Abstract
Progressive myoclonus epilepsy (PME) is a syndrome characterized by myoclonic seizures (lightning-like jerks), generalized convulsive seizures, and varying degrees of neurological decline, especially ataxia and dementia. Previously, we characterized three pedigrees of individuals with PME and ataxia, where either clinical features or linkage mapping excluded known PME loci. This report identifies a mutation in PRICKLE1 (also known as RILP for REST/NRSF interacting LIM domain protein) in all three of these pedigrees. The identified PRICKLE1 mutation blocks the PRICKLE1 and REST interaction in vitro and disrupts the normal function of PRICKLE1 in an in vivo zebrafish overexpression system. PRICKLE1 is expressed in brain regions implicated in epilepsy and ataxia in mice and humans, and, to our knowledge, is the first molecule in the noncanonical WNT signaling pathway to be directly implicated in human epilepsy.
Introduction
More than a dozen clinico-molecular forms of progressive myoclonus epilepsy (PME) are known, including Unverricht-Lundborg disease (MIM 254800 resulting from CSTB mutations [MIM 601145]), Lafora disease (MIM 254780 resulting from EPM2A [MIM 607566] or NHLRC1 [MIM 608072] mutations), the family of neuronal ceroid lipofuscinoses (with a variety of molecular defects including PPT1 [MIM 256730], CLN4 [MIM 204300], and CLN5 [MIM 256731] mutations), and myoclonic epilepsy with ragged red fibers (MERFF [MIM 545000] with mitochondrial t-RNA mutations). Previously, we characterized three families with individuals affected with PME and ataxia but normal brain imaging, where either clinical features or linkage mapping excluded known PME loci.1–3
This report identifies a mutation in PRICKLE1 (MIM 608500) in all three of these pedigrees. PRICKLE1 is part of the noncanonical or planar cell polarity (WNT/PCP) pathway, in which some WNT family members activate a β-CATENIN (CTNNB1 [MIM 116806])-independent pathway.4 In Drosophila and vertebrates, the WNT/PCP pathway likely regulates cell polarization.5 Depleting Prickle genes in the zebrafish embryo alters the convergent-extension movements essential for gastrulation and disrupts normal calcium signaling.6–8 PRICKLE1 is part of a gene family encoding proteins containing a highly conserved PET domain, which mediates Prickle1-protein-binding interactions.6,9–11 Prickle1 was discovered independently based on its ability to bind and functionally interact with the RE1-SILENCING TRANSCRIPTION FACTOR (REST [MIM 600571], which was thus separately named Rilp, for REST/NRSF interacting LIM domain protein), an essential regulator of neural genes.12,13 The PRICKLE1 mutation identified in this study is located in the PET domain and disrupts the PRICKLE1 and REST interaction in vitro and alters the normal function of PRICKLE1 in an in vivo zebrafish overexpression system.
Material and Methods
Subjects
Clinical details of the three pedigrees were previously described;1–3 pedigree B was subsequently expanded with eight more affecteds identified in three nuclear families. Clinical studies were approved by the Institutional Review Boards of the Tel Aviv Sourasky Medical Center and the Jordan University of Science and Technology. Informed consent was obtained from participating subjects and their legal guardians. The control brain specimens were obtained from a 60-year-old male with cirrhosis who died suddenly of atherosclerotic heart disease, after exemption by the Institutional Review Board of the University of Iowa and within guidelines established by Iowa statute.
Fine Mapping and Haplotyping
Microsatellite markers within the chromosome 12 pericentromeric linkage region were selected from the Marshfield human linkage map. Genotyping of 47 individuals from 3 families (Figure 1) was performed by the Australian Genome Research Facility. Marker order is based on the current human sequence map (NCBI Build 36.3).
Figure 1.
