Abstract
To identify and validate genes associated with bone mineral density (BMD), which is a prominent osteoporosis risk factor, we tested 379,319 SNPs in 1000 unrelated white U.S. subjects for associations with BMD. For replication, we genotyped the most significant SNPs in 593 white U.S. families (1972 subjects), a Chinese hip fracture (HF) sample (350 cases, 350 controls), a Chinese BMD sample (2955 subjects), and a Tobago cohort of African ancestry (908 males). Publicly available Framingham genome-wide association study (GWAS) data (2953 whites) were also used for in silico replication. The GWAS detected two BMD candidate genes, ADAMTS18 (ADAM metallopeptidase with thrombospondin type 1 motif, 18) and TGFBR3 (transforming growth factor, beta receptor III). Replication studies verified the significant findings by GWAS. We also detected significant associations with hip fracture for ADAMTS18 SNPs in the Chinese HF sample. Meta-analyses supported the significant associations of ADAMTS18 and TGFBR3 with BMD (p values: 2.56 × 10−5 to 2.13 × 10−8; total sample size: n = 5925 to 9828). Electrophoretic mobility shift assay suggested that the minor allele of one significant ADAMTS18 SNP might promote binding of the TEL2 factor, which may repress ADAMTS18 expression. The data from NCBI GEO expression profiles also showed that ADAMTS18 and TGFBR3 genes were differentially expressed in subjects with normal skeletal fracture versus subjects with nonunion skeletal fracture. Overall, the evidence supports that ADAMTS18 and TGFBR3 might underlie BMD determination in the major human ethnic groups.
Main Text
Osteoporosis (MIM 166710) is the most common metabolic skeletal disease; it is estimated that over 200 million people worldwide have osteoporosis.1 It is mainly characterized by low bone mass and microarchitectural deterioration of bone tissue, with the consequent increase in fragility and susceptibility to fractures.2 Bone mineral density (BMD) has a high heritability, ∼70%, and it is an important measurable risk factor for osteoporotic fractures, because these fractures can develop with even mild stress and trauma when BMD has decreased to the threshold point.3 Consequently, BMD is the predominant surrogate phenotype used in studying osteoporosis.
So far, several genes for osteoporosis have been established, mostly through the large-scale meta-analyses launched by the GENOMOS consortium. The examples include the associations of Estrogen Receptor-α (ESR1 [MIM 133430]) PvuII and XbaI SNPs with fracture risk, the association between the Cdx2 polymorphism of Vitamin D Receptor (VDR [MIM 601769]) and vertebral fracture risk, the association between the Sp1 polymorphism of Collagen Type I α-1 (COL1A1 [MIM 120150]) and BMD, and the associations between Low Density Lipoprotein Receptor-Related Protein 5 (LRP5 [MIM 603506]) SNPs and BMD.4–8 However, the majority of the genetic factors that influence BMD variation remain unknown.9,10
The goal of this study was to identify, by use of a genome-wide association study (GWAS) and replication approaches, genes influencing human BMD variation at the hip and spine, the clinically most important skeletal sites. The clinical characteristics of participants in five independent cohorts—the white U.S. GWAS sample (n = 1000), the white U.S. family sample (n = 1972), the Chinese hip fracture (HF) sample (n = 700), the Chinese BMD sample (n = 2995), and the Tobago cohort of African origin (n = 908 men)—are described in Tables 1–5. Except for the white U.S. family sample, all samples were population-based. The publicly shared Framingham 550K GWAS data from the family-based Framingham Osteoporosis Study (n = 2953 white subjects) were also analyzed for the replication SNPs.
Table 1.
Characteristics of White U.S. GWAS Sample
| Trait | Male |
Female |
||
|---|---|---|---|---|
| ≤50 years (n = 250) | >50 years (n = 251) | Premenopause (n = 249) | Postmenopause (n = 250) | |
| Age (years) | 33.44 (9.66) | 67.33 (6.74) | 33.97 (8.45) | 66.36 (5.67) |
| Height (cm) | 180.00 (6.78) | 175.67 (6.63) | 165.38 (6.13) | 162.22 (6.43) |
| Weight (kg) | 88.03 (15.35) | 90.04 (14.47) | 70.74 (16.51) | 71.71 (15.10) |
| Spine BMD (g/cm2) | 1.054 (0.123) | 1.085 (0.203) | 1.045 (0.118) | 0.944 (0.102) |
| Hip BMD (g/cm2) | 1.066 (0.146) | 1.010 (0.141) | 0.953 (0.123) | 0.861 (0.135) |
n = 1000, data are shown as mean (SD).
Table 2.
Characteristics of White U.S. Family Sample
| Trait | Sons (n = 246) | Daughters (n = 895) | Fathers (n = 318) | Mothers (n = 513) |
|---|---|---|---|---|
| Age (years) | 38.7 (11.0) | 39.1 (10.3) | 63.6 (9.9) | 62.3 (10.6) |
| Height (cm) | 179.1 (7.5) | 164.8 (6.1) | 176.3 (7.1) | 162.5 (6.3) |
| Weight (kg) | 90.1 (17.3) | 71.7 (16.0) | 90.7 (16.1) | 73.14 (14.9) |
| Spine BMD (g/cm2) | 1.06 (0.14) | 1.05 (0.13) | 1.06 (0.14) | 0.97 (0.16) |
| Hip BMD (g/cm2) | 1.06 (0.13) | 0.97 (0.13) | 1.01 (0.17) | 0.88 (0.14) |
n = 1972, data are shown as mean (SD).
Table 3.
Characteristics of Chinese HF Sample
| Case | Control | |
|---|---|---|
| Number | 350 | 350 |
| Sex ratio (M/F) | 124/226 | 173/177 |
| Age (years) | 69.35 (7.41) | 69.54 (6.09) |
| Weight (kg) | 59.15 (12.05) | 59.61 (10.84) |
| Height (cm) | 162.84 (8.31) | 159.41 (9.20) |
n = 700, data are shown as mean (SD).
Table 4.
Characteristics of Chinese BMD Sample
| Trait | Male |
Female |
||
|---|---|---|---|---|
| ≤50 years (n = 1298) | >50 years (n = 139) | Premenopause (n = 1149) | Postmenopause (n = 369) | |
| Age (years) | 26.78 (4.53) | 65.59 (8.59) | 27.38 (6.30) | 60.50 (8.02) |
| Height (cm) | 169.92 (5.57) | 166.95 (5.61) | 158.71 (5.11) | 156.26 (5.48) |
| Weight (kg) | 63.37 (9.00) | 70.22 (9.61) | 51.40 (6.71) | 59.70 (8.94) |
| Spine BMD (g/cm2) | 0.98 (0.11) | 0.94 (0.15) | 0.94 (0.10) | 0.82 (0.14) |
| Hip BMD (g/cm2) | 0.98 (0.13) | 0.91 (0.13) | 0.88 (0.10) | 0.81 (0.13) |
n = 2995, data are shown as mean (SD).
