Abstract
Low lean body mass (LBM) is related to a series of health problems, such as osteoporotic fracture and sarcopenia. Here we report a genome-wide association (GWA) study on LBM variation, by using Affymetrix 500K single-nucleotide polymorphism (SNP) arrays. In the GWA scan, we tested 379,319 eligible SNPs in 1,000 unrelated US whites and found that two SNPs, rs16892496 (p = 7.55 × 10−8) and rs7832552 (p = 7.58 × 10−8), within the thyrotropin-releasing hormone receptor (TRHR) gene were significantly associated with LBM. Subjects carrying unfavorable genotypes at rs16892496 and rs7832552 had, on average, 2.70 and 2.55 kg lower LBM, respectively, compared to those with alternative genotypes. We replicated the significant associations in three independent samples: (1) 1488 unrelated US whites, (2) 2955 Chinese unrelated subjects, and (3) 593 nuclear families comprising 1972 US whites. Meta-analyses of the GWA scan and the replication studies yielded p values of 5.53 × 10−9 for rs16892496 and 3.88 × 10−10 for rs7832552. In addition, we found significant interactions between rs16892496 and polymorphisms of several other genes involved in the hypothalamic-pituitary-thyroid and the growth hormone-insulin-like growth factor-I axes. Results of this study, together with the functional relevance of TRHR in muscle metabolism, support the TRHR gene as an important gene for LBM variation.
Main Text
Loss and function impairment of skeletal muscle in the elderly is related to a series of diseases or health problems, such as sarcopenia,1 mobility limitation, osteoporosis (MIM 166710), higher risk of fracture, impaired protein balance, dyslipidemia (MIM 151660), obesity (MIM 601665), insulin resistance, overall frailty, and increased mortality.2,3 Lean body mass (LBM) measured by dual energy X-ray absorptionmetry (DXA) is a good index for quantity and quality of skeletal muscle.4 LBM is under strong genetic determination with heritability ranging from 52% to 84%.5–7 However, specific genes underlying variation in LBM are largely unknown.
Here we report a genome-wide association (GWA) study for LBM by using Affymetrix 500K SNP arrays in a sample of 1000 unrelated US whites. The interested associations were further replicated in three independent samples, including an unrelated US white sample comprising 1488 subjects, 593 nuclear families comprising 1972 US whites, and an unrelated Chinese sample comprising 2955 subjects.
All our study subjects of US whites were identified from an established cohort containing ∼6000 subjects recruited from the Midwestern US. This cohort was initiated to search quantitative trait loci and/or genes underlying body compositions (bone mass for osteoporosis, fat body mass for obesity, and lean body mass for sarcopenia studies). The inclusion and exclusion criteria have been described in detail elsewhere.8 The Chinese sample contains 2955 healthy Chinese Han adults living in Changsha City, China. The same inclusion and exclusion criteria used for the white subjects were applied in the recruitment of the Chinese sample. All studies were approved by local Institutional Review Boards. Signed informed-consent documents were obtained from all study participants.
LBM and fat body mass were measured with Hologic DXA 4500 machines (Hologic Inc., Bedford, MA) for all the study samples. Anthropometric measures and a structured questionnaire covering lifestyle, diet, family information, medical history, etc. were obtained for all the study subjects. Body mass index (BMI) was calculated as the ratio of weight to square of height in unit of kg/m2. The basic characteristics of all the four study samples are provided in Table 1.
Table 1.
