Abstract
Low plasma levels of carotenoids and tocopherols are associated with increased risk of chronic disease and disability. Because dietary intake of these lipid-soluble antioxidant vitamins is only poorly correlated with plasma levels, we hypothesized that circulating carotenoids (vitamin A-related compounds) and tocopherols (vitamin E-related compounds) are affected by common genetic variation. By conducting a genome-wide association study in a sample of Italians (n = 1190), we identified novel common variants associated with circulating carotenoid levels and known lipid variants associated with α-tocopherol levels. Effects were replicated in the Women's Health and Aging Study (n = 615) and in the α-Tocopherol, β-Carotene Cancer Prevention (ATBC) study (n = 2136). In meta-analyses including all three studies, the G allele at rs6564851, near the β-carotene 15,15′-monooxygenase 1 (BCMO1) gene, was associated with higher β-carotene (p = 1.6 × 10−24) and α-carotene (p = 0.0001) levels and lower lycopene (0.003), zeaxanthin (p = 1.3 × 10−5), and lutein (p = 7.3 × 10−15) levels, with effect sizes ranging from 0.10–0.28 SDs per allele. Interestingly, this genetic variant had no significant effect on plasma retinol (p > 0.05). The SNP rs12272004, in linkage disequilibrium with the S19W variant in the APOA5 gene, was associated with α-tocopherol (meta-analysis p = 7.8 × 10−10) levels, and this association was substantially weaker when we adjusted for triglyceride levels (p = 0.002). Our findings might shed light on the controversial relationship between lipid-soluble anti-oxidant nutrients and human health.
Introduction
There is evidence that genetic variation affects the production and activity of antioxidant enzymes, such as superoxide dismutases (SOD), and self-synthesized antioxidant molecules, such as glutathione.1,2 Whether genetic variation also affects the concentration and activity of extrinsic dietary antioxidants, such as carotenoids and tocopherols, has been little studied.
Carotene and structurally related compounds serve as the precursors for vitamin A and play important roles in the immune response, vision, and cellular differentiation.3 Plants are capable of de novo synthesis of a variety of carotenoid pigments, but animals need to get the essential carotenoids from their diet and modify them through a limited repertoire of metabolic transformations. The six carotenoids most abundant in human serum are β-carotene, α-carotene, β-cryptoxanthin, lycopene, lutein, and zeaxanthin.4 In observational studies, high circulating levels of carotenoids protect against age-related decline of muscle strength, physical and cognitive disability, and chronic morbidity.5–9 Thus, carotenoids are frequently used as dietary supplements in the hope of maintaining health and preventing diseases such as cancer and coronary heart disease. Unfortunately, large clinical trials have been unable to show benefits for the formulations tested, and some suggest that megadose supplementation might be harmful10 For example, in the ATBC trial, administration of carotenoids to male smokers was associated with excess risk of lung cancer.11–13 Other observational studies show that a high intake of fruit and vegetables is protective against inflammation, cardiovascular disease, decline in physical performance, and mortality.14–21
Vitamin E comprises eight related tocopherols and tocotrienols, which are fat-soluble vitamins with antioxidant properties. α-tocopherol has the highest bioavailability and is the most studied tocopherol. Many consider α-tocopherol to be the most important lipid-soluble antioxidant and claim that it protects cell membranes from oxidation by reacting with lipid radicals produced in the lipid peroxidation chain reaction.22 A low α-tocopherol serum level strongly predicts disability in older persons23, correlates with frailty24 and poor cognitive function25 and is linked with the characteristic mild proinflammatory state often present in older persons.26
The correlation between vitamin intake assessed by dietary questionnaire and plasma levels is moderate for carotenoids27,28 and very poor for α-tocopherol.29 Interestingly, changes in serum level after ingestion of a standard dose of carotenoids is extremely variable between individuals but relatively stable across multiple experiments conducted in the same individual.30 Numerous factors influence absorption, digestion, transportation, storage, chemical transformation, and excretion of these compounds.31 Although few if any genetic studies attempting to identify heritability in these processes have been performed, it is reasonable to presume that genetic variation could affect all these mechanisms.
In an attempt to identify genetic factors affecting carotenoids and tocopherol levels, we performed a genome-wide association study (GWAS) of the participants of the InCHIANTI study, which was conducted on a representative population in the Tuscany Chianti area (Italy). Main findings were replicated in the Women's Health and Aging Study (WHAS), a study of disability in older women in Baltimore, MD, and the α-Tocopherol, β-Carotene Cancer Prevention (ATBC) Study, a Finnish cancer-prevention trial of α-tocopherol and β-carotene supplementation.
Material and Methods
Sample Description
The InCHIANTI study is a population-based study of the older population living in the Chianti region of Tuscany, Italy. The details of the study have been previously reported.32 In brief, 1616 residents were selected from the population registry of two small towns, Greve in Chianti and Bagno a Ripoli. The participation rate was 90% (n = 1453), and the subjects ranged between 21 and 102 years of age. Overnight fasting blood samples were utilized for genomic DNA extraction and measurement of carotenoids, tocopherols, and other chemicals. The study protocol was approved by the Italian National Institute of Research and Care of Aging Institutional Review.
Overall, 1307 participants donated a blood sample, and DNA was extracted for 1242 of them. There were 85 parent-offspring pairs, six sibling pairs, and two half-sibling pairs documented. We investigated any further familial relationships by using genome-wide SNP data and PLINK software and calculated an inflation factor with PLINK software as previously described.33 All lambda inflation factors were very low (<1.06) and so did not appreciably alter association statistics. We excluded individuals who had less than 97% completeness in genotyping (n = 12) and who did not have measures of carotenoids and/or tocopherols (n = 39). To estimate the ethnicity of each of the InCHIANTI samples, we used the first principal components from the EIGENSTRAT analysis of a set of 42,048 independent QC-ed SNPs. This revealed that all individuals were of European ancestry. The final sample size was 1191.
We used data from two independent studies to replicate the most significant findings obtained in the InCHIANTI GWAS: The first study was the Women's Health and Aging Studies (WHAS) I and II, which are companion studies of community-dwelling older women and were conducted in Baltimore.34 WHAS I was a prospective cohort of women age 65 and older and was designed to sample the one-third of women who were most disabled in a 12-zip-code area of Baltimore. After screening, 1,002 of 1,409 eligible women consented to participate, and 78% of whom consented to baseline blood draw. WHAS II sampled the two-thirds of women, age 70–79, who were the least disabled in the population. Four hundred and four women in WHAS I and 211 in WHAS II were of European ancestry and had DNA available. We performed statistical analysis on WHAS I and II separately; we then meta-analyzed the results and checked for heterogeneity between the two branches of the study.
The second study population was a subset of the ATBC study, a Finnish randomized controlled clinical trial of the effect of β-carotene and α-tocopherol supplementation on cancer risk in adult smokers. From 1985 to 1988, 29,133 male smokers aged 50 to 69 years were recruited from southwestern Finland (n = 290 406) and randomized to one of four intervention regimens. The individuals who participated in this study were a subset of 2136 participants in the placebo arm.12
Measures of retinol were also available from 362 participants of European ancestry from the Baltimore Longitudinal Study of Aging, conducted by the National Institute on Aging. Volunteers age 20 years or older join the study for life. Extensive evaluations are performed approximately every two years and include the collection of a blood sample.
Study protocols of the InCHIANTI, BLSA, WHAS, and ATBC studies complied with the Declaration of Helsinki and were approved by the relevant institutional review boards. In all studies eligible subjects consented to participate after receiving a comprehensive description of the survey procedures and risks associated with them.