Table 5.
Characteristics of Tobago Cohort of African Origin
| Age (yr) | 56.2 (9.6) |
| Height (cm) | 175.5 (6.9) |
| Weight (kg) | 84.47 (14.79) |
| Total body BMD (g/cm2) | 1.27 (0.11) |
| Spine BMD (g/cm2) | 1.12 (0.16) |
| Hip BMD (g/cm2) | 1.15 (0.14) |
| Femoral neck BMD (g/cm2) | 1.01 (0.15) |
| Trochanter BMD (g/cm2) | 0.89 (0.13) |
| Intertrochanteric BMD (g/cm2) | 1.34 (0.16) |
| Ward's Triangle BMD (g/cm2) | 0.85 (0.19) |
n = 908, all of which are men. Data are shown as mean (SD).
During the discovery phase, we carried out a GWAS using the Affymetrix Gene Chip Human Mapping 500K Array Set. We successfully genotyped a total of 379,319 single-nucleotide polymorphisms (SNPs) in the white GWAS sample (1000 subjects), recruited from Midwestern U.S. for BMD analyses. The subject-recruitment procedures, standard examinations of BMD and related phenotypes, genotyping with Affymetrix 500K Array, genotyping quality control, and SNP-exclusion procedures have been detailed elsewhere.11–13 In the GWAS sample, the hip and spine BMD data were adjusted by significant covariates, including age, sex, height, and weight, and analyzed with allelic and haplotypic association tests (haplotype trend regression [HTR]14) implemented in HelixTree 5.3.1. The association analyses were conducted in (1) the total sample, (2) the male and female subgroups, (3) the premenopausal white females, and (4) the postmenopausal white females, each group analyzed separately.
Given the LD among SNPs across the whole genome, the Bonferroni correction could be considered overly conservative; therefore, we adopted the pointwise GWAS significance threshold proposed elsewhere,15 ∼4.2 × 10−7. The genewise approach used in calculating this threshold took into account recent estimates of the total number of genes in the human genome. Because 20 GWA tests (men/women/total samples; hip/spine BMDs; premenopausal/postmenopausal female samples; single-SNP testing/sliding-window testing) were conducted, the pointwise GWAS significance threshold that we used here was 2.1 × 10−8 (Bonferroni adjustment of 4.2 × 10−7).
EIGENSTRAT16 software was used for guarding against spurious associations due to potential population stratification. The LD patterns of the implicated genes were analyzed and plotted with the use of the Haploview program17 with the HapMap data from the International HapMap project. The FASTSNP program was used for predicting the function of the SNPs of interest.18
We used five independent samples for replication. The first was a white U.S. family sample comprising 1972 white individuals, from 593 nuclear families, who were recruited and phenotyped in the same way as were those in the white U.S. GWAS sample.11,12 Genotyping was performed by KBioscience (Herts, UK) via the technology of competitive allele-specific PCR (KASPar), which is detailed at the company's website. Five SNPs of interest (Table 6) were successfully genotyped. The replication rate (duplicate concordance rate) was 99.7% for the genotyping in the white family sample, and the average call rate was 97.8%.
Table 6.
Summary of Association Results in GWAS and Replication Studies
| SNP | US White GWAS Sample p Value | US White Family Sample p Value | Framingham Sample (White) p Value | Chinese HF Sample p Value | Chinese BMD Sample p Value | Tobago BMD Sample (African) p Value |
|---|---|---|---|---|---|---|
| ADAMTS18rs16945612 | Hip BMD/female sample:5.75 × 10−7 (allele)4.61 × 10−4 (haplotype) | Hip BMD:1.6 × 10−2Spine BMD:3.5 × 10−3 | Hip BMD: 6 × 10−3 | Hip fracture: 1.9 × 10−2 | Hip BMD:9 × 10−3 Spine BMD: 1 × 10−2 | Hip BMD: 1.7 × 10−1 Trochanter BMD: 3.2 × 10−2 |
| ADAMTS18rs11859065 | Hip BMD/female sample:1.28 × 10−6 (allele)4.91 × 10−4 (haplotype) | Hip BMD:1.65 × 10−2 Spine BMD: 3.5 × 10−3 | Hip BMD: 5.5 × 10−3 | Hip fracture: 1.9 × 10−2 | Hip BMD:9 × 10−3 Spine BMD: 1 × 10−2 | NAa |
| ADAMTS18rs11864477 | Hip BMD/female sample:4.17 × 10−7 (allele)2.90 × 10−5 (haplotype) | Hip BMD:1.65 × 10−2 Spine BMD: 3.5 × 10−3 | Hip BMD: 6.5 × 10−3 | Hip fracture: 1.9 × 10−2 | Hip BMD:9 × 10−3 Spine BMD: 1 × 10−2 | NAa |
| ADAMTS18rs11860781 | Hip BMD/female sample:2.03 × 10−6 (allele)5.78 × 10−4 (haplotype) | Hip BMD: 1 × 10−2 | Hip BMD: 1.0 × 10−2 | Hip fracture: 1.7 × 10−1 | Hip BMD: 1 × 10−1 | NAa |
| TGFBR3rs17131547 | Spine BMD/total sample:3.91 × 10−4 (allele)3.47 × 10−8 (haplotype) | Spine BMD: 1 × 10−2 | Spine BMD: 3 × 10−2 | NAb | NAb | Spine BMD: 6.7 × 10−3 Total body BMD: 3.1 × 10−2 |
| Hip BMD: 1.1 × 10−3 | ||||||
| Femoral Neck BMD: 5.4 × 10−3 | ||||||
| Intertrochanter BMD: 4.93 × 10−4 | ||||||
| Trochanter BMD: 1.3 × 10−2 | ||||||
| Ward's triangle BMD: 2.82 × 10−4 |
rs11859065, rs11864477 and rs11860781 was not genotyped in Tobago sample considering the redundancy in genotyping since they are all in strong LD with rs16945612 in the HapMap African sample.
rs17131547 was monomorphic in Chinese.
The second replication cohort was the Framingham sample from the Framingham Osteoporosis Study,19 which has been detailed before.20,21 Genotype and phenotype data were downloaded from the dbGaP database. Data download and usage was authorized by the SHARe data-access committee. We have the data on 2953 phenotyped white subjects, 448 from the original cohort (160 men and 288 women) and 2505 from the offspring cohort (1114 men and 1391 women). The original-cohort participants had BMD measurements calculated via a dual X-ray absorptiometry machine (Lunar DPX-L), performed at the hip and spine during exam 24. The offspring-cohort participants were scanned with the same machine at exam 6 or 7. The Framingham sample was genotyped with the use of approximately 550,000 SNPs (Affymetrix 500K mapping array plus Affymetrix 50K supplemental array). The genotype data for the five SNPs of interest (Table 6) were analyzed for BMD associations.