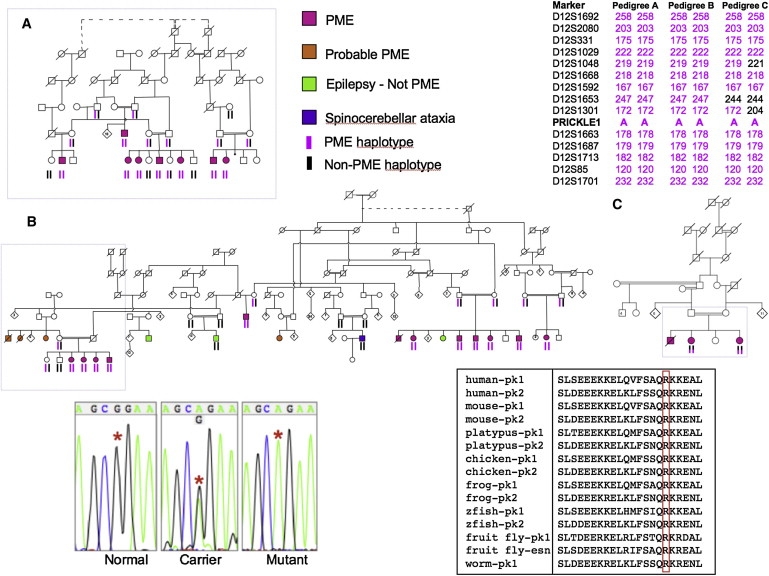
Pedigrees of the Affected Families, Representative Sequences, and Evolutionary Comparison of the Altered PRICKLE1 Amino Acid
Nine nuclear families from three pedigrees including 23 subjects with progressive myoclonus epilepsy and ataxia (pink symbol). Boxes on pedigrees indicate individuals previously reported by Berkovic et al.1 (A), El-Shanti et al.2 (B), and Straussberg et al.3 (C). Dotted lines indicate individuals believed to be related, but the exact relationship was unknown. Subjects who probably had the familial syndrome but were not personally examined are shown in orange. Shared chromosome 12 haplotypes of affected subjects are shown on the top right. Haplotypes were remarkably stable within nuclear families and extended pedigrees. Individuals with epilepsy or ataxia, clinically distinct from the familial syndrome (green and purple symbols), did not share the haplotypes or have the PRICKLE1 mutation. Representative DNA sequence chromograms from normal, carrier, and affected (mutant) individuals are in the bottom left panel with the red asterisks denoting the position of the abnormal nucleotide. Amino acid sequence alignment surrounding the altered amino acid for PRICKLE proteins in multiple species. pk1, Prickle1 protein; pk2, Prickle2 protein; esn, espinas protein; zfish, zebrafish. Accession numbers for the protein sequences are: human-pk1, NP_694571; human-pk2, NP_942559; mouse-pk1, NP_001028389; mouse-pk2, NP_001074615; platypus-pk1, XP_001505284; platypus-pk2, XP_001508261; chicken-pk1, XP_416036; chicken-pk2, XP_001234704; frog-pk1, NP_001016939; frog-pk-2, NP_001103517; zfish-pk1, NP_899185; zfish-pk2, NP_899186; fruit fly-pk1, NP_724534; fruit fly-esn, CAB64381; worm-pk1, NP_741435. The amino acid altered in the families and the corresponding amino acid in Prickle proteins from other species are boxed in red.
Resequencing
PRICKLE1 amplicons (Table S2 available online) were sequenced with an automated ABI sequencer with dye terminator chemistry. After DNA amplification, unincorporated PCR primers and dNTPs in the sample were removed prior to sequencing by isolation of the desired band in a 2% agarose gel, followed by column purification. Sequences were analyzed with the computer program PHRED, which calls the bases, and PHRAP that assembled the sequence on a PC.
Control Genotyping
The 1054 individuals from the HGD-CEPH panel and the 300 Middle Eastern individuals were genotyped with the Taqman (ABI) assay on an ABI 7900 HT Fast Real Time PCR machine with the following primers according to manufacturer's instructions: PR1ex4-ex4F, GAAAAAAGAGTTGCAGGTGTTCAGT; PR1ex4-ex4R, TTAATTGTTCCTCTTCCCAGTGCTT; PR1ex4-ex4V1, VIC, CTCAGCGGAAGAAA; PR1ex4-ex4M1, FAM, CTCAGCAGAAGAAA.