Genotyping
In InCHIANTI and BLSA, genome-wide genotyping was performed with the Illumina Infinium HumanHap550 genotyping chip (chip versions 1 and 3) as previously described.33 The exclusion criteria for SNPs were minor allele frequency < 1% (n = 25,422), genotyping completeness < 99% (n = 23,610), and Hardy-Weinberg equilibrium p < 0.0001 (n = 517). We genotyped SNPs that reached GWAS significance in the InCHIANTI study in the WHAS by using Taqman technology.35 Genotyped samples included 5% blind duplicates, and genotypes were consistent across these duplicates. All SNPs had a call rate > 98% and were in Hardy-Weinberg equilibrium (p > 0.05). In the ATBC study, genotyping was performed with Sequenom's iPLEX chemistry according to the manufacturer's protocol (Sequenom, San Diego, CA). The samples included 2% known duplicates, 5% blind duplicates, and 2% water controls. Genotyping success rate was > 97% for all SNPs, and all were in Hardy-Weinberg equilibrium (p > 0.05). Genotypes for all known and blind duplicate pairs were consistent. Genotyping of APOA5 variants not captured on the Infinium chip was performed in the InCHIANTI samples with Taqman technology.
Carotenoids and Tocopherols Assay
In all three studies blood samples were collected by venipuncture, and aliquots of serum or plasma were stored at −70°C and never thawed before carotenoids and vitamin E assays. In both InCHIANTI and WHAS, carotenoids, retinol, and tocopherols were measured via high-performance liquid chromatography (HPLC).36,37 Retinol concentration was also obtained from the HPLC chromatogram. The method used in the InCHIANTI study and in the WHAS subset evaluated at the 1-year follow-up allowed for separate measures from lutein and zeaxanthin,36 whereas the method used in WHAS at baseline (WHAS I) did not allow separation of the peaks for lutein and zeaxanthin.37 Within-run and between-run coefficients of variation were, respectively, 7.3% and 9.6% for α-carotene, 4.5% and 5.4% for β-carotene, 2.7% and 3.5% for β-cryptoxanthin, 2.6% and 7.1% for lutein, 6.2% and 6.8% for zeaxanthin, 7.5% and 7.8% for lycopene, 3.3% and 2.8% for retinol, 4.1% and 9.7% for α-tocopherol, and 5.1% and 6.9% for γ-tocopherol. β-carotene, α-tocopherol, and retinol were measured in baseline samples from the ATBC study with a previously described reverse-phase liquid chromatography method.38 In the BLSA, only an HPLC measure of retinol was available.39
Statistical Analysis
Measures of carotenoids and vitamin E were appropriately transformed when non-normally distributed (see Table 1). For retinol we used an inverse normal transformation in each study to allow meta-analysis across studies that had used different units. Full details of genome-wide association tests are previously described.33 In brief, for each autosomal SNP for each of the vitamin measures, we performed linear regression analyses by using PLINK software with age and sex as covariates and assuming additive allele effects with one degree of freedom. We checked for possible statistical over-inflation due to relatedness or residual population admixture by using an inflation factor, generated with EIGENSTRAT, for each trait (see Table 1).
Table 1.
Summary Details of Carotenoid and Tocopherol Levels
| Study or Vitamin Measure | Units | N | Mean (SD) | Transformation Used | Lambda Inflation Factor |
|---|---|---|---|---|---|
| InCHIANTI | |||||
| β-carotene | μmol/liter | 1191 | 0.41 (0.27) | Natural log | 1.034 |
| α-carotene | μmol/liter | 1191 | 0.057 (0.051) | Natural log | 1.018 |
| β-cryptoxanthin | μmol/liter | 1191 | 0.21 (0.16) | Natural log | 1.018 |
| Lycopene | μmol/liter | 1191 | 0.68 (0.33) | Square rootb | 1.03 |
| Zeaxanthin | μmol/liter | 1191 | 0.063 (0.024) | Natural logc | 1.016 |
| Lutein | μmol/liter | 1191 | 0.38 (0.15) | Square-root | 1.026 |
| Retinol | μmol/liter | 1191 | 1.92 (0.50) | Natural log | 1.03 |
| α-tocopherol | μmol/liter | 1191 | 33.44 (7.65) | Noned | 1.054 |
| γ-tocopherol | μmol/l | 1191 | 2.20 (0.96) | Natural log | 1.024 |
| WHAS I | |||||
| β-carotene | μmol/liter | 404 | 0.42 (0.38) | Natural log | N/A |
| α-carotene | μmol/liter | 404 | 0.099 (0.091) | Natural log | N/A |
| β-cryptoxanthin | μmol/liter | 404 | 0.14 (0.15) | Natural log | N/A |
| Lycopene | μmol/liter | 404 | 0.56 (0.31) | None | N/A |
| Zeaxanthin/ Lutein | μmol/liter | 404 | 0.34 (0.16) | Square root | N/A |
| Luteina | μmol/liter | 289 | 0.17 (0.08) | Square root | N/A |
| Zeaxanthina | μmol/liter | 289 | 0.066 (0.029) | Square root | N/A |
| Retinol | μmol/liter | 404 | 0.94 (0.35) | Inverse normal | N/A |
| α-tocopherol | μmol/liter | 404 | 23.42 (8.79) | Natural log | N/A |
| WHAS II | |||||
| β-carotene | μmol/liter | 211 | 0.57 (0.54) | Natural log | N/A |
| α-carotene | μmol/liter | 211 | 0.143 (0.137) | Natural log | N/A |
| β-cryptoxanthin | μmol/liter | 211 | 0.16 (0.11) | Natural log | N/A |
| Lycopene | μmol/liter | 211 | 0.71 (0.34) | None | N/A |
| Zeaxanthin/ Lutein | μmol/liter | 211 | 0.43 (0.20) | Square root | N/A |
| Retinol | μmol/liter | 211 | 1.06 (0.36) | Inverse normal | N/A |
| α-tocopherol | μmol/liter | 211 | 24.25 (8.77) | Natural log | N/A |
| BLSA | |||||
| Retinol | ug/ml | 362 | 0.63 (0.63) | Inverse normal | N/A |
| ATBC | |||||
| β-carotenee | μmol/liter | 2126 | 0.40 (0.30) | Natural log | N/A |
| Retinol | ug/liter | 2136 | 585.5 (122.2) | Inverse normal | N/A |
| α-tocopherolf | μmol/liter | 2131 | 27.10 (7.22) | Natural log | N/A |
N/A = not available.
Lutein and zeaxanthin separated in WHAS I, round 2.
Square-root transformation was used for GWAS but none was used for meta-analysis purposes or presentation in Table 4.
Natural-log transformation was used for GWAS, but square-root transformation was used for meta-analysis purposes and presentation in Table 4.
No transformation was used for GWAS, but natural-log transformation was used for meta-analysis purposes and presentation in Table 4.
Converted from μg/liter with the conversion factor of 53.27. μg/liter values were transformed and used in analyses, the results of which are in Table 2.
Converted from μg/liter with the conversion factor of 44.17. μg/liter values were transformed and used in analyses, the results of which are in Table 4.
We performed analysis of the WHAS, ATBC, and BLSA data to mirror the analysis and replicate the major findings in the InCHIANTI population. We carried out age- and sex-adjusted multiple linear regressions that related each of the seven most highly associated SNPs with each one of the vitamin measures under an additive genetic model assumption. We performed meta-analyses of results across different studies by weighting each study with an inverse variance method as implemented in STATAv10 (StataCorp. 2007. Stata Statistical Software: Release 10. College Station, TX: StataCorp LP).
