Abstract
Cleft lip with or without cleft palate (CL/P) is a complex trait with evidence that the clinical spectrum includes both microform and subepithelial lip defects. We identified missense and nonsense mutations in the BMP4 gene in 1 of 30 cases of microform clefts, 2 of 87 cases with subepithelial defects in the orbicularis oris muscle (OOM), 5 of 968 cases of overt CL/P, and 0 of 529 controls. These results provide confirmation that microforms and subepithelial OOM defects are part of the spectrum of CL/P and should be considered during clinical evaluation of families with clefts. Furthermore, we suggest a role for BMP4 in wound healing.
Main Text
Isolated clefts of the lip with or without cleft palate (CL/P [MIM 119530]) comprise about 70% of all children born with an orofacial cleft. Cases of overt CL/P display a range of severity, from notches in the vermillion to complete bilateral clefts of the lip and palate.1,2 Also observed are more subtle expressions of the CL/P phenotype that are sometimes termed “microforms” and typically involve small defects of the lip, alveolar arch, or scar-like ridges above the lip.3,4 Such microforms extend to the muscle fibers of the superior orbicularis oris muscle (OOM).5,6 Microform cleft lip (congenital healed cleft lip [MIM 600625]) is a rarely reported birth defect that is suggested to occur in 0.06 /10,000 live births.7 Even more subtle than the visible microforms are subepithelial defects of the OOM. Martin et al.8 observed such defects in 18-week-old fetuses and developed a method to visualize OOM defects by using ultrasonography.9 A significant increase in the frequency of OOM defects has been reported in people who are related to individuals with CL/P but who do not have overt clefts.9 The fact that subepithelial defects of the OOM are part of the spectrum of orofacial cleft expression was also confirmed by Neiswanger et al.10 Furthermore, histologic studies of OOMs visualized as abnormal by ultrasound showed both disorganized OOM fibers and excess connective tissue in comparison to normal OOMs.11
Informative mouse models for cleft lip and cleft palate have aided the search for genes involved in human CL/P12–14, as have rare Mendelian CL/P forms that are close phenocopies of isolated CL/P.15,16 The current study was motivated by a mouse model in which a conditional knockout for Bmp4 (MIM 112262) had an unusual “healed” cleft-lip phenotype17; Liu et al.17,18 found that all embryos had bilateral cleft lip at 12 days after conception but that by 14.5 days only 22% still exhibited cleft lip. They hypothesized that many of the initial cleft lips had healed in utero. In this report we evaluate the BMP4 gene in individuals born with overt CL/P, microform CL/P, and subepithelial OOM defects.
Clinical aspects of the sample collection have been described elsewhere.10,19,20 The University of Iowa Institutional Review Board (IRB) gave approval for sample collection (approval numbers 199804081, 200003065, and 200109094) in conjunction with local approval in the Philippines, Colombia, and Children's Hospital and Regional Medical Center, Seattle, Washington. The Ohio State University approved sample collection in Ohio (approval number 98H0041). The University of Pittsburgh approved sample collection in Pittsburgh, Spain, Guatemala, and the Philippines (approval numbers 0405013, 0511198, and 0607057) after approval by the IRBs of the local sites. The ethics committee of the School of Dentistry, Aichi-Gakuin University, approved the sample collection in Mongolia (approval number 11) in conjunction with local approval in Mongolia. DNA samples from 30 individuals with microform cleft lip (27 Filipinos, two Americans, and one Colombian), from 87 cases with OOM defects (nine Filipinos, 31 Americans, 34 Guatemalans, and 13 Europeans), from 968 cases with overt CL/P (537 Filipinos and 431 Mongolians) and from 529 controls (345 Filipinos, 90 Mongolians, and 94 Europeans) were sequenced according to published protocols.16 Five primer pairs (Table S2) were used for examining all four BMP4 exons and 50 bp of flanking sequence (NP_001193). M.L.M., A.C.L., A.H., A.R.V., J.C.M., or S.D.H. reviewed subjects with CL/P to exclude any with syndromic features. OOM defects were assessed by high-resolution ultrasound according to the methods of Neiswanger et al.10 (Figures 1A and 1B). Note that all cases and controls are unrelated; only one cleft or OOM defect case was taken from any particular family.
Figure 1.