The entire PRICKLE1 gene was also directly resequenced in an additional 288 individuals, including 96 individuals of Jordanian-Palestinian ancestry.
Immunohistochemistry and Immunostaining Protocols
For mouse sections and HeLa cells, the solution used was PBS 1× Dulbecco's (pH 7.4) and the blocking solution for rabbit antibodies used was serum-free blocking media (Dako, X0909). For mouse combined with rabbit antibodies, we used M.O.M. Mouse Ig Blocking Reagent (Vector, BMK-2202); add 2 drops of stock solution to 2.5 ml of PBS. For human sections, the solution used was PBS 1X Dulbecco's (pH 7.4) and the blocking solution was serum-free blocking media (Dako, X0909).
Antibodies
Rabbit polyclonal antibody to mouse Prickle1 was produced with amino acids 808–822 by A.G.B. and with amino acids 339–514 by D.A. Specificity for Prickle1 and not Prickle2 was confirmed by immunostaining of myc-tagged cDNAs encoding Prickle1 or Prickle2 proteins into HeLa cells (data not shown). NeuN (Chemicon), GFAP (Dako), myc 9E10 (Sigma), and GFAP (Santa Cruz) antibodies were used at a 1:400–1:500 dilutions for immunohistochemistry and immunoblotting. Mouse and rabbit antibodies were diluted in M.O.M. Diluent (600 μl of protein concentrate stock solution added to 7.5 ml of PBS [Vector, BMK-2202]). Sequential rabbit antibodies were diluted in PBS 1X. Secondary antibodies were diluted 1/500 in blocking for mouse+rabbit antibodies or in PBS for rabbit antibody. We used the goat anti-rabbit IgG (Fab′)2-Alexa Fluor 568 conjugated antibody (Invitrogen, A21069) for the anti-GFAP antibody and the goat anti-mouse IgG (Fab′)2-Alexa Fluor 568 conjugated Ab (Invitrogen, A11019) for anti-NeuN antibody. We used the goat anti-rabbit IgG (Fab′)2-Alexa Fluor 488 conjugated Ab (Invitrogen, A11070) for the Prickle1 antibodies. The nuclear counterstain To-Pro3 (Invitrogen, T3605) was diluted 1/2000 in PBS 1X or DAPI. Slides were mounted with Vector Labs, H-1000.
Staining Protocol, Mouse Sections
Procedure. Procedures were performed at room temperature or as indicated. Sections were deparaffinized with autostainer program #3 and rehydrated with ddH2O. To perform antigen retrieval by microwave, citrate buffer (pH 6.0) was prewarmed for ∼30 s in a Teflon Coplin jar, and slides were added under the following conditions: restriction temperature, +95°C; wattage, #6 (601 W); pulse 5 min, hold 5 min (no microwave applied), pulse 5 min, then slides were left in the dish to cool to room temperature for about 20 min. Next, slides were washed in PBS 1X (pH 7.4), 3x3 min. Sections were permeabilized with 0.1% Triton X-100 in PBS for 10 min, washed in PBS 1X (pH 7.4), 3x3 min. Blocking was performed for rabbit and mouse primary antibodies together by applying the working solution of M.O.M. mouse Ig blocking reagent X1 hr, then PBS 3x3 min, then working solution M.O.M. diluent for 5 min.
Primary Antibodies. Serum-free blocking media was added for 1 hr, then primary antibodies were added at the following dilutions: rabbit anti-prickle 1 antibodies 1/250-1/375 were added with mouse anti-NeuN (Chemicon, MA13377) 1/500 or rabbit anti-prickle 1 affinity-purified antibody 1/250-1/375 was added followed by rabbit anti-GFAP Ab (Dako, Z0344) 1/1000 and stored in a slide folder at +4°C.
Sequential Stainings. The following variation was used. Sections were blocked (serum-free protein block) for 30 min, then primary antibody (anti-GFAP antibody) was added for 1 hr, then washed with PBS 1X (pH 7.4), 6 times for 3 min, and then the secondary antibody-conjugated fluorophore was added (in the dark) for 30 min. The slide was then washed with PBS 1X (pH 7.4), 6x for 3 min. Nuclear counterstain (in the dark) was then added for 5 min. Slides were then rinsed in PBS 1X and mounted in VectaShield and stored in slide-folder at +4°C. Nuclei were viewed in the Far red channel (647 nm Ex). Anti-Prickle1 antibodies were viewed in the green channel (488 nm Ex), and anti-GFAP or anti-NeuN antibodies were viewed in red channel (568 nm Ex).