Results
Mean values of the major carotenoids and α-tocopherol measured in the InCHIANTI, BLSA, and ATBC studies are reported in Table 1. The results of the genome-wide association study performed in the InCHIANTI samples for the seven main carotenoids and the two main tocopherols, limited to the SNPs that yielded association with a p < 1 × 10−5, are reported in Table S1 in the Supplemental Data. Five SNPs were associated with β-carotene and/or lutein plasma levels, and two SNPs were associated with α-tocopherol with a p value < 5 × 10−7, a threshold value previously used by other GWAS studies.40 There were no associations at a p value < 5 × 10−7 for any of the other six measures. Results were substantially unchanged when the analysis was repeated with carotenoid levels averaged across baseline and carotenoids measured at the 3 year and 6 year follow-up instead of with only baseline values (data not shown). Subsequent analyses focused on the five SNPs associated with β-carotene or lutein plasma levels and the two SNPs associated with α-tocopherol levels.
Of the SNPs associated with carotenoid levels, rs6513787 maps to chromosome 20, whereas rs6420424, rs8044334, rs11645428, and rs6564851 all map to a 23 kb region of chromosome 16 that includes the Polycystic kidney disease 1-like 2 gene and is 7.7 kb 5′ from the β-carotene 15,15′-monooxygenase 1(BCMO1) gene. BCMO1 catalyzes the first step in the conversion of dietary provitamin carotenoids to vitamin A in the small intestine40–43 and so represents an excellent candidate.
Of the SNPs associated with α-tocopherol levels, rs12272004 maps to chromosome 11 close to the APOA5 gene and is highly correlated with the APOA5 coding variant “S19W” (linkage disequilibrium r2 = 0.88); rs2903269 occurs on chromosome 4 more than100 kbp from the nearest known gene.
We successfully genotyped seven SNPs in the WHAS study (five at p < 5 × 10−7 with β-carotene levels and two at p < 5 × 10−7 with α-tocopherol levels) and six SNPs in the ATBC study (four at p < 5 × 10−7 with β-carotene levels—rs6420424 could not be fitted into the same iPLEX pool as the others—and two at p < 5 × 10−7 with α-tocopherol levels). Associations between the five carotenoid-relevant SNPs and β-carotene plasma levels across the different studies are described in Table 2. Results for the four SNPs in or close to the BCMO1 gene on chromosome 16 (Figure 1) confirmed a robust association with β-carotene plasma levels in both the WHAS and ATBC studies (Table 2). A meta-analysis across studies yielded p values all lower than 1 × 10−12. These four SNPs were all in linkage disequilibrium with each other (r2 ranged from 0.22–0.55). The association between rs6513787 on chromosome 20 and β-carotene levels was not replicated in the WHAS (p = 0.34) or ATBC (p = 0.13) (Table 2).
Table 2.
SNPs Associated with β-carotene at p < 5 × 10−7 in the InCHIANTI GWAS and Follow-up Results in the WHAS and the ATBC Study
| Study | SNP | Allelesa | Chromosome | MAF | N | Effect (95% Confidence Intervals)a | Standard Error | p Value |
|---|---|---|---|---|---|---|---|---|
| inCHIANTI | rs6513787 | C/A | 20 | 0.08 | 1190 | −0.222 (−0.306, −0.137) | 0.043 | 3.6 × 10−7 |
| inCHIANTI | rs6420424 | G/A | 16 | 0.34 | 1191 | 0.152 (0.103, 0.201) | 0.025 | 1.9 × 10−9 |
| inCHIANTI | rs8044334 | T/G | 16 | 0.24 | 1189 | 0.161 (0.106, 0.216) | 0.028 | 1.3 × 10−8 |
| inCHIANTI | rs11645428 | G/A | 16 | 0.47 | 1190 | −0.118 (−0.165, −0.072) | 0.024 | 8.3 × 10−7 |
| inCHIANTI | rs6564851 | T/G | 16 | 0.36 | 1171 | 0.160 (0.112, 0.208) | 0.024 | 9.7 × 10−11 |
| WHASc | rs6513787 | C/A | 20 | 0.05 | 576 | −0.100 (−0.307, 0.107) | 0.106 | 0.34 |
| WHASc | rs6420424 | G/A | 16 | 0.49 | 602 | 0.165 (0.082, 0.248) | 0.042 | 1.0 × 10−4 |
| WHASc | rs8044334 | T/G | 16 | 0.36 | 597 | 0.169 (0.084, 0.255) | 0.044 | 1.1 × 10−4 |
| WHASc | rs11645428 | G/A | 16 | 0.33 | 599 | −0.196 (−0.279, −0.113) | 0.042 | 3.7 × 10−6 |
| WHASc | rs6564851 | T/G | 16 | 0.48 | 602 | 0.199 (0.116, 0.282) | 0.042 | 2.6 × 10−6 |
| ATBC | rs6513787 | C/A | 20 | 0.04 | 2126 | −0.03 (−0.13, 0.07) | 0.05 | 0.13 |
| ATBC | rs6420424b | G/A | 16 | N/A | N/A | N/A | N/A | N/A |
| ATBC | rs8044334 | T/G | 16 | 0.35 | 2129 | 0.07 (0.034, 0.114) | 0.02 | 0.0003 |
| ATBC | rs11645428 | G/A | 16 | 0.28 | 2129 | −0.12 (−0.160, 0.075) | 0.022 | 7.4 × 10−8 |
| ATBC | rs6564851 | T/G | 16 | 0.39 | 2108 | 0.13 (0.092, 0.171) | 0.020 | 1.1 × 10−10 |
| Meta-analysis | rs6513787 | C/A | 20 | 3892 | −0.136 (−0.197, −0.075) | 0.031 | 1.4 × 10−5 | |
| Meta-analysis | rs6420424 | G/A | 16 | 1793 | 0.155 (0.11, 0.198) | 0.022 | 6.5 × 10−13 | |
| Meta-analysis | rs8044334 | T/G | 16 | 3915 | 0.109 (0.079, 0.139) | 0.015 | 9.3 × 10−13 | |
| Meta-analysis | rs11645428 | G/A | 16 | 3918 | −0.129 (−0.159, −0.099) | 0.015 | 1.5 × 10−17 | |
| Meta-analysis | rs6564851 | T/G | 16 | 3881 | 0.149 (0.120, 0.177) | 0.015 | 1.6 × 10−24 | |
N/A = not available, MAF = minor allele frequency.
Effect direction refers to second allele.
rs6420424 failed genotyping in the ATBC study.
Meta-analysis results of WHAS I and II are given.
Figure 1.

Associations between SNPs in and around the BCMO1 Gene and β-Carotene Levels
Red triangles represent the –log10 p values from all SNPs passing QC in the GWAS InCHIANTI study. The stars represent the three-study meta-analysis p values of the four SNPs taken forward into the replication studies.
The SNP rs6564851 showed the strongest association with plasma β-carotene; it had a p value of 1.6 × 10−24 in the meta-analysis, and participants with a G allele (the derived allele; the T allele is shared with chimps and other primates) had an average β-carotene level 0.26 (0.18–0.34) standard deviations (SD) higher than participants with a T allele. Haplotype-based analysis, using combinations of two, three, and four SNPs from the BCMO1 region on chromosome 16, did not reveal any associations stronger than those revealed by single-SNP-based analyses: the best association was between a two-SNP haplotype formed by rs6420424 and rs6564851 (p = 7.8 × 10−11). The SNP rs6420424 showed single-SNP effect sizes similar to those of rs6564851 but was not genotyped in the large ATBC study, and so we focused on rs6564851. The rs6564851 SNP was also significantly associated with plasma levels of other carotenoids both in the InCHIANTI study and in WHAS (Table 3). The G allele was associated with 0.28 (0.20–0.36) SD lower lutein, 0.13 (0.05–0.21) SD higher α-carotene and, respectively, 0.15 (0.07–0.23) and 0.10 (0.02–0.18) SD lower zeaxanthin and lycopene. There was no evidence of association between rs6564851 and either β-cryptoxanthin or retinol in any of the individual studies or the meta-analysis.
