Abstract
The Phoenicians were the dominant traders in the Mediterranean Sea two thousand to three thousand years ago and expanded from their homeland in the Levant to establish colonies and trading posts throughout the Mediterranean, but then they disappeared from history. We wished to identify their male genetic traces in modern populations. Therefore, we chose Phoenician-influenced sites on the basis of well-documented historical records and collected new Y-chromosomal data from 1330 men from six such sites, as well as comparative data from the literature. We then developed an analytical strategy to distinguish between lineages specifically associated with the Phoenicians and those spread by geographically similar but historically distinct events, such as the Neolithic, Greek, and Jewish expansions. This involved comparing historically documented Phoenician sites with neighboring non-Phoenician sites for the identification of weak but systematic signatures shared by the Phoenician sites that could not readily be explained by chance or by other expansions. From these comparisons, we found that haplogroup J2, in general, and six Y-STR haplotypes, in particular, exhibited a Phoenician signature that contributed > 6% to the modern Phoenician-influenced populations examined. Our methodology can be applied to any historically documented expansion in which contact and noncontact sites can be identified.
Main Text
The Phoenicians were a distinctive and independent civilization that dominated the Mediterranean Sea during the first millennium BCE, emerging from a coastal section of the Eastern Mediterranean, including the four main Bronze Age maritime cities of Tyre, Sidon, Byblos, and Arwad and located in the modern countries of Lebanon and southern Syria. From here, their maritime expertise allowed them to establish a trading empire throughout the Mediterranean and beyond.1–6 Their strategy included the establishment of settled colonies, foremost among which was Carthage in modern Tunisia, and many trading posts, where they stayed for shorter periods4 (Figure 1A). Their activities were recorded by contemporary writers, including the Egyptians, the Greeks, Biblical sources, Strabo, Pliny the Elder, and Avienus, and the remains of their cities and trading goods have been documented extensively by archaeologists.6 Thus, we have a good understanding of their origins and spread from historical sources.
Figure 1.
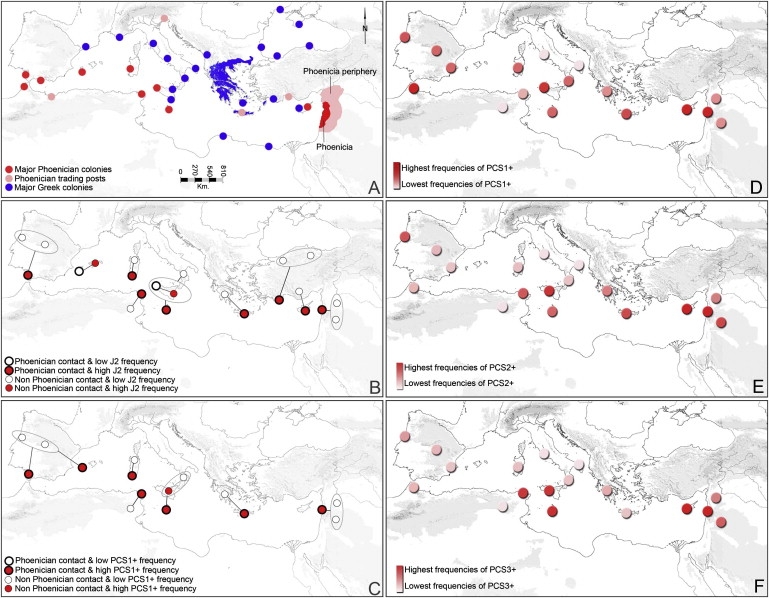
Geographical context of the Phoenician and Greek expansions
(A) Maritime expansions of the Phoenicians (11th century BCE) and Greeks. Red: Phoenicia, Phoenician colonies; pink: Phoenician trading posts; blue: Greece and Greek colonies.
(B) J2 haplogroup frequency comparisons between Phoenician contact regions (thick borders) and nearby non-contact regions (thin borders). Lines indicate paired haplogroup comparisons between two sites. An ellipse indicates a site with multiple population samples. Colored circles indicate the higher haplogroup J2 frequency site in each pair.
(C) Phoenician Colonization Signal 1 (PCS1+) haplotype frequency comparisons between Phoenician contact regions (thick borders) and nearby non-contact regions (thin borders). Lines indicate paired haplotype comparisons between two sites. An ellipse indicates a site with multiple population samples. Colored circles represent the higher PCS1+ frequency site in each pair.
(D–F) Geographical distribution of the PCS1+ (D), PCS2+ (E), and PCS3+ (F) haplotypes in the Mediterranean region. The PCS+ central haplotypes are shown in Table 2. Higher color intensities indicate higher haplotype frequencies; absolute frequencies are given in Table 3. Note the highly enriched coastal and island distribution of these haplotypes and the prominence of all in the Levant.
We set out to complement this historical information by searching for Phoenician genetic traces within modern populations. We chose the nonrecombining region of the Y chromosome for this purpose, because its male specificity means that it would have been carried by the predominantly male Phoenician traders, and its high level of geographical differentiation provides the greatest chance of recognizing colonization events.7 Human genetic history, however, can be viewed as a palimpsest, in which multiple events from different times but with similar geographical patterns are superimposed. Expansions from the Eastern Mediterranean could include the initial peopling by modern humans during the Paleolithic era, the subsequent Neolithic-era transition originating in the Fertile Crescent ∼8000 BCE, and later events, such as the Greek expansion or the Jewish Diaspora. All of these, and possibly additional events unrecorded in history, could result in broadly similar genetic patterns with an origin in or near the Levant and decreasing gradients toward the west. Several previous studies have identified Y-chromosomal types showing gradients originating in the Near East8–11 and have sometimes linked them to the Phoenicians,12 but further work is needed to distinguish between the general pattern and the specific Phoenician contribution.
Therefore, we have developed a strategy for identifying a geographical genetic pattern associated with a specific historical expansion, rather than an overall geographical gradient. The key to this was the use of historically documented locations of greater or lesser contact—in our case, Phoenician locations—matched approximately for distance from the source. Such paired locations would be expected to share general genetic patterns, reflecting the sum of multiple events, but to differ specifically in their Phoenician genetic influence if genetic transfer had taken place. Other historical expansions would have involved different locations of greater and lesser contact and so would not have produced a distinct geographically detailed signal in the same populations at this fine level of resolution. To assess the significance of any pattern that we might detect, we developed a two-fold analytic approach: first, a statistical component, the investigation of whether such a pattern might have originated by chance; and second, an empirical component, the application of the same analytical strategy to additional data sets not expected to differ in their Phoenician influence, representing instead the general Neolithic spread or the independent Greek expansion. Haplotypes that would not be expected to exhibit the specific short-ranged variational features by chance and that did not correspond to other known expansions could be considered as potentially Phoenician. With the very active intervening history, we cannot reasonably expect to identify a statistically significant signature linking the Phoenician homeland to every identified colonization region. However, colonization is expected to have produced a noisy but systematic trace of signatures. This study presents a method that identifies significant geographical preponderance of such signatures in order to decipher the genetic palimpsest.