Basic Characteristics of the Study Samples
| Sample and Trait | Males | Females |
|---|---|---|
| Initial GWA study | n = 492 | n = 481 |
| Age (years) | 50.52 (18.89) | 50.13 (17.13) |
| Height (m) | 1.78 (0.070) | 1.64 (0.065) |
| Lean body mass (kg) | 63.67 (8.22) | 43.49 (6.69) |
| Fat body mass (kg) | 23.46 (8.88) | 26.92 (10.33) |
| BMI (kg/m2) | 28.92 (4.30) | 27.29 (5.98) |
| Replication study in US white population | n = 659 | n = 829 |
| Age (years) | 63.18 (10.27) | 61.71 (10.72) |
| Height (m) | 1.77 (0.06) | 1.62 (0.07) |
| Lean body mass (kg) | 65.09 (9.21) | 45.16 (6.84) |
| Fat body mass (kg) | 25.06 (8.04) | 28.78 (9.79) |
| BMI (kg/m2) | 29.19 (4.50) | 27.83 (5.65) |
| Replication study in Chinese population | n = 1437 | n = 1518 |
| Age (years) | 30.53 (12.54) | 35.44 (15.74) |
| Height (m) | 1.69 (0.06) | 1.58 (0.05) |
| Lean body mass (kg) | 51.80 (5.76) | 37.39 (4.21) |
| Fat body mass (kg) | 11.33 (5.22) | 15.56 (5.07) |
| BMI (kg/m2) | 22.24 (2.90) | 21.38 (3.15) |
| Replication study in US white families | n = 570 | n = 1402 |
| Age (years) | 52.89 (16.22) | 47.45 (15.24) |
| Height (m) | 1.78 (0.08) | 1.64 (0.06) |
| Lean body mass (kg) | 65.88 (9.37) | 45.72 (7.13) |
| Fat body mass (kg) | 23.24 (8.92) | 26.01 (10.42) |
| BMI (kg/m2) | 28.83 (6.30) | 26.84 (5.91) |
Note: Data presented are unadjusted means (SD).
For the GWA scan, genotyping was performed for 1000 US whites with the GeneChip Human Mapping 500K Array Set (Affymetrix, Santa Clara, CA) by the Vanderbilt Microarray Shared Resource (Vanderbilt University Medical Center, Nashville, TN) with the standard protocol recommended by the manufacturer. Genotyping calls were determined with the DM algorithm9 with a 0.33 p value setting as well as with the BRLMM algorithm.10 DM calls were used for quality control of the genotyping experiment, and unsatisfactory arrays were subject to regenotyping. Eventually, 997 subjects who had at least one array (Nsp or Sty) reaching 93% call rate were retained. Because of missing data of LBM or applied covariates among the 997 subjects, the effective sample size for the GWA scan is 973. The average call rate for the 973 analyzed subjects reached >95%. BRLMM calls were used for the association analyses. The final average BRLMM call rate in the GWA cohort reached 99.27%. Out of the initial full set of 500,568 SNPs, we discarded 32,961 SNPs with SNP-wised call rate <95%, 36,965 SNPs with allele frequencies deviating from Hardy-Weinberg equilibrium (HWE) (p < 0.001), and 51,323 SNPs with minor allele frequency (MAF) <1%. Therefore, the final SNP set for the GWA scan contained 379,319 SNPs, yielding a genomic marker spacing of ∼7.9 kb on average.
For replication studies in the US white samples, genotyping was performed by KBioscience (Herts, UK) with the technology of competitive allele-specific PCR (KASPar). The replication rate (duplicate concordance rate) was 99.7% for genotyping and the average call rate was 97.8%. For the replication study in the Chinese sample, SNP genotyping was performed with a primer-extension method with MALDI-TOF mass spectrometry on a MassARRAY system as suggested by the manufacturer (Sequenom, Inc., San Diego, CA). The replication rate is 99.2% and the average call rate is 97.2%.
To control potential population stratification that may lead to spurious association results, we used the software Structure 2.211 to investigate the population structure of the GWA cohort. With 200 randomly selected unlinked SNPs, all the 973 subjects were tightly clustered together under all the three assumed number of population strata (i.e., k = 2, 3, or 4), suggesting no significant population substructure.