Table 3.
Associations of rs6564851, the SNP Most Strongly Associated with β-Carotene, with Other Carotenoids and Retinol
| Allelesb | N | Effect (95% Confidence Intervals) | Standard Error | p Value | |
|---|---|---|---|---|---|
| InCHIANTI | |||||
| α-carotene | T/G | 1171 | 0.089 (0.035, 0.143) | 0.027 | 0.001 |
| Lutein | T/G | 1171 | −0.033 (−0.043, −0.024) | 0.005 | 1.6 × 10−11 |
| Zeaxanthin | T/G | 1171 | −0.007 (−0.011, −0.003) | 0.002 | 3.3 × 10−4 |
| Lycopene | T/G | 1171 | −0.033 (−0.061, −0.006) | 0.014 | 0.016 |
| β-cryptoxanthin | T/G | 1171 | 0.0027 (−0.032, 0.085) | 0.030 | 0.37 |
| Retinol | T/G | 1171 | −0.018 (−0.005, 0.042) | 0.012 | 0.13 |
| WHAS | |||||
| α-carotene | T/G | 602 | 0.091 (0.003, 0.185) | 0.048 | 0.057 |
| Luteina | T/G | 283 | −0.029 (−0.044, −0.015) | 0.007 | 0.0001 |
| Zeaxanthina | T/G | 283 | −0.015 (−0.024, −0.005) | 0.005 | 0.003 |
| Lycopene | T/G | 602 | −0.03 (−0.063, 0.003) | 0.017 | 0.07 |
| β-cryptoxanthin | T/G | 602 | −0.021 (−0.110, −0.068) | 0.045 | 0.64 |
| Lutein/Zeaxanthina | T/G | 602 | −0.038 (−0.053, −0.023) | 0.008 | 5.4 × 10−7 |
| Retinol | T/G | 615 | −0.034 (−0.067, −0.0001) | 0.017 | 0.046 |
| ATBC | |||||
| Retinol | T/G | 2118 | 0.004 (−0.014, 0.022) | 0.009 | 0.66 |
| BLSA | |||||
| Retinol | T/G | 362 | 0.141 (−0.0054-0.2869) | 0.074 | 0.06 |
| Meta-analysis | |||||
| α-carotene | T/G | 1773 | 0.089 (0.043, 0.136) | 0.024 | 0.0001 |
| Lutein | T/G | 1454 | −0.032 (−0.040, −0.024) | 0.004 | 7.3 × 10−15 |
| Zeaxanthin | T/G | 1454 | −0.008 (−0.012, 0.004) | 0.002 | 1.3 × 10−5 |
| Lycopene | T/G | 1773 | −0.032 (−0.053, −0.011) | 0.011 | 0.003 |
| β-cryptoxanthin | T/G | 1773 | 0.012 (−0.037, 0.061) | 0.025 | 0.62 |
| Retinol | T/G | 4266 | −0.007 (−0.020, 0.006) | 0.007 | 0.27 |
In the baseline tests in WHASI and WHASII, lutein and zeaxanthin could not be separated upon assay. Separated lutein and zeaxanthin assays were performed in WHAS I, round 2 follow-up only.
Betas all refer to the rs6564851 G allele.
Associations between the two α-tocopherol-related SNPs and α-tocopherol plasma levels across the different studies are described in Table 4. Results for rs12272004 confirmed a robust association with α-tocopherol plasma levels in both the WHAS and the ATBC study (Table 4). Associations between rs2903269 and α-tocopherol plasma levels did not replicate. In the meta-analysis, the A allele of rs12272004 was associated with 0.07 SD higher α-tocopherol, with a p value of 7.8 × 10−10 (Table 4). Because rs12272004 is highly correlated with the S19W variant in the APOA5 gene, which is known to alter chylomicrons and triglyceride concentrations44, and because it is known that vitamin E levels are strongly affected by circulating lipids, we used InCHIANTI data to adjust the analysis for triglyceride levels. The strength of the association was halved (p = 0.002, effect size on natural logged α-tocopherol levels per A allele = 0.055 [0.02, 0.091]).
Table 4.
SNPs Associated with α-Tocopherol at p < 5 × 10−7 in the InCHIANTI GWAS and Follow-up Results in the WHAS and ATBC Studies
| Study | SNP | Alleles | CHR | MAF | N | Effect | Standard Error | p Value |
|---|---|---|---|---|---|---|---|---|
| inCHIANTI | rs12272004 | C/A | 11 | 0.05 | 1170 | 0.11(0.066, 0.15) | 0.022 | 3.9 × 10−7 |
| inCHIANTI | rs2903269 | C/T | 4 | 0.18 | 1180 | 0.062(0.039, 0.085) | 0.012 | 1.7 × 10−7 |
| WHAS | rs12272004 | C/A | 11 | 0.06 | 588 | 0.121(0.048, 0.194) | 0.037 | 0.001 |
| WHAS | rs2903269 | C/T | 4 | 0.19 | 601 | 0.04(−0.015, 0.095) | 0.028 | 0.160 |
| ATBC | rs12272004 | C/A | 11 | 0.07 | 2133 | 0.047(0.017, 0.077) | 0.015 | 0.0019 |
| ATBC | rs2903269 | C/T | 4 | 0.30 | 2130 | 0.009(−0.014, 0.020) | 0.009 | 0.76 |
| Meta-analysis | rs12272004 | C/A | 11 | 3891 | 0.072(0.049, 0.095) | 0.012 | 7.8 × 10−10 | |
| Meta-analysis | rs2903269 | C/T | 4 | 3911 | 0.034(−0.01, 0.08) | 0.016 | 0.14a | |
Random-effects meta-analysis was used because of highly significant heterogeneity.
We next examined whether other SNPs previously shown to alter lipid levels are associated with α-tocopherol levels (Table 5). We used the list of lipid variants reported in recent genome-wide association studies.45 Where we had not directly typed the relevant SNP as part of the GWAS, we used a closely correlated “proxy” SNP. The association between a SNP in strong linkage disequilibrium with the APOA5 S19W variant and α-tocopherol levels led us to directly type the second variant in this gene cluster known to influence triglyceride levels, rs662799 (−1131C/T). This variant is not in strong linkage disequilibrium with S19W46 and was not captured on the genome-wide chip. We found evidence that three variants associated with HD -cholesterol, two variants with LDL cholesterol, and four variants with triglycerides were individually associated with α-tocopherol levels (p < 0.05). By far the strongest effects were between the two triglyceride-raising alleles in the APOA5 gene cluster and raised α-tocopherol levels.
Table 5.