The third replication cohort was a Chinese HF sample, recruited from Xi'an City and neighboring areas in China. The sample consisted of 350 unrelated patients with osteoporotic HF and 350 unrelated controls without HF. The subject recruitment and experimentation procedures (including genotyping) have been described by Yang et al.,13 who called the same cohort a “Chinese GWA sample.” The genotype data for the four SNPs of interest (Table 6) were analyzed for HF associations.
The fourth replication sample, the Chinese BMD sample, comprised 2955 Chinese adults living in Changsha City of China. The subject-recruitment criteria were the same as those adopted for the white U.S. samples. BMD was measured with the same model Hologic 4500W machines (Hologic, Bedford, MA, USA) under the same strict protocols applied for the white U.S. samples. The coefficient of variation (CV) values of the dual-energy X-ray absorptiometry (DXA) measurements for hip and spine BMDs were approximately 1.01% and 1.33%, respectively. Genotyping was performed with the use of a primer-extension method, with MALDI-TOF mass spectrometry for multiplexed genotyping of SNPs on a MassARRAY system, performed as suggested by the manufacturer (Sequenom, San Diego, CA) and well described by Braun et al.22 Four SNPs of interest (Table 6) were successfully genotyped. The replication rate is 99.2%, and the average call rate is 97.2%.
The fifth replication sample was the Tobago BMD sample, comprising 908 men of West African ancestry who were randomly selected from a large population-based study of BMD among 2501 men aged 40 and older on the Caribbean island of Tobago.23 This sample was of West African ancestry with low non-African admixture (6% non-African).24 The detailed recruitment scheme and phenotyping procedures have been described elsewhere.23 The two SNPs of interest (Table 6) were successfully genotyped with the fluorogenic 5′-nuclease TaqMan allelic-discrimination assay system (Applied Biosystems, Foster City, CA). The assays were performed under standard conditions on a 7900HT real-time PCR instrument. All genotype calls were determined by two independent investigators, and only concordant calls were used. The genotyping consensus rate, based on approximately 8% blind replicate genotypes, was 99.7%. The average completeness of genotyping for the two markers was 98.3%.
In all studies, informed consent was obtained from participants and studies were approved by the local institutional review boards or ethical committees.
Statistical analyses for replication samples involved the following: (1) In the white U.S. family sample and the Framingham Osteoporosis Study sample, we conducted the family-based association test (FBAT)25 for the SNPs of interest for their association with the BMD residuals adjusted by significant covariates, including age, sex, height, and weight. (2) In the Chinese HF sample, the genotype distributions of ADAMTS18 SNPs between fracture and nonfracture groups were analyzed with logistic regression models controlling for age, sex, height, and weight as covariates. (3) In the Chinese BMD sample, the statistical procedures were the same as those used for the white U.S. GWAS sample. (4) In the Tobago cohort comprising only men, SNPs were tested for association with BMD via linear regression as a test for an additive association between the number of copies of the minor allele and BMD. The models were adjusted for age, weight, and height. Analyses were performed with SAS version 9.1 (SAS Institute, Cary, NC, USA). The magnitude and direction of SNP effects were estimated by a linear-regression model for random samples and by a quantitative transmission-disequilibrium test (QTDT) for family samples.
Finally, the meta-analyses for the significant SNPs from (1) all of the three white BMD samples (white U.S. GWAS, white U.S. family, and Framingham samples) and (2) all of the five BMD samples from different ethnic origins (GWAS, white U.S. family, Framingham, Chinese BMD, and Tobago samples) were conducted, respectively, with the weighted z score-based meta-analysis approach26,27 (weighted by the square root of the sample size of each combining sample) used for quantification of the overall evidence for association with BMD variation.
The initial GWAS results were as follows: For the single-SNP allelic analyses, we did not find any significant associations with hip/spine BMD in either the total GWAS sample or the male subsample at the genome-wide threshold of 2.1 × 10−8. In females, although no GWAS-level significant association with BMD was found, the SNP rs11864477 in the ADAMTS18 gene (ADAM metallopeptidase with thrombospondin type 1 motif, 18 [MIM 607512]) associated with hip BMD (p = 4.17 × 10−7, Figure 1A, Table 6) at the closest level to the genome-wide significance threshold. In the sliding-window haplotype analyses, the most significant haplotype for spine BMD almost hit the GWAS-significance cutoff. This haplotype was located in the TGFBR3 gene (transforming growth factor, beta receptor III [MIM 600742]) (p = 3.47 × 10−8; Figure 1B, Table 6).
Figure 1.
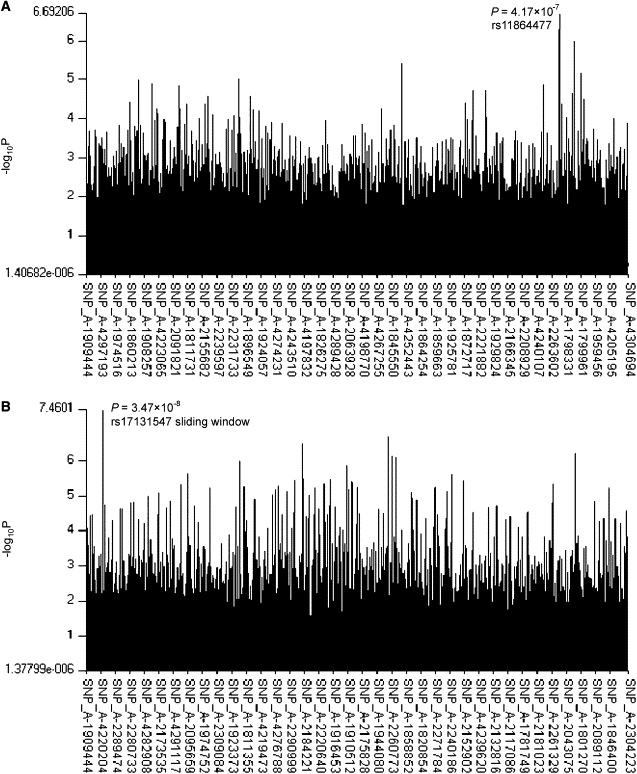
Genome-wide Association Results for BMD in the White U.S. GWAS Sample
(A) Genome-wide association results for hip BMD in the female sample by the single-SNP approach.
(B) Genome-wide association results for spine BMD in the total sample by the 5-SNP-size sliding-window approach.
