong selection of the different alleles of the p53 codon 72 locus in populations living in different latitudinal regions, especially because the latitude-dependent distribution of the genotypes was observed in the same ethnic group.
It had been hypothesized that the latitude-dependent variation of p53 Arg72 could be due to varying average UV radiation exposure of populations from different latitudes.11 To test this, we collected data regarding the average intensities of UV radiation over the years 1971 to 2000 from the National Meteorological Information Centre of China, for all of the regions that the studied populations inhabit. Surprisingly, the frequency of p53 Arg72 did not correlate with the average intensity of UV radiation for either the 67 populations as a whole (r = 0.232, p = 0.059) or for the 17 Han Chinese populations (r = 0.130, p = 0.619) (Table 1; Table S3). These data suggested that the latitude-dependent variation of the alleles of the p53 codon 72 locus was not due to varying average UV radiation levels.
Another p53-activating stress that significantly varies with latitude is temperature. Both cold and warm temperature fluctuations have been shown to elicit a p53-dependent cellular stress response.16,17 To explore whether average temperature differences could explain the latitude dependence of the p53 Arg72 genotype, we collected both the average winter (average temperature in January) and summer (in July) temperatures from 1951 to 2006 for all regions studied, from the National Meteorological Information Centre of China. Interestingly, we observed that low average winter temperature showed an association with high frequency of p53 Arg72 (r = −0.67, p < 0.01 for the 67 populations, and r = −0.77, p < 0.01 for the 17 Han Chinese populations, two-tailed t test) (Table 1, Figures 2A and 2B; Table S2, Figure S2), whereas no correlation was observed for summer temperature (r = −0.237, p = 0.054, two-tailed t test; Figure S3). Correlation analysis of the environmental factors indicated that latitude was highly associated with winter temperature (r = −0.96, p < 0.01, two-tailed t test). To provide further supporting statistical evidence, we performed the Bonferroni correction and the p values were still significant (Table S2). We also performed partial correlation analysis to test the multifactor effect, in which the observed correlation of p53 Arg72 with winter temperature still held when other environmental factors were used as control variables (Table S4). Together, these data suggest that the latitude-dependent difference in the distribution of p53 Arg72 is not due to the variation of UV radiation, but to the variation of winter temperature.
Figure 2.
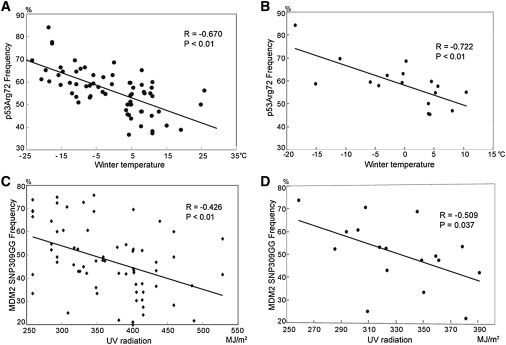
The Correlation of the p53 Arg72 and MDM2 SNP309GG Frequencies with Winter Temperature and UV Radiation
(A) The correlation of p53 Arg72 allele frequency with winter temperature in the 67 populations.
(B) The correlation of p53 Arg72 allele frequency with winter temperature in the 17 Han Chinese populations.
(C) The correlation of MDM2 SNP309GG frequency with UV radiation in the 67 populations.
(D) The correlation of MDM2 SNP309GG frequency with UV radiation in the 17 Han Chinese populations.
The MDM2 SNP309 G allele has opposing activities on the p53 pathway to that of p53 Arg72. We therefore tested the influence of winter temperature on the Mdm2 SNP309 locus by genotyping the same populations studied above (Figure S4). In contrast to the p53 codon 72 data, the occurrence of the Mdm2 polymorphism was not found to correlate with winter temperature, irrespective of the population studied (r = 0.104, p = 0.404 for the 67 populations) (Table 1; Figure S4).