Effects of Known Lipid SNPs on α-Tocopherol Levels
| SNP | Nearest Gene(s) | Allele | Allele Frequency | Genotyped Proxy SNP Used (r2) | Effect (95% Confidence Intervals) | p Value |
|---|---|---|---|---|---|---|
| HDL-Lowering Variants | ||||||
| rs4149268 | ABCA1 | C | 0.27 | −0.32 (−0.96 to 0.31) | 0.32 | |
| rs1800775 | CETP | C | 0.43 | −0.86 (−1.49 to −0.23) | 0.007 | |
| rs2144300 | GALNT2 | C | 0.42 | rs10779835 (0.97) | −0.28 (−0.89 to 0.33) | 0.37 |
| rs1800588 | LIPC | C | 0.74 | −1.08 (−1.82 to −0.34) | 0.004 | |
| rs2156552 | LIPG | A | 0.80 | rs4939883 (0.95) | −0.97 (−1.88, −0.057) | 0.04 |
| rs328 | LPL | C | 0.88 | −0.55 (−1.41 to 0.31) | 0.21 | |
| rs2238104 | MVK-MMAB | C | 0.56 | −0.21 (−0.83 to 0.42) | 0.52 | |
| LDL-Raising Variants | ||||||
| rs693 | APOB | A | 0.49 | 0.72 (0.11 to 1.33) | 0.021 | |
| APOE | ɛ4 | 0.09 | 0.68 (−0.58 to 1.93) | 0.29 | ||
| rs646776 | CELSR2-PSRC1-SORT1 | T | 0.71 | 0.44 (−0.34 to 1.22) | 0.27 | |
| rs12654264 | HMGCR | T | 0.42 | rs6896136 (0.87) | 0.31 (−0.30 to 0.92) | 0.32 |
| rs6511720 | LDLR | G | 0.10 | −1.23 (−2.14 to −0.32) | 0.008 | |
| rs11206510 | PCSK9 | T | 0.85 | 0.49 (−0.28 to 1.27) | 0.21 | |
| Triglyceride-Raising Variants | ||||||
| rs1748195 | ANGPTL3 | C | 0.68 | rs1167998 (1.00) | −0.69 (−1.36 to −0.028) | 0.041 |
| rs662799 | APOA1-C3-A4-A5 | G | 0.02 | 2.38 (0.05, 0.019) | 7.3 × 10−7 | |
| rs3135506 | APOA1-C3-A4-A5 | G | 0.06 | rs12272004 (0.88) | 2.94 (1.63 to 4.26) | 1.2 × 10−5 |
| rs693 | APOB | A | 0.49 | 0.72 (0.11 to 1.33) | 0.021 | |
| rs2144300 | GALNT2 | C | 0.42 | rs10779835 (0.97) | 0.28 (−0.33 to 0.89) | 0.37 |
| rs780094 | GCKR | T | 0.38 | −0.28 (−0.89 to 0.34) | 0.38 | |
| rs328 | LPL | C | 0.88 | −0.55 (−1.41 to 0.31) | 0.21 | |
| rs17145738 | MLXIPL | C | 0.88 | rs2240466 (1.00) | 0.04 (−1.13 to 1.22) | 0.94 |
| rs17321515 | TRIB1 | A | 0.60 | rs6982636 (1.00) | −0.32 (−0.95 to 0.30) | 0.31 |
Discussion
By analyzing GWAS data from a representative sample of the Italian population and conducting a replication study with two other populations, we found that common variation near the gene encoding the enzyme β-carotene 15,15′-monooxygenase (BCMO1) is highly associated with plasma levels of several carotenoids. We also found that the polymorphism rs12272004, a SNP that is highly correlated with the S19W variant in the APOA5 gene, significantly affects plasma concentrations of α-tocopherol. This effect is probably mediated or confounded by increased circulating lipids.
Genetic Effect on Carotenoid Plasma Levels
The G allele of rs6564851 near the BCMO1 gene was associated with significantly higher β-carotene levels and, to a lesser extent, α-carotene plasma levels. The discovery InCHIANTI sample is likely to overestimate effect sizes, but based on the larger of the two replication samples, ATBC, rs6564851 explains 1.9% of the variance in β-carotene levels. This effect size is large in comparison to the effects conferred by common SNPs on other human traits such as height47 and body-mass index48, although it is similar to effect sizes seen for other circulating biomarkers.33 Based on data from European individuals in HapMap, rs6564851 is in linkage disequilibrium with SNPs at r2 > 0.2 across a region of 45 kbp stretching from 53 to 7 kbp 5′ of the BCMO1 gene. BCMO1 is a 15-15′ dioxygenase that catalyzes the first step in the conversion of dietary provitamin A carotenoids to vitamin A in the small intestine.41,42 Of the carotenoids measured in our study, β-carotene, α-carotene, and β-cryptoxanthin are considered potential vitamin A precursors because they contain at least one unsubstituted β-ionone ring and a polyene side chain attached with at least five conjugated double bonds (Figure 2). The 15-15′ central cleavage performed by BCMO1 transforms one β-carotene molecule into two molecules of retinal (Figure 3).41,42,49 The same reaction can generate one molecule of retinal from each molecule of α-carotene and β-cryptoxanthin (Figure 3). Given these metabolic pathways, our findings suggest the G allele of rs6564851, or an allele in strong linkage disequilibrium, causes reduced BCMO1 activity, which results in higher circulating levels of unconverted β-carotene and, to a lesser extent, α-carotene. Consistent with this theory, in our meta-analysis the effect of the G allele on β-cryptoxanthin was slightly positive, although not statistically significant. The description of a rare missense mutation (threonine to methionine) in BCMO1 is also consistent with our findings. This mutation silences as much as 90% of the BCMO1 enzymatic activity and results in hypercarotenaemia and hypovitaminosis A.50 There is no strong evidence that rs6564851 influences expression levels of the BCMO1 gene (p > 0.001), at least in lymphoblastoid cell lines, but this is not surprising because BCMO1 expression is very limited, if present at all, in lymphocytes.
Figure 2.

Molecular Structure of Carotenoids Measured in the Study
β-carotene, α-carotene, and β-cryptoxanthin are considered potential vitamin A precursors because they contain at least one unsubstituted β-ionone ring and a polyene side chain attached to at least five conjugated double bonds. Note that the portion of the molecule shaded in gray is “equivalent” to retinol.
Figure 3.
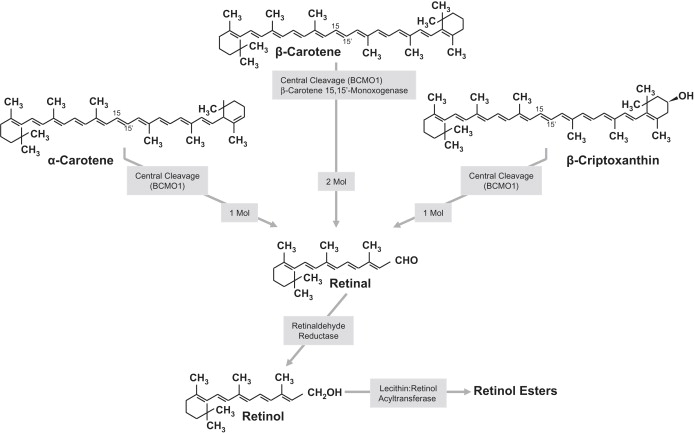
BCMO1-Catalyzed Central Cleavage of β-Carotene Forms Two Molecules of Retinal, Whereas the Cleavage of α-Carotene or β-cryptoxanthin Forms Only One Molecule of Retinal
Within the enterocytes, retinal is transformed into retinol, and then retinyl-esters, before being packed into nascent chylomicrons and transported through the lymphatic system.