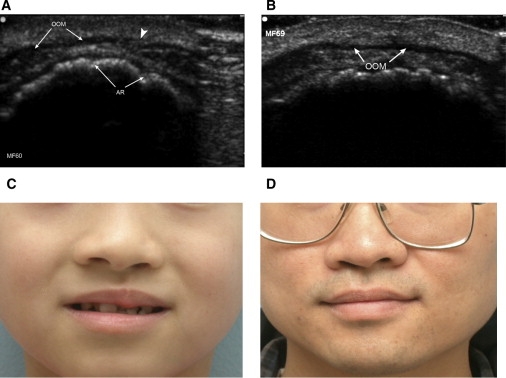
Subepithelial and Microform Defects
Ultrasound Images of the Proband S91C, Who Has an Orbicularis Oris Muscle Defect, and Her Husband
Ultrasound images are taken in the transverse plane; the top of the image corresponds to anterior structures; OOM = orbicularis oris muscle; AR = alveolar ridge. The arrowhead in (A) points to the discontinuity in the OOM of the proband (A). In contrast, note the continuous appearance of the husband's OOM (B). (C) A child (A346V) has left microform CLP. (D) His father (A346V) has subtle right microform cleft lip and a “heart-shaped” bifid uvula. See Table S1 for a summary of the cases and controls sequenced for BMP4.
Table S1 in the Supplemental Data summarizes the origins of the cases and controls studied. The mutations identified in the cases and their relatives are provided in Table S2 and depicted in Figure 2. No mutations were found in the controls. Five of the total 968 cases (0.51%) with overt nonsyndromic CL/P were found to have previously unreported missense or nonsense mutations. Two of the total 87 cases with OOM defects and one of the 30 cases with microform CL/P had novel missense mutations. One common missense-encoding SNP (p.A152V, c.455 C > T, SNP rs17563) was found in both cases and controls, with a borderline-significant difference in frequency (the frequency of the C allele for the case and control is 0.20 and 0.26, respectively, and that for the T allele for the case and control is 0.80 and 0.74, respectively) (p = 0.04).
Figure 2.
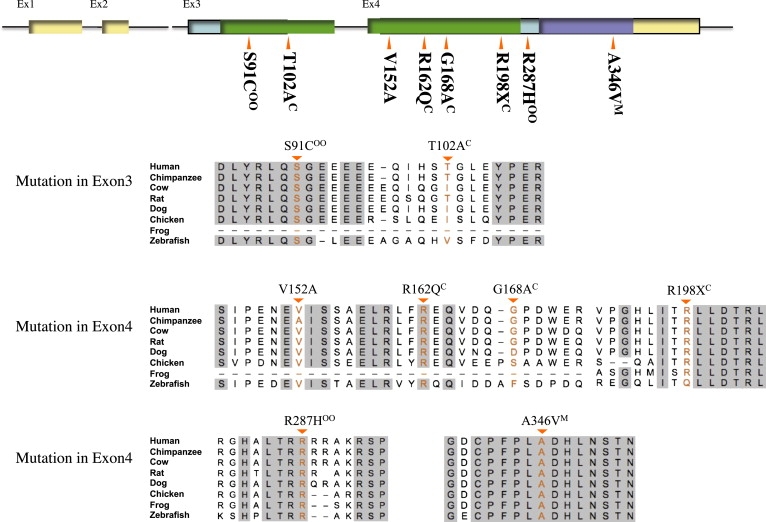
Comparative Amino Acid Sequence Alignment of Vertebrate BMP4 and Mutations of BMP4 in Humans
The top row shows a schematic diagram of the human BMP4 gene with the positions of exons and their translated protein domains. Both exon 3 and exon 4 are in the translated region. Filled green is the TGF beta propeptide domain and violet is the TGF beta domain. Gray background in the alignment indicates regions of complete amino acid identity across species. Comparative amino acid alignment follows the order of Homo sapiens (Human [NP_001193]), Pan troglodytes (Chimpanzee [XP_509954]), Bos taurus (Cow [NP_001039342]), Rattus norvegicus (Rat [NP_036959]), Canis familiaris (Dog [XP_851628]), Gallus gallus (Chicken [NP_990568]), Xenopus tropicalis (Frog [NP_001017034]), and Danio rerio (Zebrafish [NP_571417]). C is Cleft lip and palate; oo is orbicularis oris defect; M is Microform cleft.