Staining Protocol, Human Sections
Procedure. Procedures were performed at room temperature or as indicated. Sections were deparaffinized (with an autostainer) and rehydrated with ddH2O. Antigen retrieval was performed by microwave staining. Citrate buffer (pH 6.0) was prewarmed in the microwave for ∼30 s in a Teflon Coplin jar. Slides were then added with temperature restriction: +95°C, wattage: #6 (601 W), pulse 5 min, hold 5 min (no microwave applied), pulse 5 min. Slides were then cooled to room temperature for about 20 min, washed in PBS 1X (pH 7.4), 3x for 3 min, and permeabilized with 0.1% Triton X-100 in PBS for 10 min, washed in PBS 1X (pH 7.4), 3x3 min blocked with serum-free blocking media for 1 hr. Primary antibodies were added for 1 hr, then washed with PBS, 6x for 3 min, secondary antibodies were added in the dark for 1 hr, then washed with PBS, 6x for 3 min. Slides were mounted with VectaShield and sections were stored in a slide folder at +4°C.
Primary Antibodies. Primary antibodies were diluted in PBS 1X. Rabbit anti-prickle 1 antibody 1/250-1/375 and mouse anti-NeuN (Chemicon, MA13377) 1/500 were used together, and rabbit anti-prickle 1 antibody 1/250-1/375 and rabbit anti-GFAP Ab (Dako, Z0344) 1/1000 were used sequentially. Goat anti-rabbit IgG (Fab′)2-Alexa Fluor 488 conjugated antibody (Invitrogen, A11070) secondary antibody was diluted 1/500 in PBS. To-Pro3 (Invitrogen, T3605) was diluted 1/2000 in PBS 1X and was used as the nuclear counterstain. Slides were mounted with VectaShield (Vector Labs, H-1000).
Sequential Stainings. After the first primary and secondary antibodies were applied, slides were blocked (serum-free protein block) for 30 min, and then the second primary antibody was applied for 1 hr, then the slides were washed in PBS 1X (pH 7.4), 6x for 3 min, and then the second secondary antibody-fluorophore conjugated was added in the dark for 30 min, and then washed in PBS 1X (pH 7.4), 6x for 3 min. Nuclear counterstain was then added (in the dark) for 5 min, and slides were rinsed with PBS 1X, and mounted in VectaShieldand and stored in a slide-folder at +4°C. Nuclei are viewed in the far red channel (647 nm Ex), anti-Prickle1 antibodies are viewed in the green channel (488 nm Ex), and anti-NeuN or GFAP antibodies are viewed in red channel (568 nm Ex).
HeLa Cell Staining
Mouse monoclonal anti-Myc 9E10 (Sigma) 1/1000 was added (EGFP viewed directly in green channel), then PBS was added 6 times for 3 min followed by secondary antibodies in the dark for 1 hr, then PBS 6 times for 3 min, and slides were then mounted with VectaShield and double labeled sections were stored with Ms + Rb Abs in slide folder at +4°C.
Confocal Microscopy
All confocal microscopy was performed on a Zeiss 510 Confocal Microscope at the University of Iowa or on a Leica confocal microscope at Stanford University. The thickness of the confocal images and all details of exposure time are embedded within the digital photographs used in this manuscript and available upon request.