In spite of the strong suggestion for reduced BCMO1 activity, we found no effect of rs6564851 on retinol plasma concentration. This is not entirely surprising. The retinal produced by the central cleavage of vitamin A precursors is efficiently transformed to retinol by the retinaldehyde reductase and esterified within enterocytes by lecithin:retinol acyltransferase.51 Retinyl esters are transported by nascent chylomicrons through the lymphatic system and taken up by hepatocytes, where they are rehydrolyzed into retinol and then either secreted by hepatocytes into the circulation or stored in hepatic stellate (Ito) cells, in a ratio aimed at maintaining a stable retinol plasma level.49 Thus, plasma retinol level is not a sensitive indicator of vitamin A and only changes when liver reserves of vitamin A are severely depleted. Future studies should quantify the effect of rs6564851 on vitamin A status by using isotope dilution techniques to measure total body stores of vitamin A.52 The effect of the rs6564851 polymorphism may be particularly important in persons who are at high risk of vitamin A deficiency; such persons include individuals with cystic fibrosis and malabsorption syndromes and preschool children in developing countries.53
A puzzling finding of this study is that participants with a G allele in rs6564851 had significantly lower levels of lycopene, lutein, and zeaxanthin plasma concentrations. Lycopene, lutein and zeaxanthin are synthesized only by plants and mammals need to consume them in their diet. Because of their molecular structure, there is no reason to believe that these carotenoids are a direct target for BCMO1. However, it has been proposed that carotenoids antagonize absorption of each other, suggesting that uptake by intestinal cells is a protein-transport facilitated process.54 Thus, it may be hypothesized that, through a still-unknown mechanism, higher α- and β-carotene plasma levels directly affect absorption, cell transport, and bio-availability of other carotenoids that are not vitamin A precursors.
The G allele of rs6564851, or some other allele in strong linkage disequilibrium, may affect health negatively by reducing the availability of vitamin A but also positively by increasing the plasma concentration of powerful antioxidant molecules. A number of studies have shown that higher plasma concentrations of antioxidant carotenoids are associated in a protective manner against the development of chronic diseases and disability.5–9 Future studies in large representative populations should test the hypothesis that long-term exposure to higher carotenoid concentrations caused by the rs6564851 G allele is associated with lower risk of health-related outcomes, including lower risk of cancer, cardiovascular disease, and physical disability. There was no association between the rs6564851 G allele and grip strength (p = 0.42, N = 983) or walk speed (p = 0.47, N = 705) in the InCHIANTI study, but larger studies are likely to be needed to demonstrate such an association. The variant rs6564851 is not captured well by SNPs available in the publicly available WTCCC data—the best proxy, rs7199144, has an r2 of 0.47 with rs6564851 in European samples and is not associated with any of the seven diseases studied as part of the WTCCC project (p > 0.05).
Genetic Effect on α-Tocopherol Plasma Levels
We found that the A allele of rs12272004 was strongly and consistently associated with higher plasma α-tocopherol levels. Vitamin E is a fat-soluble vitamin synthesized by plants, and its intake depends strictly on dietary content. Vitamin E absorption and distribution follows processes similar to those utilized during fatty acid digestion and metabolism. Vitamin E molecules enter enterocytes through lipid-rich micelles and are packaged into chylomicrons in combination with high-density lipoproteins and low-density lipoproteins and released into circulation. Because known variants of the APOA5 gene cause hypertriglyceridemia and hyperchylomicronemia,44 it is not surprising that the rs12272004 SNP, which is highly correlated with the S19W APOA5 SNP, appears to affect tocopherol levels. This is consistent with previous data showing that, in type 2 diabetes, a polymorphism of the APOA5 gene (rs662799 T→C) affects both VLDL triglycerides and tocopherol plasma levels.55 Also consistent with our interpretation, many of the SNPs that in the literature were found to affect circulating lipids were significantly associated with α-tocopherol levels in our study. Finally, after we adjusted our analysis for triglycerides, the association was substantially attenuated. Our data add to the literature by showing that the effect of APOA5 on α-tocopherol is not limited to diabetic patients and that APOA5 rs2272004 has one of the strongest genetic effects on α-tocopherol levels across the genome. The effect is specific for α-tocopherol and does not include γ-tocopherol. However, the concentration of γ-tocopherol is much higher than that of α-tocopherol, and the resulting lower precision and reliability of the measure may have affected our findings.
Projecting the effect of rs2272004 on health is difficult. Elevation in circulating triglycerides increases cardiovascular risk, but an incremented level of α-tocopherol may have a beneficial effect. For example, we have previously demonstrated that high α-tocopherol levels are protective against disability and frailty in the older population.23
Conclusions
In this study we found strong evidence that common polymorphisms near the BCMO1 gene are associated with substantial and significant increases in antioxidant carotenoid plasma levels. It is likely that the same genetic variation also affects vitamin A synthesis, although this effect could not be confirmed with the methodology used in our study and should be tested in future research. We also found that a SNP close to the APOA5 gene is associated significantly with differential levels of α-tocopherol, possibly because of its effect on circulating triglycerides and chylomicrons. Curiously, in both cases the identified SNPs affect human health in ways that are both potentially positive and negative. Whether positive or negative aspects prevail should be tested in the context of future longitudinal studies. Finally, the common variants associated with carotenoid levels could be used in Mendelian randomization studies,56 that aim to dissect the causal directions of associations between carotenoid levels and secondary phenotypes.
Supplemental Data
Supplemental Data include two tables and are available with this article online at http://www.ajhg.org/.
Supplemental Data
Web Resources
The URL for data presented herein is as follows:
Center for Statistical Genetics, http://www.sph.umich.edu/csg/liang/asthma/
Acknowledgments
The InCHIANTI study was supported as a “targeted project” (ICS110.1\RS97.71) by the Italian Ministry of Health and in part by the National Institute on Aging (contracts N01-AG-916413, N01-AG-821336). The ATBC study was supported by the US Public Health Service contracts N01-CN-45165, N01-RC-45035, and N01-RC-37004 from the National Cancer Institute, Department of Health and Human Service. This research was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health. A portion of that support was through an R&D contract with the MedStar Research Institute. We would like to thank Dr. Arun Barua (Iowa State University) for precious help in interpreting our findings in the context of carotenoid metabolism in humans. We acknowledge the important contribution of Leena Peltonen Palotie of the Finnish Public Health Institute, who made genotyping the ATBC study possible. The WHAS was supported by a MERIT AWARD from the National Institute on Aging, Pathogenesis of Physical Disability in Aging Women (R37 AG19905 and R01 AG027012) and the Johns Hopkins Claude D. Pepper Older Americans Independence Center (P30 AG021334).