In order to apply this strategy, we therefore needed to (1) choose suitable population sample sites for investigating Phoenician and other expansions, (2) generate or identify from available sources Y-chromosomal data sets from the chosen sites, (3) develop our test methodology, and, finally, (4) consider the broader significance of any signals that emerged from the chosen population sites.
When choosing populations, we considered that trade-driven colonization would have mediated the genetic legacy of the Phoenician expansion. Minor colonization sites were established for the servicing of ships en route, as well as for connecting with and guarding interests in foreign trade centers. This servicing was necessary for the expansion of trade throughout the Mediterranean basin with the maritime technology of the first millennium BCE and established the regional variations that we seek to detect. Carthage emerged as the dominant Central Mediterranean colony, connecting western-metals trade to the rest of the wealthy Mediterranean trading sites. Opportunity for establishing Phoenician colonization was greatest and most lasting in minimally occupied regions. Documented major colonies and trading posts are summarized in Figure 1A. We constructed pairs of testing sites generally orthogonal to the anticipated background of the Neolithic gradient originating in the Levant, resulting in localized groups of tests. The Phoenician-influenced regions selected were, thus, the coastal Lebanese Phoenician Heartland and the broader area of the rest of the Levant (the “Phoenician Periphery”); then Cyprus and South Turkey; then Crete; then Malta and East Sicily; then South Sardinia, Ibiza, and Southern Spain; and, finally, Coastal Tunisia and cities like Tingris in Morocco (Figures 1B and 1C). For each, we identified nearby sites of lesser or no Phoenician contact. Examples of the comparisons used thus include heartland versus periphery, colony versus trading center, and trading center versus noncontact sites.
In addition, we sought to discriminate Phoenician candidate lineages from those spread by other colonization expansions affecting many of the same islands and regions. We constructed a Neolithic-expansion test set by choosing paired sites from the region, both of which lacked known Phoenician contact, and comparing the site closer to the Levant with that farther away (Table 1 and Table S3, available online). The colonization by Greeks and later groups occurred largely into regions still unoccupied by the Phoenicians, yielding colonial segregation; Greek sites are also shown in Figure 1A. We wished to design similar tests to evaluate a potential signature of the Jewish Diaspora. This, however, proved problematic. At the time of the Roman destruction of Herod's Temple in 70 CE, there were already more Jews living outside than within Israel.13 The dispersals through time and space were complex, with communities being established and dispersed, sometimes on multiple occasions. It was, thus, difficult to identify any locality where significant Jewish settlement did not occur for at least some period.14 Therefore, our approach was not suitable for identifying lineages associated with the Jewish Diaspora, which has already been well studied with the use of other approaches.15
Table 1.
Y-SNP Haplogroup Colonization-Site Gradient Tests with Aggregate Scores for Phoenician Colonies, Neolithic Backgrounda, and Greek Colonies
| Tests | E3b | G | I | J∗(xJ2) | J2 | K2 | L | R1a | R1b |
|---|---|---|---|---|---|---|---|---|---|
| Phoenician Test Sites | |||||||||
| Heartland versus Periphery | 0.574 | 0.986 | 0.012 | 1.000 | 0.011 | 0.137 | 0.0002 | 0.894 | 0.003 |
| −1 | −1 | +1 | −1 | +1 | +1 | +1 | −1 | +1 | |
| Homeland versus Levant | 0.968 | 0.833 | 0.260 | 1.000 | 0.000 | 0.188 | 0.0002 | 0.033 | 0.001 |
| −1 | −1 | +1 | −1 | +1 | +1 | +1 | +1 | +1 | |
| Cyprus versus S. Turkey | 0.249 | 1.0 | 0.337 | 0.963 | 0.150 | 0.586 | − | 0.957 | 0.973 |
| +1 | −1 | +1 | −1 | +1 | +1 | − | −1 | −1 | |
| S. Turkey versus N. Turkey | 0.278 | 0.860 | 0.794 | 0.233 | 0.449 | 0.541 | 1.000 | 0.194 | 0.417 |
| +1 | −1 | −1 | +1 | +1 | +1 | −1 | +1 | +1 | |
| Lowland Crete versus Lasithi Plateau | 0.211 | 0.685 | 0.436 | 0.858 | 0.000 | 0.905 | − | 0.988 | 1.000 |
| +1 | −1 | +1 | −1 | +1 | −1 | − | −1 | −1 | |
| Crete versus Greece | 0.994 | 0.783 | 0.989 | 0.624 | 0.0003 | 0.338 | − | 0.897 | 0.145 |
| −1 | −1 | −1 | +1 | +1 | +1 | − | −1 | +1 | |
| Malta versus Sicily | 1.000 | 0.561 | 0.434 | 1.000 | 0.016 | 1.000 | − | 0.083 | 0.653 |
| −1 | 0 | +1 | −1 | +1 | −1 | − | +1 | −1 | |
| W. Sicily versus E. Sicily | 0.962 | 0.035 | 0.0711 | 0.0893 | 0.973 | 0.814 | − | 0.666 | 0.131 |
| −1 | +1 | +1 | −1 | −1 | −1 | − | 0 | +1 | |
| Sicily versus S. Italy | 0.816 | 0.936 | 0.376 | 0.573 | 0.208 | 0.570 | − | 0.788 | 0.692 |
| −1 | −1 | +1 | +1 | +1 | +1 | − | −1 | +1 | |
| S. Sardinia versus N. Sardinia | 0.736 | 0.935 | 0.123 | 0.141 | 0.206 | − | − | 0.677 | 0.561 |
| −1 | −1 | +1 | +1 | +1 | − | − | +1 | −1 | |
| Ibiza versus Mallorca & Minorca | 0.846 | 0.046 | 0.956 | 1.000 | 0.842 | 0.000 | − | 1.000 | 0.941 |
| −1 | +1 | −1 | −1 | −1 | +1 | − | −1 | −1 | |