Differences between groups were assessed with Fisher's exact tests (see Web Resources). The frequency of BMP4 mutations was greater for all cases (overt CL/P plus microforms plus OOM defects) than for all controls (8/1085 = 0.74% versus 0/529 = 0%) but was not statistically significant (p = 0.06). Notably, the BMP4 mutation frequency for overt CL/P cases alone was not significantly greater than for controls (5/968 = 0.52% versus 0/529, p = 0.17), whereas the frequency for microform plus OOM cases was significantly greater than for controls (3/117 = 2.56% versus 0/529, p = 0.006). Furthermore, the BMP4 mutation frequency in overt CL/P cases was significantly less than the frequency in microform plus OOM cases (5/968 versus 3/117, p = 0.04).
Comparative amino acid sequence analysis (Figure 2) showed complete conservation of the amino acid sequence in eight species (human, chimpanzee, cow, rat, dog, chicken, frog, and zebrafish) at positions p.S91C (c.271 A > T), p.R162Q (c.485 G > A), p.R287H (c.860 G > A), and p.A346V (c.1037 C > T). The missense mutation S91C was predicted to be possibly damaging by POLYPHEN (see Web Resources)21, and the R162Q and A346V variants are predicted to be intolerant amino acid changes by SIFT (see Web Resources).22 In addition, one nonsense mutation, p.R198X (c.592 C > T), was identified. The microform case with the missense mutation A346V is shown along with his son in Figures 1C and 1D. The father (1D) has a bifid uvula (MIM 192100) and microform cleft lip and the son with the same mutation has a microform cleft lip and a cleft palate. Two other cases with missense mutations (S91C and R287H) were parents who had OOM defects only and a child who had unilateral cleft lip and palate and who had the same mutation as the affected parent. The OOM defect for the S91C variant is shown in Figure 1A. The SIFT intolerant amino acid change, R162Q, was found in a child with cleft lip and palate who had a father with the R162Q variant. The father himself, diagnosed as unaffected by routine clinical observation, did not have an ultrasound evaluation. Each of the parent-to-child transmissions has a more severe phenotype in the child, which is suggestive of anticipation, but the numbers are small and the cases biased in ascertainment so that these findings are only suggestive of an increase in severity with each passing generation.
Three other variants were identified. The first, V152A, is a previously reported SNP (rs17563). We found a borderline difference in case and control frequencies for this SNP, and a recent report has shown a frequency difference for the same associated allele in a Chinese cleft population23 consisting of 184 cases and 205 controls. The second variant, p.G168A (c.503 G > C), was seen in a proband with cleft lip and palate and in a clinically unaffected mother who did not have an OOM ultrasound evaluation. The third variant, p.T102A (c.304 A > G), was seen in two patients with cleft lip and palate (one Filipino and one Mongolian). In both families, one parent of the case had the T102A variant and no overt cleft but did not have an OOM ultrasound (summarized in Table S2).
In this report a mutation search within BMP4 in cases with overt CL/P, microforms, and subepithelial OOM defects found that 3 of 117 microform or OOM defect cases had a missense mutation predicted to be disruptive by in silico protein modeling. This was also observed in 5 of 968 overt CL/P cases, whereas no mutations were found in 529 controls. In three of the OO/microform cases, the mutations identified segregated from a parent with an OOM defect or microform to a child with an overt cleft. The frequency of BMP4 mutations in microform and OOM defect cases was significantly higher than in controls (p = 0.006), but the frequency in overt CL/P cases did not reach significance (p = 0.17).
A recent report demonstrates that disruptions in BMP4 and Hedgehog signaling result in craniofacial developmental anomalies of the brain, eye, and digits (MCOPS6 [MIM 607932]).24 They demonstrate ocular clefting-like phenotypes (coloboma [MIM 120200]) in some cases and found both frameshift and missense BMP4 mutations that generate a more severe and general phenotype than the mutations reported here. In addition both SHH [MIM 600725]25 and PTCH1 [MIM 601309]26 have been associated with clefting in mice and humans.
The results reported here are consistent with findings in the Bmp4 mouse model17,18, suggesting either that amino acid alterations in BMP4 result in delayed lip closure (resulting in the appearance of a healed scar) or that actual healing of the cleft has occurred through an unknown mechanism. The mouse model is a conditional knockout of craniofacial tissues and so is not directly analogous to the heterozygous amino acid variants observed here, but the striking phenotypic overlap of the unusual clinical appearances adds support to the connections between the genotypes and phenotypes observed. Bmp signaling has been implicated in dorsal closure in Drosophila, a model system for wound healing. Our finding that BMP4 has a role in microform and subepithelial clefting is consistent with the speculation that a conserved genetic pathway might be involved in both wound healing and CL/P.27 Identifying factors that modulate wound healing in the embryo would afford opportunities both for identification of groups at high risk for poor wound healing (where surgical approaches or timing might be influenced) and for the use of these factors to enhance the wound repair itself.