The MDM2 SNP309 genotypes were then analyzed for their association with average UV radiation intensity, another environmental factor that is known to be latitude dependent. Strikingly, UV radiation strength showed a significant negative correlation with the frequency of the MDM2 SNP309 G/G genotype, whereby high frequency of the G/G genotype associated with low UV radiation strength (r = −0.43, p < 0.01 for the total 67 populations, and r = −0.51. p = 0.037 for the 17 Han Chinese populations, two-tailed t test; Table 1, Figures 2C and 2D; Table S2, Figures S4 and S5). Partial correlation analysis confirmed this result and ruled out the influence of the other environmental factors (Table S4). Thus, the variation of this p53-activating stress correlates not with alleles of the p53 codon 72 locus, but with the frequency of the other functional p53 pathway polymorphism, MDM2 SNP309, even though latitude dependence of this locus was not demonstrated in these populations. These results suggest that although both p53 Arg72 and MDM2 SNP309 G/G polymorphisms affect p53 activities, they have different sensitivities to different environmental stress. Collectively, this supports the hypothesis that the alleles of both loci are under selective pressure from at least one environmental exposure known to activate the p53 cellular stress response.
To further investigate whether the two polymorphisms had undergone natural selection, we sequenced the 1.74 kb region (5 exons and 4 introns spanning from exon 2 to exon 6) of p53 flanking the codon 72 site in 182 randomly selected East Asian individuals, as well as 20 African individuals for control. A total of 10 variants (including nine SNPs and one 16 bp insertion) were identified in the 1.74 kb region (Table S5). The population and haplotype structure-based Fs test18 was conducted, in which East Asians showed a strong signature of natural selection (Fs = −6.67, p = 0.034) when compared with Africans (Fs = −2.7, p = 0.11). Interestingly, when the East Asians were separated into southern and northern groups, we observed a strong signature of selection in the northern group (Fs = −6.28, p = 0.025), but not in the southern group (Fs = −3.04, p = 0.13). The suggested selection was also supported by the Z test (performed with SPSS 13.0) in which a significant haplotype frequency difference was detected between East Asians and Africans (Z = −3.27, p = 0.001; two-tailed exact test), and the northern group (Z = −2.22, p = 0.025; two-tailed exact test) showed a stronger signature than that of the southern group (Z = −1.92, p = 0.061; two-tailed exact test). This pattern is consistent with the observed correlation of p53 codon 72 with winter temperature, because the much colder winter in the north would generate a stronger selective pressure on the northern populations. Additionally, the Hardy-Weinberg equilibrium (HWE) test indicated that 12 of the 67 populations showed deviation from HWE, again supporting the proposed natural selection on the p53 codon 72 polymorphism in East Asia. Similarly, the MDM2 SNP309 locus also deviated from Hardy-Weinberg equilibrium in 19 out of the 67 populations tested (18 with heterozygote deficit and one with heterozygote excess) (p < 0.05, Fisher's exact test; Table S6 and Figure S6). Together, these data suggest that selection pressure from winter temperatures and UV exposure on the p53 codon 72 and MDM2 SNP309 loci, respectively, are strong, and that the allelic selection is an on-going process in eastern Asian populations. A stronger correlation was not observed for the two-loci combination analysis, when compared with the single locus (Tables S7 and S8). Collectively, these data support the notion that the two loci are under selection from different environmental stresses.
In agreement with previous findings, where the frequency of the p53 Pro72 allele showed a north-south cline from 17% in Swedish Saamis to 63% in African Blacks (Nigerians),11 we found that the frequency of p53 Arg72 is associated with latitude. However, we found only a weak correlation between latitude and average UV radiation intensity in the 67 populations tested (r = 0.147, p = 0.235, two-tailed t test). Instead, we found that the correlation coefficient between latitude and average winter temperature was extremely high (r = −0.957, p < 0.01, two-tailed t test). The observed latitude-dependent frequency of p53 Arg72 is, therefore, tightly associated with winter temperature and not UV radiation.