| S. Spain versus Valencia | 0.767 | 0.317 | 0.896 | 0.738 | 0.142 | 1.000 | − | 0.738 | 0.539 |
| −1 | +1 | −1 | −1 | −1 | −1 | − | −1 | +1 | |
| Contact Spain versus Iberia | 0.879 | 0.988 | 0.259 | 0.807 | 0.176 | 0.385 | − | 0.141 | 0.197 |
| −1 | −1 | +1 | 0 | +1 | +1 | − | +1 | +1 | |
| Coastal Tunisa versus Inland Tunisia | 0.890 | − | − | − | 0.0013 | 0.863 | − | 1.000 | 0.952 |
| −1 | − | − | − | +1 | −1 | − | −1 | −1 | |
| α = 0.05 | 1.000 | 0.135 | 0.486 | 1.000 | 3.3 × 10−5 | 0.486 | 0.0073 | 0.512 | 0.153 |
| α = 0.30 | 0.839 | 0.936 | 0.579 | 0.798 | 2.5 × 10−4 | 0.420 | 0.216 | 0.839 | 0.644 |
| Δf | 0.993 | 0.987 | 0.133 | 0.927 | 0.0287 | 0.290 | 0.5 | 0.910 | 0.395 |
| Control Test Sites | |||||||||
| Turkey #5 versus Turkey #3 | 0.555 | 0.691 | 0.421 | 0.794 | 0.275 | 0.421 | 1.000 | 0.148 | 0.557 |
| +1 | −1 | +1 | +1 | +1 | +1 | −1 | +1 | +1 | |
| Turkey #8 versus Turkey #1 | 0.941 | 0.0252 | 0.866 | 0.987 | 0.982 | 0.366 | 0.601 | 0.750 | 0.315 |
| −1 | +1 | −1 | −1 | −1 | +1 | +1 | −1 | +1 | |
| Greece versus Albania | 0.578 | 0.655 | 0.988 | 0.944 | 0.735 | 0.605 | 0.605 | 0.499 | 0.157 |
| 0 | +1 | −1 | −1 | −1 | +1 | +1 | +1 | +1 | |
| Serbia versus Croatia | 0.013 | 0.724 | 1.000 | − | 0.022 | − | − | 0.484 | 0.181 |
| +1 | +1 | −1 | − | +1 | − | − | +1 | +1 | |
| Italy WCL versus Italy NWA | 0.058 | 0.760 | 0.553 | 0.580 | 0.036 | − | − | 0.963 | 0.990 |
| +1 | −1 | +1 | +1 | +1 | − | − | −1 | −1 | |
| Italy WCP versus Italy CMA | 0.007 | 0.285 | 0.316 | 0.602 | 0.987 | − | − | 0.570 | 0.725 |
| +1 | +1 | +1 | +1 | −1 | − | − | +1 | −1 | |
| Italy SLA versus Italy NEL | 0.999 | 0.671 | 0.458 | − | 0.121 | − | − | 0.471 | 0.617 |
| −1 | −1 | +1 | − | +1 | − | − | +1 | −1 | |
| Italy TLB versus Italy EBL | 0.257 | 0.244 | 0.999 | − | 0.034 | − | − | 0.840 | 0.960 |
| +1 | +1 | −1 | − | +1 | − | − | −1 | −1 | |
| S. Portugal versus N. Portugal | 0.998 | 0.377 | 0.601 | 0.031 | 0.223 | − | − | 0.246 | 0.583 |
| −1 | +1 | 0 | +1 | +1 | − | − | +1 | −1 | |
| S. Greece versus N. Greece | 0.229 | 0.511 | 0.071 | 0.974 | 0.418 | 0.694 | 1.000 | 1.000 | 0.3000 |
| +1 | +1 | +1 | −1 | +1 | 0 | −1 | −1 | +1 | |
| S. Egypt versus N. Egypt | 0.984 | 0.164 | 0.402 | 0.153 | 0.931 | 0.176 | − | 1.000 | 0.409 |
| −1 | +1 | +1 | +1 | −1 | +1 | − | −1 | +1 | |
| α = 0.05 | 0.102 | 0.432 | 0.432 | 0.337 | 0.015 | 1.000 | 1.000 | 1.000 | 1.000 |
| α = 0.30 | 0.210 | 0.828 | 0.980 | 0.748 | 0.078 | 0.760 | 1.000 | 0.887 | 0.887 |
| Δf | 0.377 | 0.113 | 0.377 | 0.363 | 0.274 | 0.688 | 0.688 | 0.500 | 0.500 |
| Greek Test Sites | |||||||||
| Greece & Crete versus Turkey #7 | 0.1951 | 0.9264 | 0.0183 | 0.9865 | 0.4270 | 0.5180 | 1.0000 | 0.0513 | 0.5364 |
| +1 | −1 | +1 | −1 | +1 | +1 | −1 | +1 | +1 | |
| Greece & Crete versus Turkey #4 | 0.7091 | 0.6836 | 0.0000 | 1.0000 | 0.3777 | 0.1891 | 1.0000 | 0.4615 | 0.3583 |
| −1 | −1 | +1 | −1 | +1 | +1 | −1 | +1 | +1 | |
| E. Sicily versus Sardinia | 0.0002 | 0.9994 | 1.0000 | 0.0272 | 0.0000 | 0.0032 | − | 0.1462 | 0.5451 |
| +1 | −1 | −1 | +1 | +1 | +1 | − | +1 | +1 | |
| E. Sicily versus W. Sicily | 0.0735 | 0.9904 | 0.9779 | 0.2649 | 0.0552 | 0.4294 | − | 0.6833 | 0.9271 |
| +1 | −1 | −1 | +1 | +1 | +1 | − | −1 | −1 | |
| Greece & Crete versus Cyprus | 0.9410 | 0.0096 | 0.1852 | 0.9881 | 0.864 | 0.717 | − | 0.0184 | 0.0184 |
| −1 | +1 | +1 | −1 | +1 | −1 | − | +1 | +1 | |
| Greece & Crete versus S. Italy | 0.9967 | 0.9712 | 0.0815 | 0.9572 | 0.0110 | 0.493 | − | 0.0144 | 0.0144 |
| −1 | −1 | +1 | −1 | +1 | +1 | − | +1 | +1 | |
| Greece & Crete versus Spain | 0.2539 | 0.0937 | 0.00142 | 0.9252 | 0.000 | 0.0000 | − | 0.0000 | 0.0000 |
| +1 | +1 | +1 | −1 | +1 | +1 | − | +1 | +1 | |
| Turkey #8 & #9 versus Lebanon | 0.9967 | 0.0008 | 0.5830 | 1.0000 | 0.7884 | 0.906 | 0.97 | 0.1899 | 0.1899 |
| −1 | +1 | −1 | −1 | −1 | −1 | −1 | +1 | +1 | |
| Turkey #8 & #9 versus Palestinian | 0.9998 | 0.0218 | 0.3905 | 1.0000 | 0.0310 | 0.711 | 0.2133 | 0.0195 | 0.0195 |
| −1 | +1 | +1 | −1 | +1 | −1 | +1 | +1 | +1 | |
| Greece versus Lebanon | 0.1653 | 0.1351 | 0.0000 | 1.0000 | 0.979 | 0.788 | 1.0 | 0.0000 | 0.0000 |
| +1 | +1 | +1 | −1 | −1 | −1 | −1 | +1 | +1 | |
| Greece versus Palestinians | 0.5177 | 0.3013 | 0.0000 | 1.0000 | 0.4529 | 0.602 | 1.0 | 0.0000 | 0.0000 |
| +1 | +1 | +1 | −1 | +1 | +1 | −1 | +1 | +1 | |
| Crete versus Lebanon | 0.9908 | 0.6702 | 0.0044 | 1.0000 | 0.0003 | 0.312 | 1.0 | 1.0 | 0.0000 |
| −1 | −1 | +1 | −1 | +1 | +1 | −1 | −1 | +1 | |
| Crete versus Palestinians | 0.9993 | 0.8484 | 0.0043 | 1.0000 | 0.0000 | 0.142 | 1.0 | 0.0000 | 0.0000 |
| −1 | −1 | +1 | −1 | +1 | +1 | −1 | +1 | +1 | |
| S. Italy versus Coastal Tunisia | 0.9358 | 0.0111 | 0.1771 | 0.1771 | 0.4996 | 0.7255 | − | 0.4253 | 0.0026 |
| −1 | +1 | +1 | +1 | +1 | −1 | − | +1 | +1 | |
| Sicily versus Coastal Tunisia | 0.9889 | 0.0430 | 0.0597 | 0.1834 | 0.1989 | 0.6332 | − | 0.4672 | 0.0018 |
| −1 | +1 | +1 | +1 | +1 | +1 | − | +1 | +1 | |
| α = 0.05 | 0.5367 | 0.0006 | 0.0000 | 0.5367 | 0.0000 | 0.1710 | 1.0000 | 0.0000 | 0.0000 |
| α = 0.30 | 0.7031 | 0.1311 | 0.0007 | 0.7031 | 0.0500 | 0.7031 | 0.9423 | 0.0037 | 0.0006 |
| Δf | 0.8491 | 0.5000 | 0.0176 | 0.9824 | 0.0037 | 0.1509 | 0.9961 | 0.0037 | 0.0005 |
Designated control.