We then analyzed the above two implicated genes in more detail. In the female sample, as shown in Figure 2 and Table 6, three other ADAMTS18 gene SNPs, which are near and in significantly strong LD with rs11864477 (pairwise r2 > 0.90), were associated with hip BMD (p values = 2.03 × 10−6, 5.75 × 10−7, and 1.28 × 10−6 for rs11860781, rs16945612, and rs11859065, respectively). These four SNPs were suggestive for hip BMD in the total sample (p values = 8.31 × 10−4, 3.84 × 10−3, 1.98 × 10−3, and 1.66 × 10−3 for rs11864477, rs11860781, rs16945612, and rs11859065, respectively). Haplotype block 12, containing rs16945612, rs11864477, and rs11859065, was suggestively significant for hip BMD in the female subgroup (p = 9.95 × 10−6) and the total sample (p = 1.9 × 10−2) (Figure 2). In the male sample, we detected a significant haplotype window that contained rs11860781 (highly correlated with rs16945612, rs11864477, and rs11859065 [r2 > 0.8]) and was suggestively associated with hip BMD (p = 6 × 10−3). Analyses stratified by menopausal status showed consistent results. All of these four highly correlated SNPs were suggestively associated with hip BMD in the postmenopausal white women (p values = 2 × 10−3 for all four SNPs) and in the premenopausal white women (p = 4 × 10−3 for rs11864477; p = 6 × 10−3 for rs16945612; p = 1 × 10−2 for rs11860781 and rs11859065). These data support the proposal that the ADAMTS18 gene contributes to the variation of hip BMD.
Figure 2.
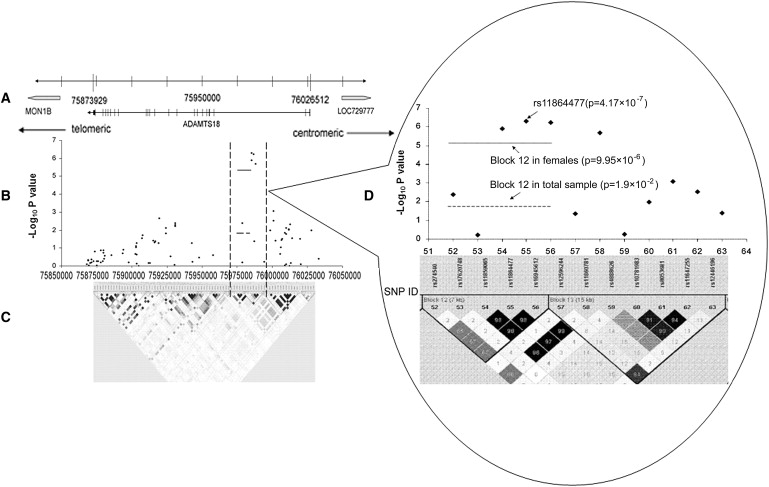
Association Signals in the ADAMTS18 Gene in 16q23
(A) Genomic locations of chromosome 16q23 genes between 75873929 and 76026512 bp.
(B) The negative log10 p values from the female sample are plotted for genotyped markers in the ADAMTS18 region.
(C) Pairwise r2 plot for the genotype data of ADAMTS18 in this study. The intensity of shading is proportional to r2. The x axis represents physical positions.
(D) Enlarged picture of the significant region in ADAMTS18. The numbers on the x axis are the SNP IDs corresponding to those in Table S1.
For the TGFBR3 gene, we found that rs17131547 was the key SNP for the most significant haplotype window (p = 3.47 × 10−8 for spine BMD association), composed of TGFBR3 SNPs—rs17131547, rs12403389, rs4658112, rs17131544, and rs2087299. Several other haplotype windows harboring rs17131547 were also suggestively significant for spine BMD (p values lie in [0.005, 0.05]). Single-SNP analyses showed that rs17131547 was suggestive for spine BMD in the total sample (p = 3.91 × 10−4; Figure 3 and Table 6), the male sample (p = 4.27 × 10−3), and the female sample (p = 3.8 × 10−2). Interestingly, no adjacent SNPs near rs17131547 were significant for spine BMD (Figure 3 and Table S2, available online). LD analyses showed that rs17131547 was an independent SNP with almost no LD with any of the other typed SNPs in the TGFBR3 gene (pairwise r2 < 0.04), suggesting the independent association of this SNP or a nearby untyped SNP with spine BMD. The detailed information and association results for both ADAMTS18 and TGFBR3 are summarized in Tables S1 and S2.
Figure 3.
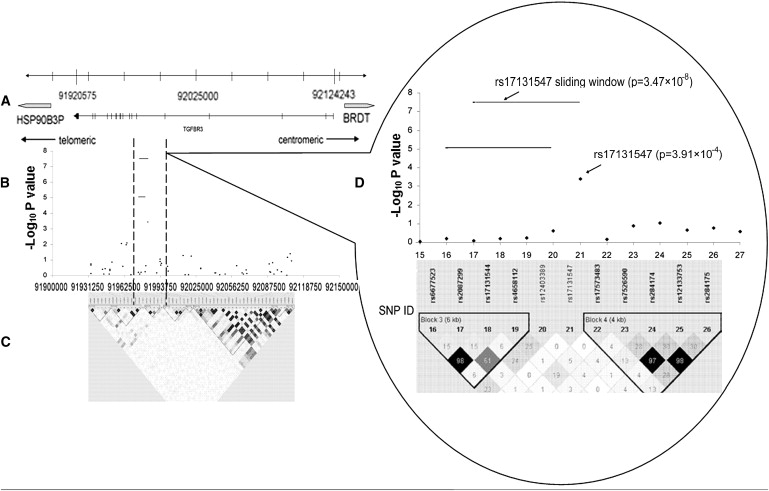
Association Signals in the TGFBR3 Gene on the Chromosome Region 1p22
(A) Genomic locations of chromosome 1p22 genes between 91900000 and 92150000 bp.
(B) The negative log10 p values from the total sample are plotted for genotyped markers in the TGFBR3 region.
(C) Pairwise r2 plot for the genotype data of TGFBR3 in this study. The intensity of shading is proportional to r2. The x axis represents physical positions.
(D) Enlarged picture of the significant region in TGFBR3. The numbers on the x axis are the SNP IDs corresponding to those in Table S2.
In the white U.S. GWAS sample, we compared raw BMD values between the groups carrying different alleles of the two most significant BMD-associated SNPs. As shown in Figure 4, subjects carrying the C allele of rs11864477 in ADAMTS18 had a significantly lower mean hip BMD value than those carrying the alternative T allele (3% lower; raw hip BMD values were 0.946 versus 0.975 g/cm2 for C versus T alleles: p = 0.006). Subjects carrying the A allele of rs17131547 in TGFBR3 had a significantly higher mean spine BMD value than those carrying the G allele (5% higher; raw spine BMD values were 1.084 versus 1.032 g/cm2 for A versus G alleles, p = 0.029). It was also estimated that in the initial GWAS sample, ADAMTS18 SNPs explained 3.77%–3.85% of hip BMD variation, whereas the TGFBR3 SNP explained 1.23% of spine BMD variation (Table 7). In all of the replication samples, the directions of SNP effects were the same as those in the white U.S. GWAS sample. The effect sizes of the SNPs of interest in all of the replication samples were smaller than those in the white GWAS sample, which could be explained by the weaker associations in the replication samples.