For both the initial GWA scan and subsequent replication studies, stepwise regression was performed to screen the effects of age, sex, age2, age-by-sex interaction, and fat body mass on LBM variation. Age, sex, and fat body mass were significant effectors (p < 0.05) and raw LBM values were adjusted for these factors. The adjusted phenotype values, if departing from normal distribution, were further subjected to BoxCox transformation to ensure the normality with the software Minitab (Minitab Inc., State College, PA). F-tests were conducted to achieve the genotype-wise association tests for the unrelated samples, whereas the FBAT method was used for association tests in the family-based sample. All the association analyses were performed with the software HelixTree 5.3.1, in which FBAT was implemented.12 To further ensure robustness of the association tests, we also used EIGENSTRAT13 to perform GWA analyses and cross-check the results against those obtained with HelixTree. EIGENSTRAT can detect and correct for potential population admixture and differences in laboratory treatment among samples.13
We adopted the conservative Bonferroni method to account for the multiple-testing problem in the GWA study. Because 379,319 eligible SNPs were eventually tested, the significance level for the GWA scan was set as 1.32 × 10−7 (i.e., 0.05/379319). The quantile-quantile (Q-Q) plotting method was used to determine the significance level for “suggestive association.” The Q-Q plot was constructed by ranking the observed p values from smallest to largest and plotting them against the same ordered p values under the null hypothesis of no association (expected p values). We calculated the expected p values for 1000 times and superimposed them in the Q-Q plot to construct a 95% concentration band after the method proposed by Stirling.14 Deviations from the band in the Q-Q plot suggest that the corresponding associations are highly likely to be true ones;15 therefore, we took the p value at the point of deviation (p < 1.26 × 10−4, Figure S1 available online) as the threshold for suggestive associations.
Point estimates and their standard errors for per-allele effect sizes of the favorable alleles of interested SNPs on LBM were calculated through linear regression analyses with SAS 9.1 (SAS Institute Inc., Cary, NC). The effect estimate and standard error of the most significant SNP for each separate association analysis in the initial GWA scan and subsequent replication studies were combined in the meta-analysis via the inverse variance method implemented in the software Comprehensive Meta Analysis.
In the GWA scan, we identified two genome-level significant SNPs (p values of 7.55 × 10−8 and 7.58 × 10−8 for rs16892496 and rs7832552, respectively), and other 146 suggestive SNPs associated with LBM (Figure 1, Table S1). The two SNPs showing genome-level significant associations with LBM are located in the only intron of the thyrotropin-releasing hormone receptor (TRHR) gene (MIM 188545) (Figure 2). These two SNPs also achieved the strongest associations with LBM in EIGENSTRAT analyses with p values of 1.42 × 10−7 and 1.79 × 10−7 for rs16892496 and rs7832552, respectively. Interestingly, 15 other SNPs of the TRHR gene also showed suggestive associations with LBM (Table 2, Figure 2). The two most significant SNPs, rs16892496 and rs7832552, are in strong linkage disequilibrium (LD) (r2 = 0.98). Compared to subjects carrying TT or GT genotypes at rs16892496, those carrying the GG genotype had 2.70 kg higher LBM, on average. Similarly, subjects carrying the TT genotype at rs7832552 had 2.55 kg higher LBM compared to subjects with CC or CT genotypes.
Figure 1.
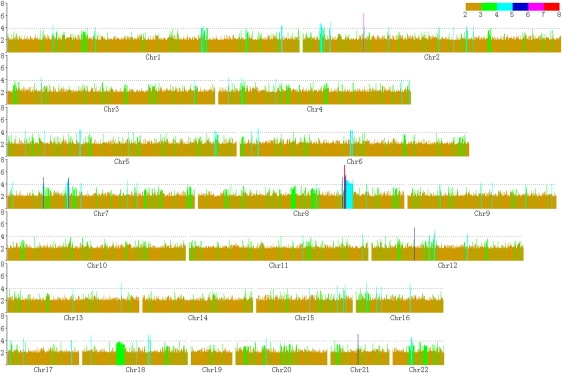
Results of the GWA Scan for Lean Body Mass
The y axis represents −log10P. Both height and colors of the bars correspond to p values of the SNPs, which are arranged along the x axis according to their physical position on chromosomes.
Figure 2.
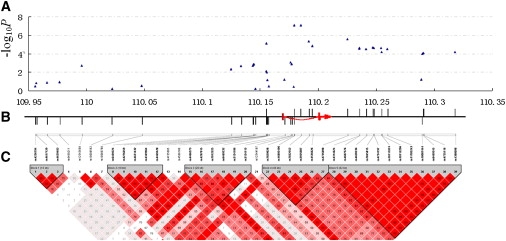
Association Signals and LD Structure of the TRHR Gene SNPs
(A) Association results for SNPs of the TRHR gene in the GWA scan. The y axis shows −log10P of the association results and the x axis shows the physical position (Mb) on chromosome 8.