Data from Lebanon were available,16 and we collected 1330 additional DNA samples from Syrian, Palestinian, Tunisian, Moroccan, Cypriote, and Maltese males with at least three generations of indigenous ancestry. Each provided information on their geographical origin and gave informed consent for this study. Samples were typed with 11 STRs and with 58 Y-SNPs as described elsewhere16 (Table S1). We augmented our collection with suitable published data on 5,899 males from 56 sites (Table S2). Desirable sites that we were unable to include in our analysis included Libya and southern France, both of which could have provided more Greek coastal-settlement sites. The Y-chromosomal data were of two types: haplogroup data based on Y-SNPs and haplotype data based on Y-STRs. Although both types are carried on the same chromosome and are correlated,17 they were analyzed separately, because they have different mutational properties and because some data sets contain only one of the two data types. A reduced set of haplogroups that captured most of the SNP information was used as previously.16 It was also necessary to develop a similar procedure for the STR information by enumerating the regions and sizes of samples captured by various combinations of STR subsets, through a process informed by association-discovery methods.18 We identified subsets containing seven STRs that maximized regional coverage and sample number, yielding the STRs DYS19, DYS389I, DYS389b (consisting of DYS389II–DYS389I), DYS390, DYS391, DYS392, and DYS393. We lost STR coverage of some regions, reducing the number of tests that were applied to the STR set. The geographical coverage of the STR samples and the SNP samples was not identical, and the regional tests that could be constructed from historical references were not identical for both genetic marker types. For example, Moroccan samples were included and tested in the STR set but not in the SNP-typed set.
The tests were constructed and validated in several ways. First, a noncontact test-pair matrix was constructed for detecting general east-to-west background variation reflecting Neolithic migrations, and the data were evaluated for significant results reflecting general non-Phoenician background variation against which the Phoenician pattern must be identified. Second, a colonization test-pair matrix for identification of gross features of the subsequent and more widespread Greek colonization event was applied. The Greek test sought to identify features typical of the Greek expansion but focused on those characteristics distinct from the Phoenician expansion. Third, the Phoenician colonization of Tunisia presented a unique test between the colonized coastal regions and interior Berber and Arab populations, because it has a different Neolithic history19 and no intervening Greek-colonization events. Additionally, the Moroccan military colonies are expected to be weaker than the major Phoenician Tunisian-trade-based colony but also to lack Greek influence.
Test-site pairs for haplogroups relevant to the Phoenician expansion are indicated in Figure 1B, and those for STR-defined haplotypes are indicated in Figure 1C. Preponderance p values representing test-pair aggregates were computed as described below, and two techniques were employed to establish these measures.
The first test was direct frequency comparison by means of the binomial sign test. Test sites were scored as positive if the contact-site frequency was larger than the noncontact-site frequency. The number of positive results, N+, out of a total of N tests expected by chance should be randomly distributed following a binomial distribution with p = 0.05, so the probability that N+ or more would have been observed by chance according to the “nonparametric” binomial sign test is
Second, we applied Fisher's exact test to determine the chances of drawing m+ or more out of t by chance given that they were taken randomly from M+ total stronger contact samples and M− total weaker contacts across the two test regions, with probability
By the probability-integral-transform theorem, the distribution of p values may itself be considered to be a uniformly distributed random variable over the interval [0,1]. At a confidence level of α, the site was considered a positive candidate if p≥ (m+) ≤ α. This would be expected to be satisfied an α fraction of the time. For an individual site, a significant (α = 0.05) or highly significant (α = 0.01) level is usually required. However, testing for randomness even with much larger values of α is possible for putatively independent sites with the use of the binomial test, in the same way that the fairness of dice or the fairness of a coin may be tested.20 Then the probability of seeing N+ or more sites by chance at significance level of α can also be tested according to the binomial test, such that .20 Even for a relatively weak α level of significance, the probability of seeing multiple sites at that level can yield a highly significant preponderant probability. Fisher's exact test tends to be more demanding for small samples, and if the sample is too small, it will never yield significant results. Yet, the number of times that relatively small samples will satisfy a weak significance of, say, α = 0.30 still provides opportunity for probing the significance of sites with such small samples and for counting their contribution in determining the overall probability of seeing a Phoenician signal by chance.
Because there is a significant chance that a haplotype existing 3000 years ago has accumulated a one-step difference in an STR (we expect 0.6 mutations per seven-STR haplotype when a rate of 6.9 × 10−4 per locus per 25 yr is used21), these one-step neighbors have been included in each set, producing what we have labeled STR+s. STR+s can contain both haplotypes deriving from mutations, which should have been included, and independent haplotypes unconnected with the migrations that we are trying to detect. Those other sources are expected to be uncorrelated and incoherent relative to the signals we seek. STR sets can be found within multiple haplogroups, so contributions from multiple haplogroups might contribute to each of the STR+ samples as well, providing further stochastic background noise. Among STR+s, test sites were excluded when gradient differences were computed if there were two or fewer total STR+ samples in both sites. If there were fewer than three total STR+ samples, the Fisher's exact probabilities were discarded, because many comparison configurations can never show significant probabilities with such small samples.
The number and relative frequency of the major haplogroups observed in the sample regions employed in this study are shown in Table S3. Table 1 represents the outcome of Phoenician-colonization tests, the Neolithic control tests, and the Greek-colonization tests. Each of the results shows the Fisher's exact test p value as a number between 0 and 1, together with frequency-difference test as +1 or −1. Aggregate scores computed on the Fisher's exact test results for thresholds α = 0.30 and α = 0.05, as well as for counts of Δf signs of frequency differences, are reported at the end of each section. The α = 0.05 results measure whether the number of strongly different gradients is significantly different than that expected by chance, whereas the α = 0.30 results reports the same for modestly preferential sites, identifying a persistent pattern of weaker signals. Although any individual signal at this lower significance level might not be significant, the signal across all sites could be. The frequency-differences test Δf seeks to report signal in cases in which the number of samples may be low but may still contribute to a preponderance of evidence.