Figure 4.
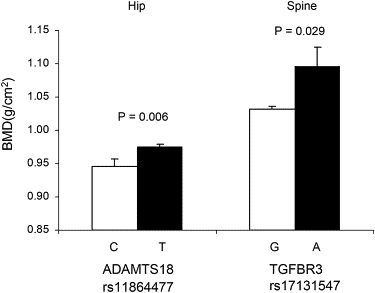
Average Raw BMD Values for Groups Stratified by Different Alleles at rs11864477 in ADAMTS18 and at rs17131547 in TGFBR3
Error bars denote standard error.
Table 7.
Association Results in the White GWAS Sample
| SNP | Effect Sizea | βb | SEc | Reference Allele | |
|---|---|---|---|---|---|
| Hip BMD/ADAMTS18 | |||||
| Female sample (GWAS) | rs16945612 | 0.03824 | 0.04428 | 0.01016 | T |
| rs11859065 | 0.03826 | 0.04429 | 0.01016 | G | |
| rs11864477 | 0.03773 | 0.04353 | 0.01006 | T | |
| rs11860781 | 0.03848 | 0.04429 | 0.01013 | A | |
| Combined white family sampled | rs16945612 | 0.01 | 0.058 | - | T |
| rs11860781 | 0.01 | 0.048 | - | A | |
| Chinese BMD sample | rs16945612 | 0.0025 | 0.06898 | 0.03108 | T |
| rs11860781 | 0.00045 | 0.0201 | 0.0209 | A | |
| Tobago BMD sample | rs16945612 | 0.00332 | 0.06131 | 0.0374 | T |
| Spine BMD/TGFBR3 | |||||
| Total sample (GWAS) | rs17131547 | 0.01231 | −0.4974 | 0.1442 | G |
| Combined white family sampled | rs17131547 | 0.012 | −0.058 | - | G |
| Tobago BMD sample | rs17131547 | 0.0025 | −0.0673 | 0.044 | G |
Magnitude and Direction of SNP effects obtained by linear regression analyses (for random samples) and QTDT (for family samples).
Effect size measured by r2.
β: Regression coefficient.
SE denotes standard error.
The effect size and direction of an interested SNP were estimated in the combined white family sample consisting of i) white U.S. family sample and ii) Framingham sample; SE of β cannot be estimated in the family samples due to the limitation of QTDT; for simplicity, the results for rs11859065 and rs11864477 were not shown in the replication samples because they were the same with the results of rs16945612 due to their high correlations.
Population stratification was unlikely to be a key confounding factor influencing BMD associations in this GWAS. This is mainly because the results from EIGENSTRAT analyses, controlling for potential population admixture or stratification, validated the significant identified BMD associations (for example, at locus rs11864477, p = 4.97 × 10−7 and 4.17 × 10−7 with and without EIGENSTRAT adjustment, respectively; and = 1.011 for BMD phenotypes, indicating no significant population stratification).
Quantile-quantile plots (Figure 5) revealed the presence of a substantial number of SNPs associated with hip and spine BMD. These results indicated that a substantial fraction of the most strongly associated SNPs could have true associations with BMD.
Figure 5.
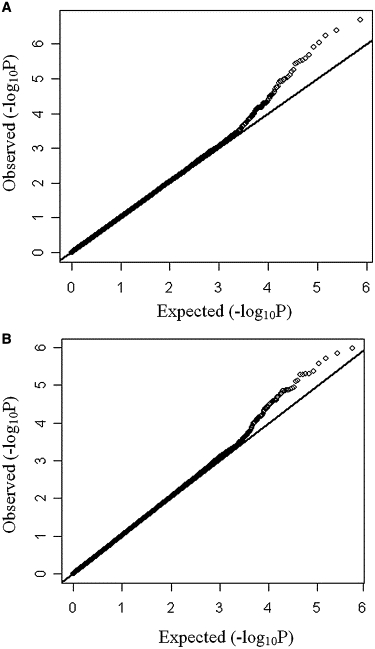
Quantile-Quantile Plots for Hip BMD and Spine BMD Associations
(A) Quantile-quantile plots for hip BMD associations in the GWAS female sample.
(B) Quantile-quantile plots for spine BMD associations in the GWAS total sample.
Axes represent the following information: y axis, observed −log10(p) values; x axis, p values expected under the null distribution for the GWAS SNPs.
Replication studies verified the significant findings of the initial GWAS. In the white U.S. family sample, the same SNPs were consistently associated with BMD variation (p values = 1.65 × 10−2 and 3.5 × 10−3 for hip and spine BMD associations, respectively, at three highly correlated ADAMTS18 SNP loci—rs16945612, rs11859065, and rs11864477; p = 1 × 10−2 for hip BMD association at rs11860781 in ADAMTS18; and p = 1 × 10−2 for spine BMD association at rs17131547 in TGFBR3; Table 6). In silico replication, again using Framingham data, showed that the four highly correlated ADAMTS18 SNPs in whites—rs16945612, rs11859065, rs11864477, and rs11860781—were associated with hip BMD variation (p values = 6 × 10−3, 5.5 × 10−3, 6.5 × 10−3, 1.0 × 10−2, respectively; Table 6), whereas the TGFBR3 SNP, rs17131547, was associated with spine BMD variation (p = 3 × 10−2, Table 6).
In the Chinese HF sample, rs16945612, rs11859065 and rs11864477, three completely correlated ADAMTS18 SNPs (pairwise r2 = 1), were significantly associated with HF (p values = 1.9 × 10−2 for all; Table 6). In the Chinese BMD sample, significant associations with hip and spine BMD variations (p values = 9 × 10−3 and 1 × 10−2, respectively; Table 6) at the same three SNPs were also detected. The significance of ADAMTS18 and TGFBR3 to BMD phenotypes was further replicated in the Tobago sample of West African ancestry (ADAMTS18-rs16945612: p = 3.2 × 10−2 for trochanter BMD; TGFBR3-rs17131547: p = 6.7 × 10−3, 3.1 × 10−2, 1.1 × 10−3, 5.4 × 10−3, 4.9 × 10−4, 1.3 × 10−2, and 2.8 × 10−4 for BMD measured at lumbar spine, total body, total hip, femoral neck, intertrochanter, trochanter, and Ward's triangle, respectively; Table 6). The other three ADAMTS18 SNPs in significant strong LD with rs16945612 were not genotyped in the Tobago sample as a result of local budgetary limits.