p53 has been shown to be activated in response to changes in temperature,16,17 and the differences in polymorphism status across latitudes illustrated in this report may reflect the need for subtle changes in the p53 pathway upon different environmental stresses. One underlying biological reason for this selection could be p53's possible role in embryo implantation. Recently, it was found that the frequency of the p53 Pro72 allele is significantly higher in women that experience recurrent implantation failure after IVF or embryo transfer treatment than those that do not.19 Mouse studies have shown that one of the key determining factors of implantation, leukemia inhibitory factor (LIF [MIM 159540]), is a transcriptional target of p53.20 Therefore, p53 influences implantation through its ability to control the expression level of LIF. To provide molecular evidence that the p53 polymorphism at codon 72 could affect the rate of implantation, we tested the ability of p53 Pro72 versus p53 Arg72 to transactivate LIF via a LIF promoter-linked luciferase reporter assay. The Soas2 cells expressing a temperature-sensitive form of p53, containing either proline or arginine at codon 72 in the p53 expression plasmid, were prepared to examine the impact of p53 codon 72 SNP upon induction of LIF. Quantitative real-time PCR was performed in triplicate and the actin gene was used as the internal control.
In agreement with the hypothesis, p53 Arg72 was clearly more active than p53 Pro72 in transactivating LIF, and this difference was found to be specific. In contrast, under the same conditions, p53 Arg72 and p53 Pro72 transactivated the p21 waf1 promoter with similar levels of activity (Figure 3). These findings support the hypothesis that selection of the p53 Arg72 allele could provide a selective advantage to populations adapting to colder climates, by reducing the risk of implantation failure.20 The reduction of conception rates in cattle under cold or heat stress is well documented.21
Figure 3.
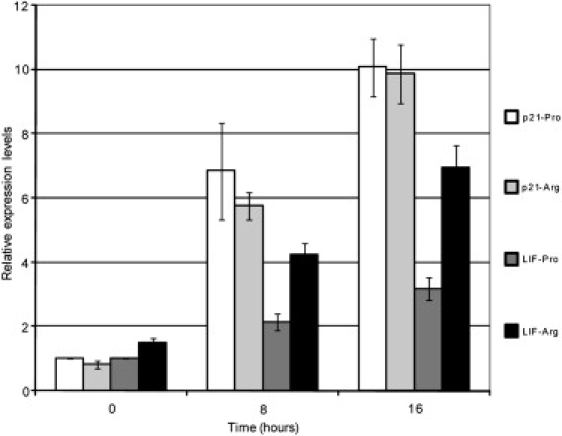
Different Activities of p53 Pro72 versus p53 Arg72 to Transactivate LIF
There is no significant difference detected for the control gene (p21). The error bars are standard errors that were calculated based on the triplicate measurements.
The observed selection for the more active p53 Arg72 allele might also be related to p53's role in cellular metabolism. p53 is activated by AMPK (MIM 602740, MIM 600497, MIM 602739) to induce cell cycle arrest in response to low glucose concentrations, through a transcriptionally dependent mechanism.22 p53 has also been shown to decrease the rate of glycolysis through induction of the TIGAR (MIM 610775)23 and SCO2 (MIM 604272) genes24 and to generate ATP via the mitochondrial pathway. It is, therefore, possible that selection of the more active p53 Arg72 allele provides a selective advantage during prolonged periods of low winter temperatures.
The p53 Arg72 isoform has also been shown to be more active than p53 Pro72 in inducing apoptosis and suppressing cellular transformation.25,26 This increase in activity results from the increased localization of p53 Arg72 to the mitochondria and is independent of transcription. Differences in the isoforms' mitochondrial localizations may also mean that the codon 72 polymorphism affects the ability of p53 to regulate mitochondrial respiration and other metabolic factors, although further studies are needed. The transcriptional activity of p53 Arg72 is also regulated by its interaction with cellular factors like iASPP (MIM 607463). p53 Arg72 shows weaker binding to iASPP and binds more efficiently to apoptotic promoters, such as Bax (MIM 600040).27 The ASPP (MIM 602143, MIM 606455) proteins alter p53's promoter selectivity for apoptotic genes, so it is conceivable that this difference in affinity may be extended to other p53-responsive promoters. Further investigation is needed to determine whether other cellular factors are involved.