It can be difficult to prove that any particular rare variant observed is etiologic in a complex trait unless those variants are strong candidates on the basis of expression and arise de novo, as was seen in a recent report suggesting that spontaneous mutations in VANGL1 (MIM 610132) could be a risk factor for neural-tube defects (NTD [MIM 182940]).28 Kryukov29 has provided empiric evidence that about 70% of missense mutations present at population frequencies of 1% or less probably contribute to the phenotype in which they are first identified. Identifying eight such rare missense variants in this cleft-related population (and identifying none in controls) would mean that there is a probability of more than 99.9% that at least one of these variants contributes to clefting. A range of other genes has been reported in which rare missense or nonsense mutations also appear to contribute to isolated CL/P.12–14,16,30–32 In this report we found a significant overrepresentation of BMP4 mutations in cases with a range of lip and OOM defects and an absence of such variants in more than 500 control samples. These findings support a role for BMP4 in the pathogenesis of isolated CL/P and/or in wound healing.
Furthermore, our observation that amino acid variations in BMP4 are associated with microform and subepithelial OOM defects provides confirmation that such subtle defects are part of the clinical spectrum of CL/P. Given that children with overt clefts had parents with microform cleft or OOM defects, genetic evaluation and counseling might now benefit from a full family phenotypic evaluation assessing such minor forms of clefts and including a high-resolution ultrasound of the upper-lip OOM. Mutations in BMP4 should therefore be considered in any family whose non-overt-cleft members exhibit microform clefts or OOM defects. Prospective evaluation of the impact of these and other subclinical phenotypes on the occurrence of cleft lip and palate will increasingly be important in genetic counseling of nonsyndromic orofacial cleft families.33
Acknowledgments
We are particularly grateful to the many patients and their families who willingly participated in this study. We were greatly assisted by many people, including those in the Department of Maxillofacial Surgery, School of Dentistry, Aichi-Gakuin University; those in the Japanese Cleft Palate Foundation (Aichi-Gakuin University, Nagoya, Japan), which is a medical organization that provides clinical care for underserved populations in Japan, Vietnam, Mongolia and Cambodia; and the many physicians and volunteers in Mongolia. We thank the outstanding technicians in our team, members of the Operation Smile Foundation for their invaluable assistance, Children of the Americas (in Guatemala), Kathy Bardi, Judith Resick, Edith Villanueva, Min Shi, Susie McConnell, Nancy Davin, and Daniel Benton. This work was supported by National Institutes of Health grants R37-DE08559 (J.C.M., M.L.M.), R01-DE016148 (M.L.M.), R21-DE016930 (M.L.M., F.W.B.D.), RO1 DE/HD012324, P50-DE016215 (J.C.M., M.L.M.), CRR/CTSA Grant UL1-RR024153 (M.L.M.), and P30 ES05605, as well as AGU High-Tech-Research Center Project, KAKENHI, Grant-in-Aid for Young Scientists (S.S.) No. 20791560 (Aichi-Gakuin). Also, this work was made possible by a donation from the Iowa Order of the Eastern Star.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Dental and Craniofacial Research or the National Institutes of Health.
Supplemental Data
Web Resources
The URLs for the data and analytic approaches presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
PolyPhen, http://genetics.bwh.harvard.edu/pph/
UCSC Genome Browser, http://genome.ucsc.edu/
Kanehisa Laboratories, http://www.kegg.org/
Preacher KJ. Calculation for the chi-square tests: An interactive calculation tool for chi-square tests of goodness of fit and independence [computer software], http://www.quantpsy.org
Note Added in Proof
In the version of this paper published online on February 26, the legend of Figure 2 contained descriptions for panels A and B, although no such panels were included in the figure. The legend has since been corrected online and in print through the removal of these panel labels. No additional text changes were made.