Plasmids
The full-length human PRICKLE1 cDNA in the PCR-bluntII-TOPO (Open Biosystems) was cloned into the EcoRI (5′) and XhoI (3′) sites of the pCS2+ vector, with the EGFP epitope added in-frame to the 3′ end of the gene by PCR site-directed mutagensis/chimeragnesis. The R104Q encoding mutation was introduced by site-directed mutagenesis, and the entire cDNA was resequenced after introduction of the mutation to insure that no additional mutations were introduced. Myc-REST was the same construct used previously to define the Prickle-REST interaction.12,13
Transfections
Transfections into HeLa cells were performed as previously described.12,13
Nuclear REST Measurement. To measure the impact of the PRICKLE1 mutation on the localization of REST, cells transfected with wild-type and myc-REST or mutant PRICKLE1 and myc-REST and quantified with a variation of the method previously reported.14,15 All GFP+/myc+ cells were counted and a score of 4 was assigned for myc nuclear fluorescence much greater than myc cytoplasmic fluorescence, 3 for myc nuclear fluorescence greater than myc cytoplasmic fluorescence, 2 for myc nuclear fluorescence equal to myc cytoplasmic fluorescence, 1 for myc nuclear fluorescence less than myc cytoplasmic fluorescence, and 0 for myc nuclear fluorescence much less than myc cytoplasmic fluorescence. The mean REST nuclear score was calculated for 100 GFP+/myc+ cells in each condition.
Coimmunoprecipitations
Coimmunoprecipitations with slight variations as previously described.12,13 In brief, myc-mouse REST and GFP-human Prickle1 plasmids were cotransfected into HeLa cells according to the Lipofectamine 2000 protocol. After 48 hr incubation, cells were lysed with 300 μl cold NET-N + Roche Protease Inhibitor. Lysates were sonicated 3 times for 4 s. 60 μl of the crude lysates were removed and mixed with 110 μl of denaturing loading buffer. The remaining lysates were precleared by adding 5 μl of anti-rabbit beads and rotating at room temperature for 20 min. The beads were removed by centrifuging samples briefly at 10,000 rpm and removing the supernatant. Precleared lysates were then mixed with 10 μl rabbit anti-GFP conjugated beads (Santa Cruz) and rotated for 2 hr at 4°C, followed by 30 min rotation at room temperature. The supernatant was removed and the beads rinsed 3 times with 500 μL of cold NET-N lysis buffer. After rinsing, 80 μl of denaturing loading buffer was added to the immunoprecipitated samples. 15 μl of IP lysates or 10 μl of crude lysates were loaded onto a 10% SDS-PAGE gel, ran for 75 min at 200V, and electroblotted overnight at 140 mA. Membrane was incubated 1:2000 anti-GFP (Roche) or anti-myc (Sigma) for 4 hr at 4°C.
PCR Conditions for Sequencing
Primers used are listed in Table S2. For exons 2–8, the mix used for PCR was: 2 μl DNA at 20 ng/μl, 2 μl 10× NH4 reaction buffer, 2 μl dNTP (5 mM), 0.75 μl MgCl2 (50 mM), 0.2 μl Biolase Taq (5 μm/μl), 0.5 μl forward primer, 0.5 μl reverse primer (20 μM per primer), add DDH2O to 20 μl. For exon 1, Invitrogen Platinum Pfx DNA polymerase was used. PCR cycling began after 10 min at 94°C as follows: (94°C, 45 s; annealing temperature, 30 s, 72°C, 45 s) ×35 cycles, followed by 72°C for 8 min, then 4°C until removed from thermocycler.
Zebrafish Constructs and Analysis
Zebrafish prickle cDNAs and wild-type and mutant (R → Q) PCR products were cloned into the Gateway vector system (Invitrogen). Both myc-tagged and untagged forms were created. Synthetic RNA was made with the mMessage machine kit (Ambion) and injected at the 1–8 cell stage. Alterations to gastrulation movements were monitored by live image capture at 20 hr postfertilization and older. Additionally, embryos were fixed at the 8–10 somite stage for whole-mount in situ hybridization with the molecular marker MyoD, which labels the midline as well as the developing somites. Equivalent protein expression was verified by myc-immunostaining and western blotting.
Statistical Analysis
The results of the zebrafish injections were subjected to the Fisher's exact test with the following contingency numbers: wild-type Zfsh pk1: 6 normal embryos, 36 defective embryos; mutant Zfish pk1: 20 normal embryos, 30 defective embryos; total: 26 normal embryos, 66 defective embryos.