The data from the CEU, CHB, and YRI HapMap samples were used to plot the LD blocks covering the four ADAMTS18 SNPs of interest (Figures S1–S3). The HapMap data corroborated that these four SNPs were highly correlated in the white and black populations, whereas in the Chinese populations, three of the four were highly correlated (rs11860781 was independent of the other three SNPs in Chinese populations).
Meta-analyses for (1) all of the white BMD samples and (2) all of the BMD samples from different ethnic origins supported the above significant associations (p values ranged from 1.19 × 10−6 to 2.13 × 10−8 for hip BMD associations at ADAMTS18 SNPs; p values = 2.56 × 10−5 and 1.49 × 10−6 for spine BMD association at rs17131547 of TGFBR3). The results of the meta-analyses are listed in Table 8.
Table 8.
Results of Meta-Analyses Using the Weighted z Score-Based Approach
| SNP | p Value of All White BMD Samples Combineda | p Value of All BMD Samples combinedb |
|---|---|---|
| ADAMTS18:rs16945612 | Hip BMD:2.84 × 10−7 | Hip BMD:2.13 × 10−8 |
| ADAMTS18:rs11859065 | Hip BMD:3.57 × 10−7 | Hip BMD:3.13 × 10−8 |
| ADAMTS18:rs11864477 | Hip BMD:2.75 × 10−7 | Hip BMD:2.48 × 10−8 |
| ADAMTS18:rs11860781 | Hip BMD:5.37 × 10−7 | Hip BMD:1.19 × 10−6 |
| TGFBR3:rs17131547 | Spine BMD:2.56 × 10−5 | Spine BMD:1.49 × 10−6 |
The z score is weighted by the square root of the sample size of each combining sample.
Including white U.S. GWAS, white U.S. family and Framingham samples (whites). N = 5925.
Including white U.S. GWAS, white U.S. family, Framingham sample, Chinese BMD and Tobago BMD samples for ADAMTS18 SNPs - rs16945612; including white U.S. GWAS, white U.S. family, Framingham sample, and Chinese BMD sample for the other three ADAMTS18 SNPs that were not genotyped in Tobago sample; while including white U.S. GWAS, white U.S. family, Framingham sample, and Tobago BMD sample for TGFBR3 - rs17131547, which is monomorphic in Chinese. N = 9,828 for ADAMTS18 SNPs - rs16945612; N = 8,920 for other ADAMTS18 SNPs; N = 6833 for the TGFBR3 SNP.
The bioinformatics analyses suggested that the allele change (T→C) at rs16945612 should generate one binding site of TEL2 (ETS Transcription Factor TEL2 [MIM 605255]). The enhanced TEL2 binding might repress ADAMTS18 expression, given that TEL2 has been identified as a transcriptional repressor.28 Particularly, TEL2 has also been shown to repress two genes (BMP-6 and RARa) that play important roles in regulating osteoblast differentiation and bone remodeling.28 Therefore, we conducted electrophoretic mobility shift assay (EMSA) to confirm the potential changes of TEL2 binding to ADAMTS18 caused by rs16945612. We made TEL2 protein preparations by transforming the pGEX-2T construct29 into Escherichia coli BL21 cells. Protein expression and extractions were performed according to manufacturer's instructions (Pharmacia, Peapack, NJ, USA). Protein concentration was measured by the BCA Protein Assay kit (Pierce, Rockford, IL, USA), with bovine serum albumin as a standard. The following double-stranded oligonucleotides were synthesized (Integrated DNA Technologies, Coralville, IA, USA) and used in EMSA: (1) the labeled wild-type TEL-2b binding-site probe, corresponding to ADAMTS18 intron3 sequences centering rs16945612 (underlined and bolded in the following sequences), prepared by annealing of the digoxigenin (DIG)-labeled oligonucleotide 5′-ACAACAACTTTATTTCTGGTCCAAG-3′ with the complementary sequence 5′-CTTGGACCAGAAATAAAGTTGTTGT-3′; (2) the mutant TEL-2b binding-site probe, prepared by annealing the DIG-labeled oligonucleotide 5′-ACAACAACTTTACTTCTGGTCCAAG-3′ with the complementary sequences 5′-CTTGGACCAGAAGTAAAGTTGTTGT-3′; and (3) the corresponding unlabeled mutant TEL2 binding-site probe. EMSA was conducted with the DIG Gel Shift kit (Roche Applied Science, Indianapolis, IN, USA). We incubated 0.8 ng of DIG-labeled probe with different amounts of nuclear extracts (0–5.7 μg) for 30 min at room temperature in a 10 μl reaction volume containing 2 μl binding buffer (5×), 1 μg poly [d(IA-T)], and 0.1 μg Poly L-lysine. For competition reactions, we used the above unlabeled mutant TEL2 binding-site probe and an unrelated oligonucleotide for competition at 125-fold molar excess of the labeled mutant TEL2 binding-site probe. For the supershift assay, we incubated 2 μg of antibodies specific for TEL2 (named α-TEL2, purchased from Abcam, CA, USA) or 2 μg of an irrelevant peptide (i.e., anti-goat IgG) with the protein-DNA complex. After incubation, the samples were separated by electrophoresis on a 6% nondenaturing polyacrylamide gel with 0.5× TBE buffer. DNA-protein complexes were electroblotted to nylon membrane (Invitrogen, Carlsbad, CA, USA) and visualized by exposure to Lumi-Film Chemiluminescent Detection Film (Roche).
Indeed, we found specific binding of TEL2 from E.coli BL21 protein extract to the mutant TEL2 binding site centering the rs16945612 minor allele “C” of ADAMTS18, confirmed by supershifting on addition of antibodies against TEL2 (Figure 6). We found no binding of the nuclear extract to the wild-type site containing the native allele “T” of rs16945612 (Figure 6). The results indicate that the T→C change at rs16945612 creates a new TEL2 binding site in the ADAMTS18 gene. In addition, the NCBI GEO expression profiles showed that the ADAMTS18 level is significantly lower in subjects with nonunion fractures (fracture that does not heal six months after injury) as compared to subjects with normal-healing fractures (Figure S4). Decreased in vivo ADAMTS18 expression might thus potentially contribute to the nonhealing of skeletal fractures. The functional evidence, together with the statistical evidence, lead us to hypothesize that allele “C” of rs16945612 might, through enhanced TEL2 binding, represses the expression of ADAMTS18 and subsequently influence the osteoporosis phenotypes.
Figure 6.