References
- 1.Gorlin R.J., Cohen M.M., Hennekam R.C.M. Orofacial Clefting Syndromes. In: Bobrow M., Harper P.S., Scriver C., editors. Syndromes of the Head and Neck. Oxford University Press; New York: 2001. pp. 850–860. [Google Scholar]
- 2.Eppley B.L., van Aalst J.A., Robrey A., Havlik R.J., Sadove A.M. The spectrum of orofacial clefting. Plast. Reconstr. Surg. 2005;115:101e–114e. doi: 10.1097/01.prs.0000164494.45986.91. [DOI] [PubMed] [Google Scholar]
- 3.Cosman B., Crikelair G.F. The minimal cleft lip. Plast. Reconstr. Surg. 1966;37:334–340. doi: 10.1097/00006534-196604000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Carstens M.H. The spectrum of minimal clefting: process oriented cleft management in the presence of an intact alveolus. J. Craniofac. Surg. 2000;11:270–294. doi: 10.1097/00001665-200011030-00013. [DOI] [PubMed] [Google Scholar]
- 5.Lehman J.A., Artz J.S. The minimal cleft lip. Plast. Reconstr. Surg. 1976;58:306–309. doi: 10.1097/00006534-197609000-00008. [DOI] [PubMed] [Google Scholar]
- 6.Heckler F.R., Oesterle L.G., Jabaley M.E. The minimal cleft lip revisited: clinical and anatomic correlations. Cleft Palate J. 1979;6:240–247. [PubMed] [Google Scholar]
- 7.Castilla E.E., Martinez-Frias M.L. Congenital healed cleft lip. Am. J. Med. Genet. 1995;58:106–112. doi: 10.1002/ajmg.1320580203. [DOI] [PubMed] [Google Scholar]
- 8.Martin R.A., Jones K.L., Benirschke K. Extension of the cleft lip phenotype: The subepithelial cleft. Am. J. Med. Genet. 1993;47:744–747. doi: 10.1002/ajmg.1320470529. [DOI] [PubMed] [Google Scholar]
- 9.Martin R.A., Hunter V., Neufeld-Kaiser W., Flodman P., Spence M.A., Furnas D., Martin K.A. Ultrasonographic detection of orbicularis oris defects in first-degree relatives of isolated cleft lip patients. Am. J. Med. Genet. 2000;90:155–161. doi: 10.1002/(sici)1096-8628(20000117)90:2<155::aid-ajmg13>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 10.Neiswanger K., Weinberg S.M., Rogers C.R., Brandon C.A., Cooper M.E., Bardi K.M., Deleyiannis F.W., Resick J.M., Bowen A., Mooney M.P. Orbicularis oris muscle defects as an expanded phenotypic feature in nonsyndromic cleft lip with or without cleft palate. Am. J. Med. Genet. A. 2007;143A:1143–1149. doi: 10.1002/ajmg.a.31760. [DOI] [PubMed] [Google Scholar]
- 11.Rogers C.R., Weinberg S.M., Smith T.D., Deleyiannis F.W.B., Mooney M.P., Marazita M.L. Anatomical basis for apparent subepithelial cleft lip: A histological and ultrasonographic survey of the orbicularis oris muscle. Cleft Palate Craniofac. J. 2008;45:518–524. doi: 10.1597/07-115.1. [DOI] [PubMed] [Google Scholar]
- 12.van den Boogaard M.J., Dorland M., Beemer F.A., van Amstel H.K. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat. Genet. 2000;24:342–343. doi: 10.1038/74155. [DOI] [PubMed] [Google Scholar]
- 13.Jezewski P.A., Vieira A.R., Nishimura C., Ludwig B., Johnson M., O'Brien S.E., Daack-Hirsch S., Schultz R.E., Weber A., Nepomucena B. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J. Med. Genet. 2003;40:399–407. doi: 10.1136/jmg.40.6.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe A., Akita S., Tin N.T., Natsume N., Nakano Y., Niikawa N., Uchiyama T., Yoshiura K. A mutation in RYK is a genetic factor for nonsyndromic cleft lip and palate. Cleft Palate Craniofac. J. 2006;43:310–316. doi: 10.1597/04-145.1. [DOI] [PubMed] [Google Scholar]
- 15.Zucchero T.M., Cooper M.E., Maher B.S., Daack-Hirsch S., Nepomuceno B., Ribeiro L., Caprau D., Christensen K., Suzuki Y., Machida J. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N. Engl. J. Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