References
- 1.Mahaney M.C., Czerwinski S.A., Adachi T., Wilcken D.E., Wang X.L. Plasma levels of extracellular superoxide dismutase in an Australian population: genetic contribution to normal variation and correlations with plasma nitric oxide and apolipoprotein A-I levels. Arterioscler. Thromb. Vasc. Biol. 2000;20:683–688. doi: 10.1161/01.atv.20.3.683. [DOI] [PubMed] [Google Scholar]
- 2.Njalsson R., Ristoff E., Carlsson K., Winkler A., Larsson A., Norgren S. Genotype, enzyme activity, glutathione level, and clinical phenotype in patients with glutathione synthetase deficiency. Hum. Genet. 2005;116:384–389. doi: 10.1007/s00439-005-1255-6. [DOI] [PubMed] [Google Scholar]
- 3.Krinsky N., Mayne S. Carotenoids in Health and Disease. Marcel Dekker; New York: 2004. H S. [Google Scholar]
- 4.Food and Nutritional Board IoM . National Accademy Press; Washington, D.C.: 2000. Dietary References Intakes for Vitamin C, Vitamin E, Selenium, and Carotenoids. [PubMed] [Google Scholar]
- 5.Bartali B., Frongillo E.A., Bandinelli S., Lauretani F., Semba R.D., Fried L.P., Ferrucci L. Low nutrient intake is an essential component of frailty in older persons. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:589–593. doi: 10.1093/gerona/61.6.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voutilainen S., Nurmi T., Mursu J., Rissanen T.H. Carotenoids and cardiovascular health. Am. J. Clin. Nutr. 2006;83:1265–1271. doi: 10.1093/ajcn/83.6.1265. [DOI] [PubMed] [Google Scholar]
- 7.Akbaraly N.T., Faure H., Gourlet V., Favier A., Berr C. Plasma carotenoid levels and cognitive performance in an elderly population: Results of the EVA Study. J. Gerontol. A Biol. Sci. Med. Sci. 2007;62:308–316. doi: 10.1093/gerona/62.3.308. [DOI] [PubMed] [Google Scholar]
- 8.Lauretani F., Semba R.D., Bandinelli S., Dayhoff-Brannigan M., Giacomini V., Corsi A.M., Guralnik J.M., Ferrucci L. Low plasma carotenoids and skeletal muscle strength decline over 6 years. J. Gerontol. A Biol. Sci. Med. Sci. 2008;63:376–383. doi: 10.1093/gerona/63.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauretani F., Semba R.D., Bandinelli S., Dayhoff-Brannigan M., Lauretani F., Corsi A.M., Guralnik J.M., Ferrucci L. Carotenoids as protection against disability in older persons. Rejuvenation Res. 2008;11:557–563. doi: 10.1089/rej.2007.0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenwald P. Beta-carotene and lung cancer: A lesson for future chemoprevention investigations? J. Natl. Cancer Inst. 2003;95:E1. doi: 10.1093/jnci/95.1.e1. [DOI] [PubMed] [Google Scholar]
- 11.The Alpha-Tocopherol, Beta Carotene Cancer Prevention Study Group The effect of vitamin E and beta carotene on the incidence of lung cancer and other cancers in male smokers. N. Engl. J. Med. 1994;330:1029–1035. doi: 10.1056/NEJM199404143301501. [DOI] [PubMed] [Google Scholar]
- 12.The Alpha-Tocopherol, Beta Carotene Cancer Prevention Study Group The alpha-tocopherol, beta-carotene lung cancer prevention study: design, methods, participant characteristics, and compliance. Ann. Epidemiol. 1994;4:1–10. doi: 10.1016/1047-2797(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 13.Virtamo J., Pietinen P., Huttunen J.K., Korhonen P., Malila N., Virtanen M.J., Albanes D., Taylor P.R., Albert P. Incidence of cancer and mortality following alpha-tocopherol and beta-carotene supplementation: a postintervention follow-up. JAMA. 2003;290:476–485. doi: 10.1001/jama.290.4.476. [DOI] [PubMed] [Google Scholar]
- 14.Bazzano L.A., He J., Ogden L.G., Loria C.M., Vupputuri S., Myers L., Whelton P.K. Fruit and vegetable intake and risk of cardiovascular disease in US adults: The first national health and nutrition examination survey epidemiologic follow-up study. Am. J. Clin. Nutr. 2002;76:93–99. doi: 10.1093/ajcn/76.1.93. [DOI] [PubMed] [Google Scholar]
- 15.Chrysohoou C., Panagiotakos D.B., Pitsavos C., Das U.N., Stefanadis C. Adherence to the Mediterranean diet attenuates inflammation and coagulation process in healthy adults: The ATTICA study. J. Am. Coll. Cardiol. 2004;44:152–158. doi: 10.1016/j.jacc.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 16.Esposito K., Marfella R., Ciotola M., Di Palo C., Giugliano F., Giugliano G., D'Armiento M., D'Andrea F., Giugliano D. Effect of a mediterranean-style diet on endothelial dysfunction and markers of vascular inflammation in the metabolic syndrome: a randomized trial. JAMA. 2004;292:1440–1446. doi: 10.1001/jama.292.12.1440. [DOI] [PubMed] [Google Scholar]
- 17.Estruch R., Martinez-Gonzalez M.A., Corella D., Salas-Salvado J., Ruiz-Gutierrez V., Covas M.I., Fiol M., Gomez-Gracia E., Lopez-Sabater M.C., Vinyoles E. Effects of a Mediterranean-style diet on cardiovascular risk factors: A randomized trial. Ann. Intern. Med. 2006;145:1–11. doi: 10.7326/0003-4819-145-1-200607040-00004. [DOI] [PubMed] [Google Scholar]
- 18.Houston D.K., Stevens J., Cai J., Haines P.S. Dairy, fruit, and vegetable intakes and functional limitations and disability in a biracial cohort: The atherosclerosis risk in communities study. Am. J. Clin. Nutr. 2005;81:515–522. doi: 10.1093/ajcn.81.2.515. [DOI] [PubMed] [Google Scholar]
- 19.Liu S., Manson J.E., Lee I.M., Cole S.R., Hennekens C.H., Willett W.C., Buring J.E. Fruit and vegetable intake and risk of cardiovascular disease: The women's health study. Am. J. Clin. Nutr. 2000;72:922–928. doi: 10.1093/ajcn/72.4.922. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Garcia E., Schulze M.B., Fung T.T., Meigs J.B., Rifai N., Manson J.E., Hu F.B. Major dietary patterns are related to plasma concentrations of markers of inflammation and endothelial dysfunction. Am. J. Clin. Nutr. 2004;80:1029–1035. doi: 10.1093/ajcn/80.4.1029. [DOI] [PubMed] [Google Scholar]
- 21.Trichopoulou A., Costacou T., Bamia C., Trichopoulos D. Adherence to a Mediterranean diet and survival in a Greek population. N. Engl. J. Med. 2003;348:2599–2608. doi: 10.1056/NEJMoa025039. [DOI] [PubMed] [Google Scholar]
- 22.Niki E., Saito T., Kawakami A., Kamiya Y. Inhibition of oxidation of methyl linoleate in solution by vitamin E and vitamin C. J. Biol. Chem. 1984;259:4177–4182. [PubMed] [Google Scholar]
- 23.Bartali B., Frongillo E.A., Guralnik J.M., Stipanuk M.H., Allore H.G., Cherubini A., Bandinelli S., Ferrucci L., Gill T.M. Serum micronutrient concentrations and decline in physical function among older persons. JAMA. 2008;299:308–315. doi: 10.1001/jama.299.3.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ble A., Cherubini A., Volpato S., Bartali B., Walston J.D., Windham B.G., Bandinelli S., Lauretani F., Guralnik J.M., Ferrucci L. Lower plasma vitamin E levels are associated with the frailty syndrome: The InCHIANTI study. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:278–283. doi: 10.1093/gerona/61.3.278. [DOI] [PubMed] [Google Scholar]