The finding that the allelic frequency of p53 codon 72 associates with winter temperature, whereas MDM2 SNP309 associates with exposure to UV, suggests that the two polymorphisms are not coselected for, and this is entirely in agreement with the biological effect they impart on the p53 pathway. The results also suggest that both alleles are functional and under selective pressure from environmental exposure. These observations are similar to those reported in a recent study of different European populations,10 where it was noted that the haplotype that harbors the functional G allele of MDM2 SNP309 deviates significantly from the standard assumptions of selective neutrality. Thus, we propose a model where the activity of p53 is critically regulated by different genetic polymorphisms in the p53 pathway, under different environmental pressures (Figures 4A and 4B).
Figure 4.
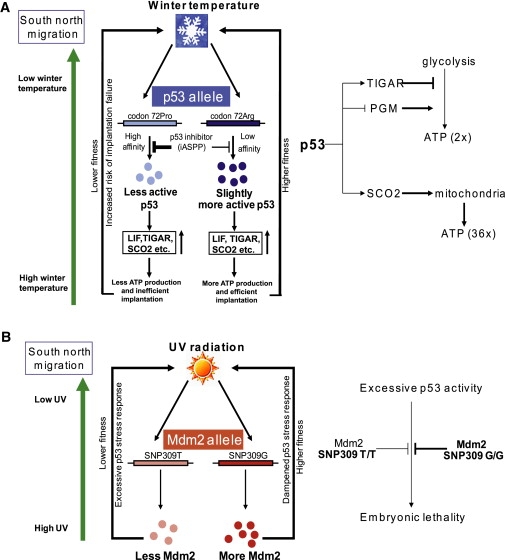
The Proposed Mechanism of Selection by Environmental Stresses on the Polymorphisms of p53 Codon 72 and MDM2 SNP309
The polymorphisms p53 codon 72 and MDM2 SNP309 are under selective pressure from the environmental stresses of winter temperature and UV radiation, respectively. The polymorphisms have an important impact upon the p53 pathway and help maintain efficient regulation under stressful conditions.
(A) In response to prolonged, cold temperatures, the p53 Arg72 allele is selected. This produces a more active form of p53, which increases the activation of genes such as LIF, SCO2, and TIGAR. p53 Arg72 may, therefore, be more efficient in regulating processes such as metabolism and embryo implantation, which would result in higher fitness levels.
(B) In contrast, the MDM2 SNP309 polymorphism is selected in areas with low UV radiation exposure. Migration of the Chinese populations northwards to areas with lower UV exposure would require less p53 in order to prevent any adverse effects of p53 hyperactivity, such as embryonic death. The MDM2 SNP309 G/G allele, which produces higher levels of Mdm2, is selected to counteract the high p53 levels, thereby conferring a selective advantage to these populations. This would prevent an overactive p53 pathway, while still enabling p53 to function as a tumor suppressor.
According to archaeological and genetic studies, the ancestral eastern Asian population of African origin arrived in eastern Asia about 60,000 years ago,28 and the major expansion of the Han Chinese populations occurred only in the past several thousand years.29 The populations, therefore, migrated from areas of high to low UV intensity, which would place less demand on the p53 pathway. This suggests that the G allele may have arisen more recently in evolution, in agreement with the recent findings of an investigation of Mdm2 polymorphisms in European, African-American, and Ashkenazi Jewish ethnic groups.10 The subsequent selection of this allele would ensure that p53 levels were kept low in those populations where UV exposure would pose no risk to the health and survival and might also provide a selective advantage to these populations when adapting to new environments, because high p53 activity is associated with death during embryonic development.30,31 The signature of positive selection in the eastern Asian populations, especially in Han Chinese (relatively homogeneous populations without deep genetic structure; Figure S7), indicates that selection on p53 codon 72 and MDM2 SNP309 has been strong, leading to the rapid formation of an allele frequency gradient across different latitudes. Another well-known case is the effect of polymorphisms of hemoglobin genes conferring resistance to malaria infection.32
Finally, a well-regulated p53 pathway has been shown to be crucial in many organisms, not only for tumor suppression but also for proper embryonic development and inflammatory responses.33,34 It is, therefore, not surprising that selective pressure from environmental stresses on functional p53 SNPs is observed in this report. It will be interesting to determine the implications of these findings in the epidemiology of cancer and other diseases.