(B) Position of the TRHR gene and the tested SNPs. The TRHR gene was represented with two red bars for exons and a red line for intron. The red arrow denotes the transcriptional direction of the TRHR gene. SNPs were illustrated by vertical lines. Lines above the horizontal axis represent SNPs showing significant and suggestive associations with LBM.
(C) The LD structure of the tested SNPs. LD pattern was analyzed in our GWA sample and plotted with the Haploview program.
Table 2.
Association Results for SNPs of the TRHR Gene in the GWA Scan
| SNP | Position | Role | Allelesa | MAFb | p Value | Effect Sizec | Favorable Alleled |
|---|---|---|---|---|---|---|---|
| rs4236794 | 109955588 | upstream | C/T | 0.37 | 0.3 | 0.043 (0.031) | T |
| rs4617129 | 109956715 | upstream | G/A | 0.33 | 0.14 | 0.034 (0.032) | A |
| rs4639483 | 109965846 | upstream | C/T | 0.38 | 0.12 | 0.055 (0.033) | T |
| rs6469211 | 109976702 | upstream | C/T | 0.34 | 0.11 | 0.038 (0.032) | T |
| rs12543698 | 109996108 | upstream | A/T | 0.2 | 1.86 × 10−3 | 0.028 (0.032) | A |
| rs10098562 | 110022238 | upstream | T/C | 0.04 | 0.6 | 0.036 (0.032) | T |
| rs10086780 | 110047705 | upstream | G/A | 0.29 | 0.28 | 0.021 (0.030) | G |
| rs6469224 | 110124863 | upstream | A/G | 0.46 | 4.69 × 10−3 | 0.086 (0.032) | G |
| rs4276659 | 110133336 | upstream | C/A | 0.44 | 1.94 × 10−3 | 0.081 (0.032) | A |
| rs4631432 | 110143381 | upstream | A/G | 0.4 | 1.73 × 10−3 | 0.096 (0.032) | G |
| rs4349964 | 110144137 | upstream | A/G | 0.41 | 1.38 × 10−3 | 0.094 (0.032) | G |
| rs4469428 | 110144284 | upstream | A/G | 0.41 | 1.23 × 10−3 | 0.082 (0.032) | G |
| rs4735085 | 110145588 | upstream | C/A | 0.32 | 0.6 | 0.033 (0.030) | A |
| rs4506180 | 110154994 | upstream | A/C | 0.5 | 7.87 × 10−3 | 0.046 (0.030) | A |
| rs4466373 | 110155042 | upstream | T/C | 0.21 | 7.41 × 10−6 | 0.061 (0.032) | T |
| rs4297015 | 110155444 | upstream | T/C | 0.18 | 9.69 × 10−3 | 0.028 (0.032) | C |
| rs7012225 | 110156031 | upstream | G/A | 0.18 | 0.07 | 0.042 (0.030) | G |
| rs6469232 | 110157417 | upstream | T/C | 0.11 | 0.3 | 0.031 (0.032) | C |
| rs3134106 | 110170951 | intron | A/C | 0.17 | 0.06 | 0.044 (0.032) | A |
| rs3134115 | 110175985 | intron | A/G | 0.4 | 8.69 × 10−4 | 0.079 (0.032) | G |
| rs12544197 | 110177001 | intron | A/G | 0.48 | 1.17 × 10−3 | 0.037 (0.032) | A |
| rs7829028 | 110178706 | intron | T/C | 0.0971 | 0.36 | 0.026 (0.032) | C |
| rs16892496 | 110179027 | intron | G/T | 0.32 | 7.55 × 10−8 | 0.107 (0.032) | G |
| rs7832552 | 110184852 | intron | T/C | 0.32 | 7.58 × 10−8 | 0.102 (0.030) | T |
| rs3925087 | 110191854 | intron | C/T | 0.36 | 4.34 × 10−6 | 0.096 (0.032) | C |
| rs4546626 | 110194938 | intron | G/T | 0.36 | 1.37 × 10−5 | 0.077 (0.032) | G |
| rs4735098 | 110225067 | downstream | G/A | 0.38 | 2.57 × 10−6 | 0.082 (0.030) | G |
| rs4314624 | 110235192 | downstream | C/G | 0.35 | 2.42 × 10−5 | 0.078 (0.032) | C |
| rs4607576 | 110235212 | downstream | T/C | 0.35 | 2.65 × 10−5 | 0.078 (0.032) | T |
| rs4628236 | 110241321 | downstream | T/C | 0.35 | 3.09 × 10−5 | 0.073 (0.032) | T |