The Neolithic control section shows nonsignificant results across all haplogroups, except for a significant J2 result in one test. The Phoenician-colony test results highlight only one haplogroup, J2, which consistently scores significantly in all three tests across the range of colonization sites (Table 1, Figure 1B). However, this haplogroup also scores significantly in Greek tests (as do some additional haplogroups; Table 1), suggesting that the same haplogroup could have been spread by several expansions, which is unsurprising considering its frequency in the Eastern Mediterranean but implies that higher phylogenetic resolution is required for identification of Phoenician-specific signals.
Table 2 shows the core STR haplotypes of the STR+ groups that we focus on, and Table S4 reports the population frequencies for these STR+s. These STR+ groups were labeled “Phoenician Colonization Signal” or PCS1+ through PCS6+. Among the total of 1268 STR+s identified, 1237 showed coverage at nine or fewer sites. From the remaining 31, several candidates—PCS1+, PCS2+, PCS4+, PCS5+, and PCS6+—were identified from their high p values (Table 3). PCS3+ scores strongly as a Phoenician-colonization candidate and is strongly associated with the SNP haplogroup E3b, but it does not show the wide geographic coverage that the other PCS+s demonstrate. It represents the strongest of the lower-coverage STR+s. Both PCS1+ and PCS2+ score well, although not as strongly as PCS3+. However, they show much broader penetration throughout the Mediterranean, and both score relatively weakly as Greek-colonization candidates. Of these, PCS1+ shows a nearly significant Greek score for α = 0.05 because of low representation in Tunisia, but it shows significant representation in Morocco, and the Greek score is simply an artifact. Both PCS1+ and PCS2+ contain multiple haplogroups, primarily J2 but including J∗(xJ2) and E3b, with PCS1+ containing the greatest diversity. The use of one-step STR+s provides the opportunity to pick up mutated descendants of those who participated in colonization, as intended, but also of those who acquired the same signature through alternative paths. As expected, those other paths have tended to degrade a systematic colonization signal, shown by the relatively weak α = 0.05 score relative to α = 0.30 in comparison with PCS3+. A “Greek Colonization Signal” STR+ group, GCS1+, was also identified, which scored weakly as a Phoenician candidate but showed a strong score on the complementary Greek test matrix.
Table 2.
Core Haplotypes Defining Y-STR Haplotype Groupsa Associated with the Phoenician or Greek Expansions
| STR+ | DYS19,DYS389I,DYS389b,DYS390,DYS391,DYS392,DYS393 |
|---|---|
| PCS1+ | 14,13,16,24,10,11,12 |
| PCS2+ | 14,14,17,23,10,11,12 |
| PCS3+ | 13,12,18,23,10,11,13 |
| PCS4+ | 14,13,17,23,10,11,12 |
| PCS5+ | 14,14,16,23,10,11,12 |
| PCS6+ | 14,13,16,22,10,11,12 |
| GCS1+ | 13,13,17,24,10,11,13 |
Designated STR+s.
Table 3.
STR+ Colonization Site Gradient Tests, with Aggregate Scores for Phoenician Colonies and Greek Colonies
| STR+ Tests | PCS1+ | PCS2+ | PCS3+ | PCS4+ | PCS5+ | PCS6+ | PCS1+ through PCS3+ | GCS1+ |
|---|---|---|---|---|---|---|---|---|
| Phoenician Test Sites | ||||||||
| Phoenician Heartland versus Phoenician Periphery | 0.425 | 0.609 | 0.922 | 0.819 | 0.467 | 0.098 | 0.606 | 0.725 |
| +1 | −1 | −1 | −1 | −1 | +1 | −1 | −1 | |
| Phoenician Heartland versus Palestinians | 0.000 | 0.078 | 0.370 | 0.156 | 0.004 | 0.014 | 0.000 | 0.983 |
| +1 | +1 | +1 | +1 | +1 | +1 | +1 | −1 | |
| Phoenician Periphery versus Palestinians | 0.000 | 0.079 | 0.080 | 0.042 | 0.012 | 0.272 | 0.000 | 0.943 |
| +1 | +1 | +1 | +1 | +1 | +1 | +1 | −1 | |
| Syria versus Palestinians | 0.241 | 0.034 | − | 0.858 | 0.247 | 0.969 | 0.022 | 0.961 |
| +1 | +1 | +1 | −1 | +1 | −1 | +1 | −1 | |
| Cyprus versus N and S Turkey | − | − | − | − | − | − | − | − |
| − | − | − | − | − | − | − | − | |
| S Turkey versus N Turkey | − | − | − | − | − | − | − | − |
| − | − | − | − | − | − | − | − | |
| Crete versus Greece | 0.093 | 0.194 | − | 0.030 | 0.257 | 0.016 | 0.040 | 0.876 |
| +1 | +1 | − | +1 | +1 | +1 | +1 | −1 | |
| Malta versus W Sicily and S Italy | 0.498 | 0.029 | − | 0.005 | − | − | 0.063 | 1.000 |
| +1 | +1 | − | +1 | − | − | +1 | −1 | |
| W Sicily versus E Sicily | − | − | − | − | − | − | − | − |
| − | − | − | − | − | − | − | − | |
| All Sicily versus All Italy | 0.926 | 0.411 | 0.248 | 0.805 | 0.850 | 0.956 | 0.623 | 0.574 |
| −1 | +1 | +1 | −1 | −1 | −1 | −1 | −1 | |
| S Spain versus Noncontact Iberia | 0.337 | 0.841 | − | 0.872 | 0.679 | 1.000 | 0.519 | 0.981 |
| +1 | −1 | − | −1 | −1 | −1 | +1 | −1 | |
| Sardinia versus Italy | 0.165 | − | − | 0.220 | 0.136 | 0.179 | 0.219 | 1.000 |
| +1 | −1 | − | +1 | +1 | +1 | +1 | −1 | |
| Sardinia versus Noncontact Iberia | 0.053 | 1.000 | − | 0.069 | 0.045 | 0.037 | 0.104 | 1.000 |
| +1 | −1 | − | +1 | +1 | +1 | +1 | −1 | |
| Coastal Tunisia versus Inland Tunisia | − | 0.177 | 0.006 | 0.021 | − | − | 0.001 | 0.304 |
| − | +1 | +1 | +1 | +1 | − | +1 | +1 | |
| All Tunisia versus Morocco | 1.000 | 0.059 | 0.000 | 0.868 | 1.000 | 1.000 | 0.245 | 0.039 |
| −1 | +1 | +1 | −1 | −1 | −1 | +1 | +1 | |
| α = 0.05 | 0.102 | 0.101 | 0.032 | 0.0022 | 0.012 | 0.012 | 0.0002 | 0.460 |
| α = 0.30 | 0.078 | 0.022 | 0.070 | 0.039 | 0.047 | 0.047 | 0.0017 | 0.986 |
| Δf | 0.033 | 0.193 | 0.063 | 0.387 | 0.274 | 0.377 | 0.019 | 0.997 |
| Greek Test Sites | ||||||||
| Crete and Greece versus Cyprus | 1.0000 | 0.9954 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.0000 |
| −1 | −1 | −1 | −1 | −1 | −1 | −1 | +1 | |
| Crete and Greece versus Sicily | 0.4261 | 0.0590 | 1.0000 | 0.3608 | 0.7137 | 0.9429 | 0.3181 | 0.0000 |
| +1 | +1 | −1 | +1 | −1 | −1 | +1 | +1 | |
| Crete and Greece versus S Italy | 0.734 | 0.046 | − | 0.490 | 0.731 | 0.869 | 0.188 | 0.000 |
| −1 | +1 | − | +1 | −1 | −1 | +1 | +1 | |