Acknowledgments
We are thankful to people who provided their DNA samples, to Maureen Murphy in the Fox Chase Cancer Center, Philadelphia, PA, who provided the Soas2-ts cells for this study, and to Hua Chen from Harvard Medical School who helped in analyzing the genetic structure of the Han Chinese populations. We are also grateful to F. Murray-Zmijewski, G. Bond, and C. Beveridge for critical reading of the manuscript. This project was supported by the National 973 project of China (2006CB701506, 2007CB815705), the CAS/SAFEA International Partnership Program for Creative Research Team, Chinese Academy of Sciences (KSCX1-YW-R-34), the National Natural Science Foundation of China (30700445, 30630013, and 30525028), the Natural Science Foundation of Yunnan Province of China, and the Ludwig Institute for Cancer Research.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
HapMap, http://www.hapmap.org/
National Meteorological Information Centre of China, http://cdc.cma.gov.cn/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
SNPs Database Resources, http://www.ncbi.nlm.nih.gov/SNP/
Accession Numbers
The identification numbers of the three newly described SNPs are ss120037575, ss120037576, and ss120037577.
References
- 1.Lu X. p53: a heavily dictated dictator of life and death. Curr. Opin. Genet. Dev. 2005;15:27–33. doi: 10.1016/j.gde.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Jin S., Levine A.J. The p53 functional circuit. J. Cell Sci. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- 3.Vousden K.H., Lu X. Live or let die: the cell's response to p53. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 4.Bond G.L., Hu W., Levine A.J. MDM2 is a central node in the p53 pathway: 12 years and counting. Curr. Cancer Drug Targets. 2005;5:3–8. doi: 10.2174/1568009053332627. [DOI] [PubMed] [Google Scholar]
- 5.Michael D., Oren M. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 6.Lozano G. The oncogenic roles of p53 mutants in mouse models. Curr. Opin. Genet. Dev. 2007;17:66–70. doi: 10.1016/j.gde.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Bond G.L., Levine A.J. A single nucleotide polymorphism in the p53 pathway interacts with gender, environmental stresses and tumor genetics to influence cancer in humans. Oncogene. 2007;26:1317–1323. doi: 10.1038/sj.onc.1210199. [DOI] [PubMed] [Google Scholar]
- 8.Murphy M.E. Polymorphic variants in the p53 pathway. Cell Death Differ. 2006;13:916–920. doi: 10.1038/sj.cdd.4401907. [DOI] [PubMed] [Google Scholar]
- 9.Bond G.L., Hu W., Bond E.E., Robins H., Lutzker S.G., Arva N.C., Bargonetti J., Bartel F., Taubert H., Wuerl P. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 10.Atwal G.S., Bond G.L., Metsuyanim S., Papa M., Friedman E., Distelman-Menachem T., Ben Asher E., Lancet D., Ross D.A., Sninsky J. Haplotype structure and selection of the MDM2 oncogene in humans. Proc. Natl. Acad. Sci. USA. 2007;104:4524–4529. doi: 10.1073/pnas.0610998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckman G., Birgander R., Sjalander A., Saha N., Holmberg P.A., Kivela A., Beckman L. Is p53 polymorphism maintained by natural selection? Hum. Hered. 1994;44:266–270. doi: 10.1159/000154228. [DOI] [PubMed] [Google Scholar]
- 12.Sjalander A., Birgander R., Saha N., Beckman L., Beckman G. p53 polymorphisms and haplotypes show distinct differences between major ethnic groups. Hum. Hered. 1996;46:41–48. doi: 10.1159/000154324. [DOI] [PubMed] [Google Scholar]
- 13.Ma H., Hu Z., Zhai X., Wang S., Wang X., Qin J., Jin G., Liu J., Wang X., Wei Q. Polymorphisms in the MDM2 promoter and risk of breast cancer: a case-control analysis in a Chinese population. Cancer Lett. 2006;240:261–267. doi: 10.1016/j.canlet.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Q.L., Yu G.R., Cai F., Liu X.A., Li Z.Q., Su W., Hui L. Spatialization research on ultraviolet radiation in China. Resources Science. 2005;27:108–113. [Google Scholar]