- 16.Riley B.M., Mansilla M.A., Ma J., Daack-Hirsch S., Maher B.S., Raffensperger L.M., Russo E.T., Vieira A.R., Dode C., Mohammadi M. Impaired FGF signaling contributes to cleft lip and palate. Proc. Natl. Acad. Sci. USA. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W., Sun X., Braut A., Mishina Y., Behringer R.R., Mina M., Martin J.F. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- 18.Liu W., Selever J., Murali D., Sun X., Brugger S.M., Ma L., Schwartz R.J., Maxson R., Furuta Y., Martin J.F. Threshold-specific requirements for BMP4 in mandibular development. Dev. Biol. 2005;283:282–293. doi: 10.1016/j.ydbio.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 19.Marazita M.L., Murray J.C., Lidral A.C., Arcos-Burgos M., Cooper M.E., Goldstein T., Maher B.S., Daack-Hirsch S., Schultz R., Mansilla M.A. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32–35. Am. J. Hum. Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray J.C., Daack-Hirsch S., Buetow K.H., Munger R., Espina L., Paglinawan N., Villanueva E., Rary J., Magee K., Magee W. Clinical and epidemiologic studies of cleft lip and palate in the Philippines. Cleft Palate Craniofac. J. 1997;34:7–10. doi: 10.1597/1545-1569_1997_034_0007_caesoc_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- 21.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng P.C., Henikoff S.T. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J.Y., Chen Y.J., Huang Y.L., Tang G.P., Zhang L., Deng B., Li M., Ma H., Luan R.S. Association of bone morphogenetic protein 4 gene polymorphisms with nonsyndromic cleft lip with or without cleft palate in Chinese children. DNA Cell Biol. 2008;27:601–605. doi: 10.1089/dna.2008.0777. [DOI] [PubMed] [Google Scholar]
- 24.Bakrania P., Efthymiou M., Klein J.C., Salt A., Bunyan D.J., Wyatt A., Ponting C.P., Martin A., Williams S., Lindley V. Mutations in BMP4 cause eye, brain, and digit developmental anomalies: Overlap between BMP4 and hedgehog signaling pathways. Am. J. Hum. Genet. 2008;82:304–319. doi: 10.1016/j.ajhg.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomason H.A., Dixon M.J., Dixon J. Facial clefting in Tp63 deficient mice results from altered Bmp4, Fgf8 and Shh signaling. Dev. Biol. 2008;321:273–282. doi: 10.1016/j.ydbio.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 26.Mansilla M.A., Cooper M.E., Goldstein T., Castilla E.E., Lopez Camelo J.S., Marazita M.L., Murray J.C. Contributions of PTCH gene variants to isolated cleft lip and palate. Cleft Palate Craniofac. J. 2006;43:21–29. doi: 10.1597/04-169R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin P., Parkhurst S.M. Parallels between tissue repair and embryo morphogenesis. Development. 2004;131:3021–3034. doi: 10.1242/dev.01253. [DOI] [PubMed] [Google Scholar]
- 28.Kibar Z., Torban E., McDearmid J.R., Reynolds A., Berghout J., Mathieu M., Kirillova I., De Marco P., Mercell E., Hayes J.M. Mutations in VANGL1 associated with neural-tube defects. N. Engl. J. Med. 2007;356:1432–1437. doi: 10.1056/NEJMoa060651. [DOI] [PubMed] [Google Scholar]
- 29.Kryukov G.V., Pennacchio L.A., Sunyaev S.R. Most rare missense alleles are deleterious in humans: Implications for complex disease and association studies. Am. J. Hum. Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vieira A.R., Avila J.R., Daack-Hirsch S., Dragan E., Felix T.M., Rahimov F., Harrington J., Schultz R.R., Watanabe Y., Johnson M. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet. 2005;1:e64. doi: 10.1371/journal.pgen.0010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiquet B.T., Lidral A.C., Stal S., Mulliken J.B., Moreno L.M., Arcos-Burgos M., Valencia-Ramirez C., Blanton S.H., Hecht J.T. CRISPLD2: A novel NSCLP candidate gene. Hum. Mol. Genet. 2007;16:2241–2248. doi: 10.1093/hmg/ddm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scapoli L., Palmieri A., Martinelli M., Vaccari C., Marchesini J., Pezzetti F., Baciliero U., Padula E., Carinci P., Carinci F. Study of the PVRL1 gene in Italian nonsyndromic cleft lip patients with or without cleft palate. Ann. Hum. Genet. 2006;70:410–413. doi: 10.1111/j.1529-8817.2005.00237.x. [DOI] [PubMed] [Google Scholar]
- 33.Weinberg S.M., Neiswanger K., Martin R.A., Mooney M.P., Kane A.A., Wenger S.L., Losee J., Deleyiannis F., Ma L., De Salamanca J.E. The Pittsburgh oral-facial cleft study: expanding the cleft phenotype. Background and justification. Cleft Palate Craniofac. J. 2006;43:7–20. doi: 10.1597/04-122r1.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