| S Italy versus N Italy | 0.939 | − | − | 0.933 | 0.951 | 0.990 | 0.620 | 0.362 |
| −1 | − | − | −1 | −1 | −1 | −1 | +1 | |
| Crete and Greece versus Malta | 0.871 | 0.908 | − | 1.000 | 0.294 | 0.443 | 0.934 | 0.000 |
| −1 | −1 | − | −1 | +1 | +1 | −1 | +1 | |
| Crete and Greece versus Iberia | 0.477 | 0.400 | − | 0.249 | 0.600 | 0.713 | 0.184 | 0.006 |
| +1 | +1 | − | +1 | +1 | −1 | +1 | +1 | |
| Turkey versus Phoenician Heartland and Periphery | − | − | − | − | − | − | − | − |
| − | − | − | − | − | − | − | − | |
| Turkey versus Syria | − | − | − | − | − | − | − | − |
| − | − | − | − | − | − | − | − | |
| Crete versus Phoenician Heartland and Periphery | 0.815 | 0.390 | 1.0 | 0.862 | 0.939 | 0.949 | 0.759 | 0.000 |
| −1 | +1 | −1 | −1 | −1 | −1 | −1 | +1 | |
| Crete versus Syria | 0.272 | 0.618 | − | 0.367 | 0.754 | 0.349 | 0.408 | 0.000 |
| +1 | −1 | − | +1 | −1 | +1 | +1 | +1 | |
| Sicily versus Tunisia | 0.014 | 0.605 | 1.000 | 0.749 | 0.053 | 0.004 | 0.906 | 0.426 |
| +1 | −1 | −1 | −1 | +1 | +1 | −1 | +1 | |
| S Italy versus Coastal Tunisia | 0.1255 | 1.0000 | 1.000 | 0.9171 | 0.4131 | 0.1802 | 0.9936 | 0.6772 |
| +1 | −1 | −1 | −1 | +1 | +1 | −1 | −1 | |
| α = 0.05 | 0.4013 | 0.3698 | 1.0000 | 1.0000 | 1.0000 | 0.4013 | 1.0000 | 0.0000 |
| α = 0.30 | 0.6172 | 0.8040 | 1.0000 | 0.9718 | 0.8507 | 0.9718 | 0.8507 | 0.0106 |
| Δf | 0.6230 | 0.7461 | 1.0000 | 0.8281 | 0.8281 | 0.8281 | 0.8281 | 0.0107 |
All of the PCS1+ through PCS3+ candidate central haplotypes are more than two steps away from each other, so the STR+s share no STR haplotypes. Therefore, their frequencies can be combined if sample counts are added together row by row to represent an aggregate PCS1+ through PCS3+ group. In general, across most geographical sites, the PCS1+, PCS2+, and PCS3+ groups combined to reinforce each other's Phoenician signals, boosting their aggregate scores but not their Greek scores (Table 3). The PCS1+, PCS2+, and PCS3+ frequencies in the Mediterranean region are represented in Figures 1D–1F.
PCS4+ through PCS6+ are all closely related to PCS1+ and PCS2+. Both PCS4+ and PCS5+ overlap PCS2+; PCS6+ does not, but shares a bridge PCS+ group (core 14,13,16,23,10,11,12) with both PCS1+ and PCS2+. Combining PCS4+ through PCS6+ with PCS1+ or PCS2+ would thus yield overcounting of some groups. Therefore, these are not included in the aggregate PCS1+ through PCS3+ group. It is notable that the range of STR+s in the cluster associated with PCS1+ and PCS2+ spans a range of five or six STR mutations, far in excess of that expected to emerge in the time since the Phoenician expansion. Although each STR+ covers geographically distinct colonies, each is rooted in the Phoenician heartland. This argues for a common source of related lineages rooted in Lebanon.
It can be deduced from the structure of the tests that admixture from other occupation of both Phoenician noncontact sites and contact sites would tend to systematically wash out the significance of a Phoenician signal throughout the range of the Phoenician Colony Specific test sites. For example, one of the five samples from Sardinia was PCS1+. Compared to Italy, at five out of 187, the probability of drawing this fraction by chance was 0.258, as reported in Table 3. If only 20% of the samples found in Italy were added to Sardinia's signal, this would have yielded two out of 47 for Sardinia, yielding a probability of 0.378, outside the α = 0.30 threshold. Likewise, 30% of the Greek contribution of Crete in PCS1+ would raise the Fisher's exact probability from 0.173 to 0.328. The fact that this dilution did not systematically destroy a preponderant Phoenician signal argues that such admixture has been low enough to allow the isolation of components that were systematically Phoenician. The results presented here suggest that any additional expansions, such as the Jewish Diaspora, and subsequent population effects showed sufficiently low admixture or drift into both colonization sites and surrounding populations for a Phoenician signal to remain significant.
Haplogroup J2, in general, and haplotypes PCS1+ through PCS6+ therefore represent lineages that might have been spread by the Phoenicians; but could the patterns that we observe be accounted for by other events, particularly the Jewish Diaspora, for which we could not develop a formal test? Note that this is a separate question from that of whether they could also have been spread by other expansions: indeed, we expect that Jews of the Diaspora carried some of the same STR+ and SNP lineages with them as did Phoenicians of Phoenician expansion. Two lines of reasoning suggest to us that we must be detecting a distinct signal. First, the frequency of Jews in the Mediterranean region over almost all of our sample sites is currently less than 0.1%, and our own collection of samples contained no individuals who identified themselves as bearing Jewish heritage in a number of sites, such as Tunisia and Morocco.22,23 Although historical admixture is expected to have occurred to some extent, recent studies tend to show strikingly low admixture in modern Jewish populations.15 Second, any such admixture is likely to have contributed to both Phoenician contact and noncontact populations and thus could not explain a systematically differential signal. The excess of J2 (Figure 1B), PCS1+ (Figures 1C and 1D), PCS2+ (Figure 1E), and PCS3+ (Figure 1F) in coastal Tunisia, the site of Carthage, compared with inland Tunisia is particularly salient, because these lineages are considerably more rare in North Africa than in Southern Europe. It also suggests that the Roman destruction of Carthage did not eliminate the Carthaginian gene pool. Further support for the PCS+ haplotypes' spread with the Phoenicians is illustrated by their generally high frequency among the Phoenician contact sites across the Mediterranean basin (Figures 1D–1F).