Results
We analyzed disease features and genetics in three consanguineous Middle-Eastern pedigrees with autosomal-recessive progressive myoclonic epilepsy syndromes with ataxia1–3 that most closely resembled Unverricht-Lundborg disease, but without mutations in CSTB. Pedigree A (Figure 1) from Northern Israel was originally reported as progressive myoclonus epilepsy (a new form of Unverricht-Lundborg disease; EPM1B) beginning between 5 and 10 years; individuals were seen in adolescence or adult life and difficulty walking prior to onset of myoclonus was reported but not directly observed.1 Part of pedigree B was seen in Jordan when subjects were young and the impressive feature was an early-onset ataxia with later myoclonus and seizures.2 Re-evaluation subsequently showed features of progressive myoclonus epilepsy that was also present in newly identified members of this pedigree residing in Jordan and Northern Israel. Similarly, pedigree C was initially regarded as a predominant ataxic syndrome when children were examined in the first decade, but later re-examination showed florid progressive myoclonic epilepsy.3 The three affected children had impaired upgaze that was also observed in some affected members of pedigrees A and B. Features of a mild sensory neuropathy seen in pedigree C were not seen in the other families.
In all three pedigrees, comprising nine nuclear families, ataxia began at 4 to 5 years and evolved into an unequivocal PME phenotype with ataxia. In many forms of PME, cognitive decline is severe and generally occurs early; however, in this disorder intellect is usually preserved. Furthermore, in all affected individuals tested, brain magnetic resonance imaging gave unremarkable results. Table 1 summarizes the clinical and molecular features compared to Unverricht-Lundborg disease.
Table 1.
Comparison of the Progressive Myoclonus Epilepsy-Ataxia Syndrome Described Here to Classical Unverricht-Lundborg Disease
| Progresssive Myoclonus Epilepsy-Ataxia Syndrome | Unverricht-Lundborg Diseasea | |
|---|---|---|
| First symptom | ataxia around age 4 yr | myoclonic or tonic-clonic seizures |
| Seizure onset: mean | 7 yr | 10–11 yr |
| Seizure onset: range | 5–10 yr | 6–16 yr |
| Progressive features | worsening myoclonus, ataxia, impaired up-gaze | worsening myoclonus, ataxia |
| Cognitive decline | mild or absent | mild or absent |
| Mode of inheritance | autosomal recessive | autosomal recessive |
| Linkage | 12p11–q13 | 21q22.3 |
| Gene | PRICKLE1 | Cystatin B |
Modified and updated from Berkovic et al.1
Previous linkage analysis mapped the disease locus in pedigree A and part of pedigree B (Figure 1) to the pericentromeric region of chromosome 12.1,2 Pedigree B was next expanded considerably, and in all three pedigrees, haplotype analysis of microsatellite markers showed an identical haplotype over 12 Mb in pedigrees A and B, whereas pedigree C shared only the distal 250,000 base pairs with pedigrees A and B (Figure 1). Pedigrees A and B shared the same family name and were thought to be distantly related but the relationship could not be established with genealogical information for 5–6 generations. Pedigree C resided in a different village, had a different family name, and elders denied a connection to the family name of pedigrees A and B and to their villages.
Resequencing the entire coding region and intron-exon boundries of 47 genes in the original linkage region (Tables S1 and S2) revealed a single, shared, missense-nucleotide change in the coding region of PRICKLE1 in all three families (c.311G → A [R104Q]; Figure 1, bottom left). The altered amino acid in PRICKLE1 lies within the highly conserved PET domain and is invariant in evolution from humans to worms and in the related PRICKLE2 and Drosophila Espinas proteins (Figure 1, bottom right). This nucleotide change segregated with the clinical phenotype in all three pedigrees and was neither present in any of 1054 individuals from the CEPH-HGD control DNA samples,16 nor found in 300 samples from unrelated Middle-Eastern individuals without epilepsy. Resequencing the entire PRICKLE1 coding region showed no other variants in another 288 control individuals without PME (including 96 Middle-Eastern individuals).