| rs7845815 | 110247020 | downstream | A/G | 0.35 | 2.04 × 10−5 | 0.075 (0.032) | A |
| rs4734197 | 110247681 | downstream | G/A | 0.34 | 2.39 × 10−5 | 0.079 (0.032) | G |
| rs10111874 | 110254194 | downstream | G/A | 0.35 | 2.35 × 10−5 | 0.075 (0.032) | G |
| rs10112296 | 110254469 | downstream | T/C | 0.35 | 6.20 × 10−5 | 0.072 (0.032) | T |
| rs11785243 | 110259359 | downstream | A/G | 0.36 | 3.12 × 10−5 | 0.075 (0.032) | A |
| rs10087444 | 110288911 | downstream | A/G | 0.44 | 0.06 | 0.035 (0.030) | G |
| rs6469245 | 110289983 | downstream | C/G | 0.41 | 9.42 × 10−5 | 0.076 (0.030) | C |
| rs4735116 | 110290780 | downstream | G/T | 0.41 | 8.51 × 10−5 | 0.075 (0.030) | G |
| rs1380098 | 110317808 | downstream | A/G | 0.41 | 6.28 × 10−5 | 0.069 (0.032) | A |
Note: The two SNPs significantly associated with lean body mass after Bonferroni correction in the GWA scan are shown in italics and bold.
The former allele represents the minor allele of each locus in our GWA sample.
Minor allele frequency calculated in our GWA sample.
Per-allele effect size of the favorable allele is expressed by beta coefficients derived from linear regression analyses.
Subjects with more favorable alleles generally have higher values of lean body mass than subjects having alternative genotypes in the population.
The replication studies largely confirmed the significant findings in the GWA scan (Table 3). The association of rs16892496 with LBM was replicated in the independent unrelated US white sample and unrelated Chinese sample (p = 0.018 and 0.013, respectively; Table 3). The SNP rs7832552 was consistently and significantly associated with LBM in all the three replication samples. Meta-analyses of the initial GWA scan and replication studies achieved the combined p values of 5.53 × 10−9 for rs16892496 and 3.88 × 10−10 for rs7832552, respectively. A forest plot for the association between rs16892496 and LBM is given in Figure 3.
Table 3.
Association Results between Lean Body Mass and the Two Significant TRHR SNPs in the GWA Scan and Three Replication Studies and Meta-analyses
| SNP | Discovery Scan (n = 973) |
Replication Studiesa |
Combinedc(n = 7415) | ||
|---|---|---|---|---|---|
| Sample 1 (n = 1488) | Sample 2 (n = 2955) | Sample 3b(n = 1972) | |||
| rs16892496 | 7.55 × 10−8 (0.107 ± 0.032) | 0.018 (0.112 ± 0.038) | 0.013 (0.074 ± 0.024) | 0.083 (0.087 ± 0.029) | 5.53 × 10−9 (0.090 ± 0.015) |
| rs7832552 | 7.58 × 10−8 (0.102 ± 0.030) | 0.0056 (0.105 ± 0.036) | 0.012 (0.076 ± 0.024) | 0.015 (0.081 ± 0.028) | 3.88 × 10−10 (0.061 ± 0.014) |
Values in the table are p values followed by the point estimates of effect sizes expressed by beta coefficients ± standard errors.
The replication studies were performed in three samples: Sample 1, the independent unrelated US white sample; Sample 2, the unrelated Chinese sample; Sample 3, the family-based US white sample.
The effect sizes for sample 3 were calculated in founders.
Accumulative p values and beta coefficients were generated under the random-effect model.
Figure 3.