The Greek expansion was not the focus of this study, but it nevertheless revealed several signals. In this case, two expansions from Western Europe that probably spread R1b chromosomes could potentially yield a “Greek” profile. According to Strabo, Brennus “the second” of the Prausi was attracted to Greece by internecine conflicts in 281 BCE. Subsequently, some of these people moved to Thracia in the north, with 20,000 of those moving to Galatia in the north-central Anatolian peninsula in 279–277 BCE.24 Subsequent European genetic transfer occurred with the Crusades16 and with European trade, leaving a general north-to-south gradient of R1b chromosomes, with a substantial concentration in Greece and Turkey, yielding a pattern that could resemble Greek colonization.
This study presents a methodology for constructing systematic tests identifying local signatures of colonization and for constructing aggregate scores measuring a consensus across all of the colonization sites. We have shown that the methodology does not produce significant signal for arbitrary sampling in noncolonization regions, and multiple markers that do not show patterns consistent with Phoenician colonization have been presented. Tests constructed to isolate Greek-colonization events from the Phoenician samples can show positive and weak scores both for Phoenician candidates and for non-Phoenician candidates, indicating that information is presented in those tests distinct from the Phoenician-colonization tests.
Application of this methodology to STR samples was more problematic as a result of prohibitively small samples at some sites and of nonuniform sample collection throughout the Mediterranean at this level of resolution, even when STR-only data were included. Smaller collections limit the statistical power to resolve signals at any of the particular sites. With the possibility of single-step STR mutations in the intervening time allowed for, identification of candidate groups (STR+s) was possible. Although true mutated descendants will systematically augment the strength of the statistical resolution, this expansion of samples will also allow inclusion of identical-by-state haplotypes with distinct histories that might even derive from other haplogroups. In conclusion, there are many ways in which a colonization signal could be diluted to undetectable levels, but statistically robust signals should represent biologically meaningful events.
We do not suggest that the Phoenicians spread only or predominantly J2 and PCS1+ through PCS6+ lineages. They are likely to have spread many lineages from multiple haplogroups, but the lineages we highlight are the most highly differentiated ones providing the most readily detectable signals. Signals can only be detected when the same or related haplotypes were transmitted to multiple locations. Because most haplotypes are rare, the use of STR+s rather than individual haplotypes, and perhaps the preferential spread of a subset of pioneering or influential Phoenician families, might have enhanced our signal. The magnitude of the Phoenician contribution to the populations investigated was estimated from the candidate STR+'s prevalence in colony versus noncolony sites. Although the total fraction of colony samples contained within the PCS1+ through PCS3+ group is around 10%, it is the fraction above background, or the difference in frequencies between contact and noncontact sites (Table S4), that actually represents Phoenician signal. The mean difference in frequency was ∼6%, providing a minimum estimate of the Phoenician input.
Given that these same lineages, including the STR haplotypes, were clearly spread in other ways as well, their identification in additional subjects would not in itself provide evidence that such people were of Phoenician descent. This, however, is a reflection of the limited phylogenetic resolution used, and it is reasonable to expect that future thorough searches for SNPs or STR combinations could lead to the discovery of rare but reliable markers of such descent. The technology for resequencing individual genomes at ever-decreasing cost makes this a realistic prospect.24
Finally, our work underscores the effectiveness of Y-chromosomal variability when combined with appropriate computational analysis for studying complex patterns of human migration, as well as the utility of wide geographical sampling with the use of a uniform marker set. This method is applicable to any type of genetic information from which descent could be inferred, such as mtDNA or autosomal regions with limited recombination, and within which enough markers are available to establish phylogeny. The numbers of sites passing at α = 0.3 and α = 0.05 levels to produce aggregates significant at the 5% level for various numbers of sites tested are outlined in Table 4. Therefore, even rather small sets at relatively low levels of significance can yield useful signal. Further applications could include systematic investigations of military expansions, such as the Greek signal, from the time of Alexander the Great in central and south Asia;25 or the Mongol signal, carried through the military and trade-regulation activities to regions from China to Moscow26 and south through North India, Iran, and Iraq. Trade and colonization without substantial military intervention also drove wealth and technological and cultural development. Examples of ways that genetic migration was mediated might include the silk and spice roads, which connected China with the Middle East through to Europe, as well as to spice sources in India and Indonesia, and the Incense Road, which connected India through the southern Arabian Peninsula. The Viking expansion involved not only raids but also significant trade and colonization, in multiple waves.27 Important African centers of trade include Timbuktu, with archaeological evidence showing that Great Zimbabwe enjoyed goods from as far away as China. To complement investigations of known migrations, our methodology could also be used to search systematically for signals of expansion within a data set, starting from each site in turn, and could thus potentially discover unrecorded migrations as well.
Table 4.
Number of Sites, k, with p Value Stronger than Significance Level α out of a Total of N Sites Tested that Are Required for the Aggregate to Pass at the 5% Significance Level
| N | k (α = 0.30) | k (α = 0.05) |
|---|---|---|
| 1 | - | 1 |
| 2 | - | 2 |
| 3 | 3 | 2 |
| 4 | 4 | 2 |
| 5 | 4 | 2 |
| 6 | 5 | 2 |
| 7 | 5 | 2 |
| 8 | 6 | 3 |
| 9 | 6 | 3 |
| 10 | 6 | 3 |
| 11 | 7 | 3 |
| 12 | 7 | 3 |
| 13 | 8 | 3 |
| 14 | 8 | 3 |
| 15 | 9 | 3 |
Acknowledgments
We thank the sample donors for taking part in this study, R. John Mitchell, Tad Schurr and other Genographic Project members for comments. We also thank Janet Ziegle and Applied Biosystems for genotyping support and Rabih Hosri for help with Figure 1. Y.X. and C.T.S. were supported by The Wellcome Trust, and M.A.J. by a Wellcome Trust Senior Fellowship in Basic Biomedical Science (057559). The Genographic Project is supported by funding from the National Geographic Society, IBM and the Waitt Family Foundation.