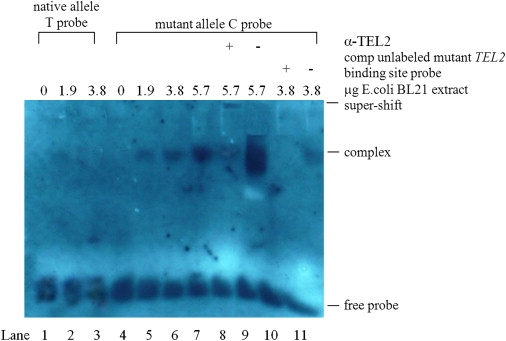
Electrophoretic Mobility Shift, Supershift, and Competition Assays with E.coli BL21 Nuclear Cell Extract and Allelic Variants of SNP rs16945612 in ADAMTS18
The 25 bp oligonucleotides containing both allelic variants of SNP rs16945612, representing native and mutated TEL-2b binding sites, were assayed with increasing (0–5.7 μg) amounts of nuclear extract of E.coli BL21 cells. Binding was not observed with oligonucleotide containing native allele T (lanes 1–3) but was present with oligonucleotide containing the mutant allele C (lanes 5–11). Binding to the mutant C allele resulted in a complex that was specifically competed by 125 × excess of unlabeled mutant TEL-2b binding-site probe containing the C allele (lane 10) but not by the same amount of unrelated oligonucleotide (lane 11). Rabbit polyclonal antibody against TEL2 (α-TEL2) revealed a much weakened band due to supershifting of the band (lane 8), whereas unrelated serum did not produce the supershift (lane 9).
TGFBR3 is the major mediator of TGF-β signaling pathways30,31 and also functions as a BMP cell-surface receptor.32 Particularly, TGFBR3 can modulate the biological function of BMP2 (bone morphogenetic protein 2 [MIM 112261]),32 which is a well-established key factor in bone biology and is significantly associated with BMD and other bone phenotypes.33 Moreover, TGFBR3 knockout mice (TBRIII−/−) demonstrated severe abnormal skeleton defects, as reported previously34 and detailed in the MGI database of The Jackson Laboratory.
Interestingly, the NCBI GEO expression profiles showed that TGFBR3 level is significantly lower in normal skeletal fracture subjects as compared to nonunion skeletal fracture subjects (Figure S5). This is opposite of the expression pattern for ADAMTS18 (Figure S4), suggesting different physiological roles for these two genes in the healing of bone fractures.
In summary, the present GWAS and multiple replication studies identified two genes, ADAMTS18 and TGFBR3, that were significantly associated with BMD variation in three major ethnic groups. The results of these studies direct attention to these two genes, which have not been well studied previously in the field of osteoporosis research. Additional molecular studies are required for defining the precise and detailed mechanisms by which these two genes contribute to BMD and osteoporosis risk.
Acknowledgments
Investigators of this work were partially supported by grants from the National Institutes of Health (R01 AR050496-01, R21 AG027110, R01 AG026564, and P50 AR055081). The study also benefited from grants from the National Science Foundation of China, the Huo Ying Dong Education Foundation, HuNan Province, Xi'an Jiaotong University, and the Ministry of Education of China. We also thank Gerard C. Grosveld for providing pGEX-2T expression constructs for EMSA analyses.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
dbGaP database, http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap
FASTSNP program, http://fastsnp.ibms.sinica.edu.tw
FBAT program, http://www.biostat.harvard.edu/∼fbat/default.html
GENOMOS Consortium, http://www.genomos.eu/
Haploview program, http://www.broad.mit.edu/mpg/haploview
The International HapMap project, http://www.hapmap.org
Jackson Laboratory MGI database, http://www.informatics.jax.org/
KBioscience genotyping protocol, http://www.kbioscience.co.uk
Mouse Genome Informatics, http://www.informatics.jax.org
NCBI GEO expression database, http://www.ncbi.nlm.nih.gov/geo
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
QTDT program, http://www.sph.umich.edu/csg/abecasis/QTDT
References
- 1.Reginster J.Y., Burlet N. Osteoporosis: a still increasing prevalence. Bone. 2006;38:S4–S9. doi: 10.1016/j.bone.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Ray N.F., Chan J.K., Thamer M., Melton L.J., III Medical expenditures for the treatment of osteoporotic fractures in the United States in 1995: report from the National Osteoporosis Foundation. J. Bone Miner. Res. 1997;12:24–35. doi: 10.1359/jbmr.1997.12.1.24. [DOI] [PubMed] [Google Scholar]
- 3.Cummings S.R., Black D. Bone mass measurements and risk of fracture in Caucasian women: a review of findings from prospective studies. Am. J. Med. 1995;98:24S–28S. doi: 10.1016/s0002-9343(05)80041-5. [DOI] [PubMed] [Google Scholar]
- 4.Macdonald H.M., McGuigan F.E., Stewart A., Black A.J., Fraser W.D., Ralston S., Reid D.M. Large-scale population-based study shows no evidence of association between common polymorphism of the VDR gene and BMD in British women. J. Bone Miner. Res. 2006;21:151–162. doi: 10.1359/JBMR.050906. [DOI] [PubMed] [Google Scholar]
- 5.Ralston S.H., Uitterlinden A.G., Brandi M.L., Balcells S., Langdahl B.L., Lips P., Lorenc R., Obermayer-Pietsch B., Scollen S., Bustamante M. Large-scale evidence for the effect of the COLIA1 Sp1 polymorphism on osteoporosis outcomes: the GENOMOS study. PLoS Med. 2006;3:e90. doi: 10.1371/journal.pmed.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ioannidis J.P., Ralston S.H., Bennett S.T., Brandi M.L., Grinberg D., Karassa F.B., Langdahl B., van Meurs J.B., Mosekilde L., Scollen S. Differential genetic effects of ESR1 gene polymorphisms on osteoporosis outcomes. JAMA. 2004;292:2105–2114. doi: 10.1001/jama.292.17.2105. [DOI] [PubMed] [Google Scholar]