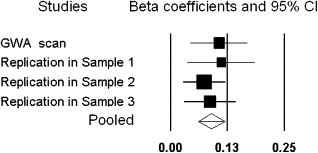
Forest Plot for the Association between rs16892496 and LBM
Black boxes denote point estimates for effect sizes (beta coefficients) in corresponding association studies, and the sizes of the box are proportional to the inverse variance weight of the estimate in the meta-analysis. Horizontal lines represent 95% confidence intervals (CIs). The diamond represents the accumulative beta coefficient computed under the random-effect model, with the 95% CI given by its width. The scale of the plot was indicated by the bottom values. Samples 1–3 represent the three samples used for the replication studies, with #1 for the unrelated US white sample, #2 for the unrelated Chinese sample, and #3 for the family-based US white sample.
TRHR encodes the thyrotropin-releasing hormone receptor, which belongs to the G protein-coupled receptor 1 family. Thyrotropic-releasing hormone (TRH) is a tripeptide (Glu-His-Pro) hormone secreted by the hypothalamus. TRH exerts its effect by binding to TRHR on the surface of pituitary thyrotrophs. The primary consequence of TRH:TRHR binding is activation of the inositol phospholipid-calcium-protein kinase C transduction pathway, which, in turn, stimulates secretion of thyroid-stimulating hormone (TSH) and prolactin (PRL). The TSH response to TRHR is the first step in the hormonal cascade of hypothalamic-pituitary-thyroid axis (HPTA) that eventually leads to the release of thyroxin, which is important in the development of vertebrate skeletal muscle.16 Mutations in the TRHR gene may decrease affinity of TRHR for TRH and result in central hypothyroidism,17 which causes impaired expression of myosin heavy chain (MHC) isoforms18 and diminished muscle cross-sectional areas.19 Furthermore, thyroxin is necessary for full anabolic action of the growth hormone-insulin-like growth factor-I (GH-IGF1) axis,20 which plays an important role in muscle protein balance and adaptative changes to load.21,22
Motivated by known physiological relevance of TRHR to the HPTA and GH-IGF1 pathways, we further performed interaction analyses for SNP rs16892496 in the TRHR gene, which showed the most significant association with LBM in the GWA scan, with 912 SNPs in other 33 genes putatively involved in the HPTA and GH-IGF1 pathways (Table S2). The most significant interaction with rs16892496 was detected for SNP rs12474719 (p = 6.04 × 10−11) of the insulin-like growth factor binding protein 5 (IGFBP5) gene. A joint modeling of rs16892496 and rs12474719 increased the LBM variation explained solely by rs16892496 from 2.29% to 2.91%.
To our knowledge, this is the first GWA study for LBM variation. We identified a significant association between the TRHR gene and LBM variation. The association findings were further supported by three independent replication studies in both US white subjects and Chinese Han population. In addition, we found that LBM variation is influenced by interactions between TRHR and several other genes involved in the HPTA and GH-IGF1 pathways.
The identified association between TRHR SNPs and LBM are unlikely to be artifacts. First, conservative Bonferroni correction was used to claim significant associations in the initial GWA scan. In addition, besides the two SNPs achieving genome-level association signal with LBM, 15 other SNPs of the TRHR gene got suggestive association signals in the GWA scan. Thus, individual genotyping errors are unlikely to be responsible for these associations. Moreover, we strictly controlled potential population stratification by using Structure and EIGENSTRAT. Genomic control analyses23 showed a small inflation factor of 1.036. Most importantly, the significant associations were replicated in three independent samples.
LBM accounts for ∼60% or more of body weight and thus may significantly contribute to variation in BMI, an index commonly used for obesity.24 Interestingly, previous linkage studies found that a locus at 8q23, which spans the TRHR gene, was linked to BMI.25,26 We speculate that the observed linkage may be partially attributable to the association between TRHR and LBM. As a support of the speculated effect of the GH-IGF1 pathway on LBM, the insulin-like growth factor 1 receptor (IGF1R) gene and the GHRH gene were also linked to fat-free mass in previous studies.27
In summary, our GWA scan and multiple replication studies, in conjunction with the known functional involvement of TRHR in muscle metabolism, suggest that polymorphisms in TRHR gene, and possibly other genes in the HPTA and GH-IGF1 pathways, significantly contribute to LBM variation. The mechanisms underlying the observed associations merit further investigation.