Genographic Consortium members: Theodore G. Schurr (University of Pennsylvania, Philadelphia, PA, USA), Fabrício R. Santos (Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil), Lluis Quintana-Murci (Institut Pasteur, Paris, France), Jaume Bertranpetit (Universitat Pompeu Fabra, Barcelona, Catalonia, Spain), David Comas (Universitat Pompeu Fabra, Barcelona, Catalonia, Spain), Chris Tyler-Smith (The Wellcome Trust Sanger Institute, Hinxton, UK), Pierre A. Zalloua (Lebanese American University, Chouran, Beirut, Lebanon), Elena Balanovska (Russian Academy of Medical Sciences, Moscow, Russia), Oleg Balanovsky (Russian Academy of Medical Sciences, Moscow, Russia), R. John Mitchell (La Trobe University, Melbourne, Victoria, Australia), Li Jin (Fudan University, Shanghai, China), Himla Soodyall (National Health Laboratory Service, Johannesburg, South Africa), Ramasamy Pitchappan (Madurai Kamaraj University, Madurai, Tamil Nadu, India), Alan Cooper (University of Adelaide, South Australia, Australia), Lisa Matisoo-Smith (University of Auckland, Auckland, New Zealand), Ajay K. Royyuru (IBM, Yorktown Heights, New York, USA), Daniel E. Platt (IBM, Yorktown Heights, New York, USA), Laxmi Parida (IBM, Yorktown Heights, New York, USA), Jason Blue-Smith (National Geographic Society, Washington, D.C., USA), David F. Soria Hernanz (National Geographic Society, Washington, D.C., USA), and R. Spencer Wells (National Geographic Society, Washington, D.C., USA).
Supplemental Data
References
- 1.Stieglitz R.R. The geopolitics of the Phoenician Littoral in the Early Iron Age. Bull. Am. Schools Orient. Res. 1990;279:9–12. [Google Scholar]
- 2.Moscati S. Weidenfeld and Nicolson Ltd.; London: 1973. The World of Phoenicians. [Google Scholar]
- 3.Markoe G. British Museum Press; London: 2000. Phoenicians. [Google Scholar]
- 4.Aubet M.E. Cambridge University Press; Cambridge: 2001. The Phoenicians and the West: Politics, Colonies and Trade. [Google Scholar]
- 5.Marston E. Benchmark Books; New York: 2002. The Phoenicians. [Google Scholar]
- 6.Harden D. Penguin Books; London: 1971. The Phoenicians. [Google Scholar]
- 7.Jobling M.A., Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat. Rev. Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- 8.Rosser Z.H., Zerjal T., Hurles M.E., Adojaan M., Alavantic D., Amorim A., Amos W., Armenteros M., Arroyo E., Barbujani G. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am. J. Hum. Genet. 2000;67:1526–1543. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semino O., Magri C., Benuzzi G., Lin A.A., Al-Zahery N., Battaglia V., Maccioni L., Triantaphyllidis C., Shen P., Oefner P.J. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the Neolithization of Europe and later migratory events in the Mediterranean area. Am. J. Hum. Genet. 2004;74:1023–1034. doi: 10.1086/386295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Semino O., Passarino G., Brega A., Fellous M., Santachiara-Benerecetti A.S. A view of the Neolithic demic diffusion in Europe through two Y chromosome-specific markers. Am. J. Hum. Genet. 1996;59:964–968. [PMC free article] [PubMed] [Google Scholar]
- 11.Semino O., Passarino G., Oefner P.J., Lin A.A., Arbuzova S., Beckman L.E., De Benedictis G., Francalacci P., Kouvatsi A., Limborska S. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell R.J., Earl L., Fricke B. Y-chromosome specific alleles and haplotypes in European and Asian populations: linkage disequilibrium and geographic diversity. Am. J. Phys. Anthropol. 1997;104:167–176. doi: 10.1002/(SICI)1096-8644(199710)104:2<167::AID-AJPA3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 13.Tripolitis A. Wm. B. Eerdmans Publishing Company; Michigan: 2001. Religions of the Hellenistic-Roman age. [Google Scholar]
- 14.Barnavi E., editor. A Historical Atlas of the Jewish People: From the Time of the Patriarchs to the Present. Schocken; New York: 1994. [Google Scholar]
- 15.Behar D.M., Garrigan D., Kaplan M.E., Mobasher Z., Rosengarten D., Karafet T.M., Quintana-Murci L., Ostrer H., Skorecki K., Hammer M.F. Contrasting patterns of Y chromosome variation in Ashkenazi Jewish and host non-Jewish European populations. Hum. Genet. 2004;114:354–365. doi: 10.1007/s00439-003-1073-7. [DOI] [PubMed] [Google Scholar]
- 16.Zalloua P.A., Xue Y., Khalife J., Makhoul N., Debiane L., Platt D.E., Royyuru A.K., Herrera R.J., Hernanz D.F., Blue-Smith J. Y-chromosomal diversity in Lebanon is structured by recent historical events. Am. J. Hum. Genet. 2008;82:873–882. doi: 10.1016/j.ajhg.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlecht J., Kaplan M.E., Barnard K., Karafet T., Hammer M.F., Merchant N.C. Machine-learning approaches for classifying haplogroup from Y chromosome STR data. PLoS Comput Biol. 2008;4:e1000093. doi: 10.1371/journal.pcbi.1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agrawal, R., and Srikant, R. (1994). Fast Algorithms for Mining Association Rules. Proceedings of the 20th International Conference on Very Large Data Bases (VLDB '94), pp. 487–499.
- 19.Arredi B., Poloni E.S., Paracchini S., Zerjal T., Fathallah D.M., Makrelouf M., Pascali V.L., Novelletto A., Tyler-Smith C. A predominantly Neolithic origin for Y-chromosomal DNA variation in North Africa. Am. J. Hum. Genet. 2004;75:338–345. doi: 10.1086/423147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sokal R.R., Rohlf F.J. W. H. Freeman; New York: 1995. Biometry: The Principles and Practice of Statistics in Biological Research. [Google Scholar]
- 21.Zhivotovsky L.A., Underhill P.A., Cinnioglu C., Kayser M., Morar B., Kivisild T., Scozzari R., Cruciani F., Destro-Bisol G., Spedini G. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanford J.V., editor. 2004 International Religious Freedom Report. U.S. Department of State; Washington: 2004. [Google Scholar]
- 23.DelaPergola S. World Jewish Population 2002. In: Singer D., Grossman L., editors. American Jewish Yearbook 2002. American Jewish Committee; NY: 2002. pp. 247–274. [Google Scholar]
- 24.Mardis E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Firasat S., Khaliq S., Mohyuddin A., Papaioannou M., Tyler-Smith C., Underhill P.A., Ayub Q. Y-chromosomal evidence for a limited Greek contribution to the Pathan population of Pakistan. Eur. J. Hum. Genet. 2007;15:121–126. doi: 10.1038/sj.ejhg.5201726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zerjal T., Xue Y., Bertorelle G., Wells R.S., Bao W., Zhu S., Qamar R., Ayub Q., Mohyuddin A., Fu S. The genetic legacy of the Mongols. Am. J. Hum. Genet. 2003;72:717–721. doi: 10.1086/367774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowden G.R., Balaresque P., King T.E., Hansen Z., Lee A.C., Pergl-Wilson G., Hurley E., Roberts S.J., Waite P., Jesch J. Excavating past population structures by surname-based sampling: the genetic legacy of the Vikings in northwest England. Mol. Biol. Evol. 2008;25:301–309. doi: 10.1093/molbev/msm255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