- 7.van Meurs J.B., Trikalinos T.A., Ralston S.H., Balcells S., Brandi M.L., Brixen K., Kiel D.P., Langdahl B.L., Lips P., Ljunggren O. Large-scale analysis of association between LRP5 and LRP6 variants and osteoporosis. JAMA. 2008;299:1277–1290. doi: 10.1001/jama.299.11.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uitterlinden A.G., Ralston S.H., Brandi M.L., Carey A.H., Grinberg D., Langdahl B.L., Lips P., Lorenc R., Obermayer-Pietsch B., Reeve J. The association between common vitamin D receptor gene variations and osteoporosis: a participant-level meta-analysis. Ann. Intern. Med. 2006;145:255–264. doi: 10.7326/0003-4819-145-4-200608150-00005. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y.J., Shen H., Xiao P., Xiong D.H., Li L.H., Recker R.R., Deng H.W. Molecular genetic studies of gene identification for osteoporosis: a 2004 update. J. Bone Miner. Res. 2006;21:1511–1535. doi: 10.1359/JBMR.051002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y.Z., Liu Y.J., Recker R.R., Deng H.W. Molecular studies of identification of genes for osteoporosis: the 2002 update. J. Endocrinol. 2003;177:147–196. doi: 10.1677/joe.0.1770147. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y.J., Liu X.G., Wang L., Dina C., Yan H., Liu J.F., Levy S., Papasian C.J., Drees B.M., Hamilton J.J. Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum. Mol. Genet. 2008;17:1803–1813. doi: 10.1093/hmg/ddn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y.Z., Wilson S.G., Wang L., Liu X.G., Guo Y.F., Li J., Yan H., Deloukas P., Soranzo N., Chinnapen-Horsley U. Identification of PLCL1 gene for hip bone size variation in females in a genome-wide association study. PLoS. ONE. 2008;3:e3160. doi: 10.1371/journal.pone.0003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang T.L., Chen X.D., Guo Y., Lei S.F., Wang J.T., Zhou Q., Pan F., Chen Y., Zhang Z.X., Dong S.S. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am. J. Hum. Genet. 2008;83:663–674. doi: 10.1016/j.ajhg.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaykin D.V., Westfall P.H., Young S.S., Karnoub M.A., Wagner M.J., Ehm M.G. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum. Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
- 15.Lencz T., Morgan T.V., Athanasiou M., Dain B., Reed C.R., Kane J.M., Kucherlapati R., Malhotra A.K. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Mol. Psychiatry. 2007;12:572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- 16.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 17.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 18.Yuan H.Y., Chiou J.J., Tseng W.H., Liu C.H., Liu C.K., Lin Y.J., Wang H.H., Yao A., Chen Y.T., Hsu C.N. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cupples L.A., Arruda H.T., Benjamin E.J., D'Agostino R.B., Sr., Demissie S., DeStefano A.L., Dupuis J., Falls K.M., Fox C.S., Gottlieb D.J. The Framingham Heart Study 100K SNP genome-wide association study resource: overview of 17 phenotype working group reports. BMC Med. Genet. 2007;8(Suppl 1):S1. doi: 10.1186/1471-2350-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannan M.T., Felson D.T., Anderson J.J. Bone mineral density in elderly men and women: results from the Framingham osteoporosis study. J. Bone Miner. Res. 1992;7:547–553. doi: 10.1002/jbmr.5650070511. [DOI] [PubMed] [Google Scholar]
- 21.Hannan M.T., Felson D.T., Dawson-Hughes B., Tucker K.L., Cupples L.A., Wilson P.W., Kiel D.P. Risk factors for longitudinal bone loss in elderly men and women: the Framingham Osteoporosis Study. J. Bone Miner. Res. 2000;15:710–720. doi: 10.1359/jbmr.2000.15.4.710. [DOI] [PubMed] [Google Scholar]
- 22.Braun A., Roth R., McGinniss M.J. Technology challenges in screening single gene disorders. Eur. J. Pediatr. 2003;162(Suppl 1):S13–S16. doi: 10.1007/s00431-003-1343-3. [DOI] [PubMed] [Google Scholar]
- 23.Hill D.D., Cauley J.A., Sheu Y., Bunker C.H., Patrick A.L., Baker C.E., Beckles G.L., Wheeler V.W., Zmuda J.M. Correlates of bone mineral density in men of African ancestry: the Tobago bone health study. Osteoporos. Int. 2008;19:227–234. doi: 10.1007/s00198-007-0450-9. [DOI] [PubMed] [Google Scholar]
- 24.Miljkovic-Gacic I., Ferrell R.E., Patrick A.L., Kammerer C.M., Bunker C.H. Estimates of African, European and Native American ancestry in Afro-Caribbean men on the island of Tobago. Hum. Hered. 2005;60:129–133. doi: 10.1159/000089553. [DOI] [PubMed] [Google Scholar]
- 25.Lange C., DeMeo D.L., Laird N.M. Power and design considerations for a general class of family-based association tests: quantitative traits. Am. J. Hum. Genet. 2002;71:1330–1341. doi: 10.1086/344696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitlock M.C. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J. Evol. Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 27.Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu X., Shin B.H., Akbarali Y., Weiss A., Boltax J., Oettgen P., Libermann T.A. Tel-2 is a novel transcriptional repressor related to the Ets factor Tel/ETV-6. J. Biol. Chem. 2001;276:9421–9436. doi: 10.1074/jbc.M010070200. [DOI] [PubMed] [Google Scholar]
- 29.Potter M.D., Buijs A., Kreider B., van Rompaey L., Grosveld G.C. Identification and characterization of a new human ETS-family transcription factor, TEL2, that is expressed in hematopoietic tissues and can associate with TEL1/ETV6. Blood. 2000;95:3341–3348. [PubMed] [Google Scholar]
- 30.Chen W., Kirkbride K.C., How T., Nelson C.D., Mo J., Frederick J.P., Wang X.F., Lefkowitz R.J., Blobe G.C. Beta-arrestin 2 mediates endocytosis of type III TGF-beta receptor and down-regulation of its signaling. Science. 2003;301:1394–1397. doi: 10.1126/science.1083195. [DOI] [PubMed] [Google Scholar]
- 31.Horner A., Kemp P., Summers C., Bord S., Bishop N.J., Kelsall A.W., Coleman N., Compston J.E. Expression and distribution of transforming growth factor-beta isoforms and their signaling receptors in growing human bone. Bone. 1998;23:95–102. doi: 10.1016/s8756-3282(98)00080-5. [DOI] [PubMed] [Google Scholar]
- 32.Kirkbride K.C., Townsend T.A., Bruinsma M.W., Barnett J.V., Blobe G.C. Bone morphogenetic proteins signal through the transforming growth factor-beta type III receptor. J. Biol. Chem. 2008;283:7628–7637. doi: 10.1074/jbc.M704883200. [DOI] [PubMed] [Google Scholar]
- 33.Styrkarsdottir U., Cazier J.B., Kong A., Rolfsson O., Larsen H., Bjarnadottir E., Johannsdottir V.D., Sigurdardottir M.S., Bagger Y., Christiansen C. Linkage of osteoporosis to chromosome 20p12 and association to BMP2. PLoS Biol. 2003;1:E69. doi: 10.1371/journal.pbio.0000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenvers K.L., Tursky M.L., Harder K.W., Kountouri N., Amatayakul-Chantler S., Grail D., Small C., Weinberg R.A., Sizeland A.M., Zhu H.J. Heart and liver defects and reduced transforming growth factor beta2 sensitivity in transforming growth factor beta type III receptor-deficient embryos. Mol. Cell. Biol. 2003;23:4371–4385. doi: 10.1128/MCB.23.12.4371-4385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