Acknowledgments
Investigators of this work were partially supported by grants from NIH (R01 AR050496-01, R21 AG027110, R01 AG026564, R21 AA015973, and P50 AR055081).The study also benefited from grants from National Science Foundation of China, Huo Ying Dong Education Foundation, Hunan Province, Xi'an Jiaotong University, and the Ministry of Education of China. The authors declare that they have no conflict of interest.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Comprehensive Meta Analysis, http://www.meta-analysis.com/
EIGENSTRAT, http://genepath.med.harvard.edu/∼reich/EIGENSTRAT.htm
Haploview program, http://www.broad.mit.edu/mpg/haploview
KBioscience genotyping technology, http://www.kbioscience.co.uk
NetAffx Analysis Center, http://www.affymetrix.com/analysis/index.affx
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
Structure, http://pritch.bsd.uchicago.edu/software.html
References
- 1.Holloszy, J.O. (1995). Workshop on sarcopenia: muscle atrophy in old age. J. Gerontol. A Biol. Sci. Med. Sci. 50 Special issue, 1–161. [PubMed]
- 2.Sipila S., Heikkinen E., Cheng S., Suominen H., Saari P., Kovanen V., Alen M., Rantanen T. Endogenous hormones, muscle strength, and risk of fall-related fractures in older women. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:92–96. doi: 10.1093/gerona/61.1.92. [DOI] [PubMed] [Google Scholar]
- 3.Karakelides H., Sreekumaran Nair K. Sarcopenia of aging and its metabolic impact. Curr. Top. Dev. Biol. 2005;68:123–148. doi: 10.1016/S0070-2153(05)68005-2. [DOI] [PubMed] [Google Scholar]
- 4.Hansen R.D., Raja C., Aslani A., Smith R.C., Allen B.J. Determination of skeletal muscle and fat-free mass by nuclear and dual-energy x-ray absorptiometry methods in men and women aged 51-84 y (1-3) Am. J. Clin. Nutr. 1999;70:228–233. doi: 10.1093/ajcn.70.2.228. [DOI] [PubMed] [Google Scholar]
- 5.Hsu F.C., Lenchik L., Nicklas B.J., Lohman K., Register T.C., Mychaleckyj J., Langefeld C.D., Freedman B.I., Bowden D.W., Carr J.J. Heritability of body composition measured by DXA in the diabetes heart study. Obes. Res. 2005;13:312–319. doi: 10.1038/oby.2005.42. [DOI] [PubMed] [Google Scholar]
- 6.Arden N.K., Spector T.D. Genetic influences on muscle strength, lean body mass, and bone mineral density: a twin study. J. Bone Miner. Res. 1997;12:2076–2081. doi: 10.1359/jbmr.1997.12.12.2076. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen T.V., Howard G.M., Kelly P.J., Eisman J.A. Bone mass, lean mass, and fat mass: same genes or same environments? Am. J. Epidemiol. 1998;147:3–16. doi: 10.1093/oxfordjournals.aje.a009362. [DOI] [PubMed] [Google Scholar]
- 8.Deng H.W., Deng H., Liu Y.J., Liu Y.Z., Xu F.H., Shen H., Conway T., Li J.L., Huang Q.Y., Davies K.M. A genomewide linkage scan for quantitative-trait loci for obesity phenotypes. Am. J. Hum. Genet. 2002;70:1138–1151. doi: 10.1086/339934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di X., Matsuzaki H., Webster T.A., Hubbell E., Liu G., Dong S., Bartell D., Huang J., Chiles R., Yang G. Dynamic model based algorithms for screening and genotyping over 100 K SNPs on oligonucleotide microarrays. Bioinformatics. 2005;21:1958–1963. doi: 10.1093/bioinformatics/bti275. [DOI] [PubMed] [Google Scholar]
- 10.Rabbee N., Speed T.P. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22:7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- 11.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange C., DeMeo D.L., Laird N.M. Power and design considerations for a general class of family-based association tests: quantitative traits. Am. J. Hum. Genet. 2002;71:1330–1341. doi: 10.1086/344696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 14.Stirling W.D. Enhancements to aid interpretation of probability plots. Statistician. 1982;31:211–220. [Google Scholar]