- 25.Cherubini A., Martin A., Andres-Lacueva C., Di Iorio A., Lamponi M., Mecocci P., Bartali B., Corsi A., Senin U., Ferrucci L. Vitamin E levels, cognitive impairment and dementia in older persons: The InCHIANTI study. Neurobiol. Aging. 2005;26:987–994. doi: 10.1016/j.neurobiolaging.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Di Iorio A., Cherubini A., Volpato S., Sparvieri E., Lauretani F., Franceschi C., Senin U., Abate G., Paganelli R., Martin A. Markers of inflammation, vitamin E and peripheral nervous system function: the InCHIANTI study. Neurobiol. Aging. 2006;27:1280–1288. doi: 10.1016/j.neurobiolaging.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talegawkar S.A., Johnson E.J., Carithers T.C., Taylor H.A., Bogle M.L., Tucker K.L. Carotenoid intakes, assessed by food-frequency questionnaires (FFQs), are associated with serum carotenoid concentrations in the Jackson heart study: Validation of the Jackson heart study delta NIRI adult FFQs. Public Health Nutr. 2008;11:989–997. doi: 10.1017/S1368980007001310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tucker K.L., Chen H., Vogel S., Wilson P.W., Schaefer E.J., Lammi-Keefe C.J. Carotenoid intakes, assessed by dietary questionnaire, are associated with plasma carotenoid concentrations in an elderly population. J. Nutr. 1999;129:438–445. doi: 10.1093/jn/129.2.438. [DOI] [PubMed] [Google Scholar]
- 29.Monge-Rojas R., Alfaro Calvo T., Nunez Rivas H. Comparison of serum concentration and dietary intake of alpha-tocopherol in a sample of urban and rural Costa Rican adolescents. Arch. Latinoam. Nutr. 2003;53:165–171. [PubMed] [Google Scholar]
- 30.Costantino J.P., Kuller L.H., Begg L., Redmond C.K., Bates M.W. Serum level changes after administration of a pharmacologic dose of beta-carotene. Am. J. Clin. Nutr. 1988;48:1277–1283. doi: 10.1093/ajcn/48.5.1277. [DOI] [PubMed] [Google Scholar]
- 31.Castenmiller J.J., West C.E. Bioavailability and bioconversion of carotenoids. Annu. Rev. Nutr. 1998;18:19–38. doi: 10.1146/annurev.nutr.18.1.19. [DOI] [PubMed] [Google Scholar]
- 32.Ferrucci L., Bandinelli S., Benvenuti E., Di Iorio A., Macchi C., Harris T.B., Guralnik J.M. Subsystems contributing to the decline in ability to walk: Bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J. Am. Geriatr. Soc. 2000;48:1618–1625. doi: 10.1111/j.1532-5415.2000.tb03873.x. [DOI] [PubMed] [Google Scholar]
- 33.Melzer D., Perry J.R., Hernandez D., Corsi A.M., Stevens K., Rafferty I., Lauretani F., Murray A., Gibbs J.R., Paolisso G. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasper J.D., Shapiro S., Guralnik J.M., Bandeen-Roche K.J., Fried L.P. Designing a community study of moderately to severely disabled older women: The women's health and aging study. Ann. Epidemiol. 1999;9:498–507. doi: 10.1016/s1047-2797(99)00026-5. [DOI] [PubMed] [Google Scholar]
- 35.Heid C.A., Stevens J., Livak K.J., Williams P.M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 36.Dorgan J.F., Boakye N.A., Fears T.R., Schleicher R.L., Helsel W., Anderson C., Robinson J., Guin J.D., Lessin S., Ratnasinghe L.D. Serum carotenoids and alpha-tocopherol and risk of nonmelanoma skin cancer. Cancer Epidemiol. Biomarkers Prev. 2004;13:1276–1282. [PubMed] [Google Scholar]
- 37.Sowell A.L., Huff D.L., Yeager P.R., Caudill S.P., Gunter E.W. Retinol, alpha-tocopherol, lutein/zeaxanthin, beta-cryptoxanthin, lycopene, alpha-carotene, trans-beta-carotene, and four retinyl esters in serum determined simultaneously by reversed-phase HPLC with multiwavelength detection. Clin. Chem. 1994;40:411–416. [PubMed] [Google Scholar]
- 38.Milne D.B., Botnen J. Retinol, alpha-tocopherol, lycopene, and alpha- and beta-carotene simultaneously determined in plasma by isocratic liquid chromatography. Clin. Chem. 1986;32:874–876. [PubMed] [Google Scholar]
- 39.Siluk D., Oliveira R.V., Esther-Rodriguez-Rosas M., Ling S., Bos A., Ferrucci L., Wainer I.W. A validated liquid chromatography method for the simultaneous determination of vitamins A and E in human plasma. J. Pharm. Biomed. Anal. 2007;44:1001–1007. doi: 10.1016/j.jpba.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodman D.S., Huang H.S. Biosynthesis of vitamin A with rat intestinal enzymes. Science. 1965;149:879–880. doi: 10.1126/science.149.3686.879. [DOI] [PubMed] [Google Scholar]
- 42.Goodman D.S., Huang H.S., Shiratori T. Mechanism of the biosynthesis of vitamin A from beta-carotene. J. Biol. Chem. 1966;241:1929–1932. [PubMed] [Google Scholar]
- 43.Lakshman M.R., Okoh C. Enzymatic conversion of all-trans-beta-carotene to retinal. Methods Enzymol. 1993;214:256–269. doi: 10.1016/0076-6879(93)14070-y. [DOI] [PubMed] [Google Scholar]
- 44.Pennacchio L.A., Olivier M., Hubacek J.A., Cohen J.C., Cox D.R., Fruchart J.C., Krauss R.M., Rubin E.M. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- 45.Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talmud P.J., Hawe E., Martin S., Olivier M., Miller G.J., Rubin E.M., Pennacchio L.A., Humphries S.E. Relative contribution of variation within the APOC3/A4/A5 gene cluster in determining plasma triglycerides. Hum. Mol. Genet. 2002;11:3039–3046. doi: 10.1093/hmg/11.24.3039. [DOI] [PubMed] [Google Scholar]
- 47.Weedon M.N., Lango H., Lindgren C.M., Wallace C., Evans D.M., Mangino M., Freathy R.M., Perry J.R., Stevens S., Hall A.S. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M., Berndt S.I., Elliott A.L., Jackson A.U., Lamina C. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2008;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blomhoff R., Green M.H., Green J.B., Berg T., Norum K.R. Vitamin A metabolism: New perspectives on absorption, transport, and storage. Physiol. Rev. 1991;71:951–990. doi: 10.1152/physrev.1991.71.4.951. [DOI] [PubMed] [Google Scholar]
- 50.Lindqvist A., Sharvill J., Sharvill D.E., Andersson S. Loss-of-function mutation in carotenoid 15,15′-monooxygenase identified in a patient with hypercarotenemia and hypovitaminosis A. J. Nutr. 2007;137:2346–2350. doi: 10.1093/jn/137.11.2346. [DOI] [PubMed] [Google Scholar]
- 51.Harrison E.H., Hussain M.M. Mechanisms involved in the intestinal digestion and absorption of dietary vitamin A. J. Nutr. 2001;131:1405–1408. doi: 10.1093/jn/131.5.1405. [DOI] [PubMed] [Google Scholar]
- 52.Ribaya-Mercado J.D., Solon F.S., Dallal G.E., Solomons N.W., Fermin L.S., Mazariegos M., Dolnikowski G.G., Russell R.M. Quantitative assessment of total body stores of vitamin A in adults with the use of a 3-d deuterated-retinol-dilution procedure. Am. J. Clin. Nutr. 2003;77:694–699. doi: 10.1093/ajcn/77.3.694. [DOI] [PubMed] [Google Scholar]
- 53.Semba R. Humana Press; Totowa, NJ: 2007. Handbook of Nutrition and Oftalmology. [Google Scholar]
- 54.During A., Hussain M.M., Morel D.W., Harrison E.H. Carotenoid uptake and secretion by CaCo-2 cells: Beta-carotene isomer selectivity and carotenoid interactions. J. Lipid Res. 2002;43:1086–1095. doi: 10.1194/jlr.m200068-jlr200. [DOI] [PubMed] [Google Scholar]
- 55.Girona J., Guardiola M., Cabre A., Manzanares J.M., Heras M., Ribalta J., Masana L. The apolipoprotein A5 gene −1131T→C polymorphism affects vitamin E plasma concentrations in type 2 diabetic patients. Clin. Chem. Lab. Med. 2008;46:453–457. doi: 10.1515/CCLM.2008.110. [DOI] [PubMed] [Google Scholar]
- 56.Timpson N.J., Lawlor D.A., Harbord R.M., Gaunt T.R., Day I.N., Palmer L.J., Hattersley A.T., Ebrahim S., Lowe G.D., Rumley A. C-reactive protein and its role in metabolic syndrome: mendelian randomisation study. Lancet. 2005;366:1954–1959. doi: 10.1016/S0140-6736(05)67786-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