- 15.Chu J.Y., Huang W., Kuang S.Q., Wang J.M., Xu J.J., Chu Z.T., Yang Z.Q., Lin K.Q., Li P., Wu M. Genetic relationship of populations in China. Proc. Natl. Acad. Sci. USA. 1998;95:11763–11768. doi: 10.1073/pnas.95.20.11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nitta M., Okamura H., Aizawa S., Yamaizumi M. Heat shock induces transient p53-dependent cell cycle arrest at G1/S. Oncogene. 1997;15:561–568. doi: 10.1038/sj.onc.1201210. [DOI] [PubMed] [Google Scholar]
- 17.Ohnishi T., Wang X., Ohnishi K., Takahashi A. p53-dependent induction of WAF1 by cold shock in human glioblastoma cells. Oncogene. 1998;16:1507–1511. doi: 10.1038/sj.onc.1201663. [DOI] [PubMed] [Google Scholar]
- 18.Fu Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kay C., Jeyendran R.S., Coulam C.B. p53 tumour suppressor gene polymorphism is associated with recurrent implantation failure. Reprod. Biomed. Online. 2006;13:492–496. doi: 10.1016/s1472-6483(10)60635-9. [DOI] [PubMed] [Google Scholar]
- 20.Hu W., Feng Z., Teresky A.K., Levine A.J. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–724. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- 21.Gwazdauskas F.C. Effects of climate on reproduction in cattle. J. Dairy Sci. 1985;68:1568–1578. doi: 10.3168/jds.S0022-0302(85)80995-4. [DOI] [PubMed] [Google Scholar]
- 22.Jones R.G., Plas D.R., Kubek S., Buzzai M., Mu J., Xu Y., Birnbaum M.J., Thompson C.B. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N., Nakano K., Bartrons R., Gottlieb E., Vousden K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 24.Matoba S., Kang J.G., Patino W.D., Wragg A., Boehm M., Gavrilova O., Hurley P.J., Bunz F., Hwang P.M. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 25.Dumont P., Leu J.I., Della Pietra A.C., 3rd, George D.L., Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 26.Thomas M., Kalita A., Labrecque S., Pim D., Banks L., Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol. Cell. Biol. 1999;19:1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergamaschi D., Samuels Y., Sullivan A., Zvelebil M., Breyssens H., Bisso A., Del Sal G., Syed N., Smith P., Gasco M. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat. Genet. 2006;38:1133–1141. doi: 10.1038/ng1879. [DOI] [PubMed] [Google Scholar]
- 28.Su B., Xiao J., Underhill P., Deka R., Zhang W., Akey J., Huang W., Shen D., Lu D., Luo J. Y-Chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am. J. Hum. Genet. 1999;65:1718–1724. doi: 10.1086/302680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen B., Li H., Lu D., Song X., Zhang F., He Y., Li F., Gao Y., Mao X., Zhang L. Genetic evidence supports demic diffusion of Han culture. Nature. 2004;431:302–305. doi: 10.1038/nature02878. [DOI] [PubMed] [Google Scholar]
- 30.Jones S.N., Roe A.E., Donehower L.A., Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 31.Montes de Oca Luna R., Wagner D.S., Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 32.Kwiatkowski D.P. How malaria has affected the human genome and what human genetics can teach us about malaria. Am. J. Hum. Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi J., Donehower L.A. p53 in embryonic development: maintaining a fine balance. Cell. Mol. Life Sci. 1999;55:38–47. doi: 10.1007/s000180050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozano G., Liu G. Mouse models dissect the role of p53 in cancer and development. Semin. Cancer Biol. 1998;8:337–344. doi: 10.1006/scbi.1998.0096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