- 15.Burton P.R., Clayton D.G., Cardon L.R., Craddock N., Deloukas P., Duncanson A., Kwiatkowski D.P., McCarthy M.I., Ouwehand W.H., Samani N.J. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsson L., Li X., Teresi A., Salviati G. Effects of thyroid hormone on fast- and slow-twitch skeletal muscles in young and old rats. J. Physiol. 1994;481:149–161. doi: 10.1113/jphysiol.1994.sp020426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collu R., Tang J., Castagne J., Lagace G., Masson N., Huot C., Deal C., Delvin E., Faccenda E., Eidne K.A. A novel mechanism for isolated central hypothyroidism: inactivating mutations in the thyrotropin-releasing hormone receptor gene. J. Clin. Endocrinol. Metab. 1997;82:1561–1565. doi: 10.1210/jcem.82.5.3918. [DOI] [PubMed] [Google Scholar]
- 18.Vadaszova A., Hudecova S., Krizanova O., Soukup T. Levels of myosin heavy chain mRNA transcripts and content of protein isoforms in the slow soleus muscle of 7 month-old rats with altered thyroid status. Physiol. Res. 2006;55:221–225. doi: 10.33549/physiolres.930861. [DOI] [PubMed] [Google Scholar]
- 19.Norenberg K.M., Herb R.A., Dodd S.L., Powers S.K. The effects of hypothyroidism on single fibers of the rat soleus muscle. Can. J. Physiol. Pharmacol. 1996;74:362–367. [PubMed] [Google Scholar]
- 20.Van den Berghe G., Baxter R.C., Weekers F., Wouters P., Bowers C.Y., Iranmanesh A., Veldhuis J.D., Bouillon R. The combined administration of GH-releasing peptide-2 (GHRP-2), TRH and GnRH to men with prolonged critical illness evokes superior endocrine and metabolic effects compared to treatment with GHRP-2 alone. Clin. Endocrinol. (Oxf.) 2002;56:655–669. doi: 10.1046/j.1365-2265.2002.01255.x. [DOI] [PubMed] [Google Scholar]
- 21.Lang C.H., Frost R.A. Role of growth hormone, insulin-like growth factor-I, and insulin-like growth factor binding proteins in the catabolic response to injury and infection. Curr. Opin. Clin. Nutr. Metab. Care. 2002;5:271–279. doi: 10.1097/00075197-200205000-00006. [DOI] [PubMed] [Google Scholar]
- 22.Gibney J., Healy M.L., Sonksen P.H. The growth hormone/insulin-like growth factor-I axis in exercise and sport. Endocr. Rev. 2007;28:603–624. doi: 10.1210/er.2006-0052. [DOI] [PubMed] [Google Scholar]
- 23.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 24.Liu X.G., Zhao L.J., Liu Y.J., Xiong D.H., Recker R.R., Deng H.W. The MTHFR gene polymorphism is associated with lean body mass but not fat body mass. Hum. Genet. 2008;123:189–196. doi: 10.1007/s00439-007-0463-7. [DOI] [PubMed] [Google Scholar]
- 25.Chagnon Y.C., Rice T., Perusse L., Borecki I.B., Ho-Kim M.A., Lacaille M., Pare C., Bouchard L., Gagnon J., Leon A.S. Genomic scan for genes affecting body composition before and after training in Caucasians from HERITAGE. J. Appl. Physiol. 2001;90:1777–1787. doi: 10.1152/jappl.2001.90.5.1777. [DOI] [PubMed] [Google Scholar]
- 26.Platte P., Papanicolaou G.J., Johnston J., Klein C.M., Doheny K.F., Pugh E.W., Roy-Gagnon M.H., Stunkard A.J., Francomano C.A., Wilson A.F. A study of linkage and association of body mass index in the Old Order Amish. Am. J. Med. Genet. C. Semin. Med. Genet. 2003;121:71–80. doi: 10.1002/ajmg.c.20005. [DOI] [PubMed] [Google Scholar]
- 27.Chagnon Y.C., Borecki I.B., Perusse L., Roy S., Lacaille M., Chagnon M., Ho-Kim M.A., Rice T., Province M.A., Rao D.C. Genome-wide search for genes related to the fat-free body mass in the Quebec family study. Metabolism. 2000;49:203–207. doi: 10.1016/s0026-0495(00)91299-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