Taken together, the high degree of conservation of the residue affected by the missense change, the absence of the identified nucleotide change in more than 1300 controls (2600 chromosomes), and the lack of other PRICKLE1 variants in nearly 300 additional controls strongly support that the PRICKLE1 gene mutation causes this autosomal-recessive, progressive myoclonus epilepsy-ataxia syndrome.
Prickle1 is expressed in multiple brain regions throughout mouse embryonic development, including regions implicated in epilepsy such as the hippocampus, cerebral cortex, and thalamus, as well as the primitive cerebellum17–20 (and data not shown). We performed immunohistochemical analysis with two different Prickle1-specific antibodies, each directed against a distinct epitope. In this way we also detected Prickle1 expression in the postnatal murine thalamus, hippocampus, cerebral cortex, and cerebellum (Figure 2). Costaining these tissues with the neuron-specific marker NeuN and the glia-specific marker GFAP demonstrated that Prickle1 is specifically expressed in neurons, but not in glia (Figures 2C, 2D, 2G, 2H, 2K, and 2L; cerebellum costaining not shown). Similarly, in human adult thalamus, hippocampus, cerebral cortex, and cerebellum, PRICKLE1 is in neurons rather than glia (Figure 3; GFAP staining not shown). These findings demonstrate that PRICKLE1 is expressed in multiple areas of the brain thought to be involved in generating seizures (neurons of thalamus, hippocampus, and cerebral cortex) and ataxia (cerebellar neurons).
Figure 2.
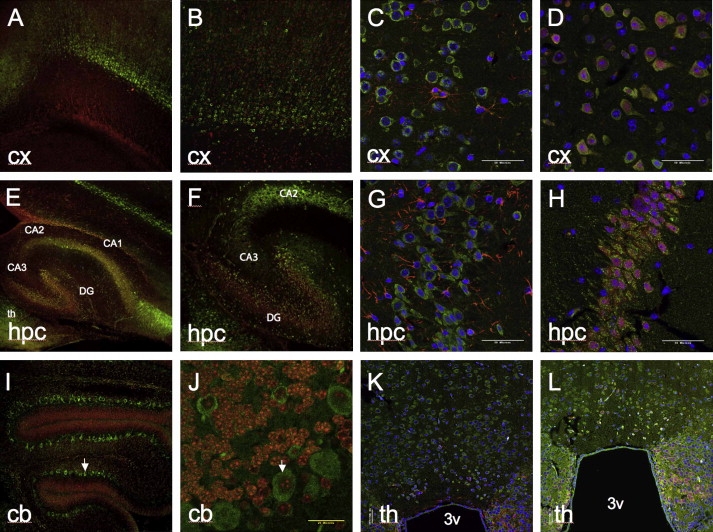
Expression of Prickle1 in Postnatal Mouse Brain
Prickle1 expression in cortex (A–D), hippocampus (E–H), cerebellum (I, J), and thalamus (K, L). For (A), (B), (E), (F), (I), and (J), Prickle1 antibody, raised against epitope corresponding to amino acids 339–514 of Prickle1, is labeled with green secondary, nuclear staining is in red. For (C), (D), (G), (H), (K), and (L), Prickle1 antibody, raised against epitope corresponding to amino acids 808–822 of Prickle1, is labeled in green. In (C), (D), (G), (H), (K), and (L), nuclear staining is in blue. In (C), (G), and (K), GFAP staining is in red. In (D), (H), and (L), NeuN, staining is in red. Arrows point to representative cerebellar Purkinje cells. 3v, third ventricle from coronal section; CA1, CA2, CA3, hippocampal Cornu Ammonis subregions; cb, cerebellum; cx, cerebral cortex; DG, dentate gyrus; hpc, hippocampus; th, thalamus. Magnification is 10× in (A), (E), and (I); 20× in (B), (F), (J), (K), and (L); 60× in (C), (D), (G), and (H). (A), (B), (E), (F), (I), and (J) are from P7 brain, (C), (D), (G), (H), (K), and (L) are from adult brain. (A)–(J) are from sagittal sections, (K) and (L) are coronal sections. A representative sample of sagittal sections from a P19 brain at multiple magnifications can be found in Figure S1.
Figure 3.
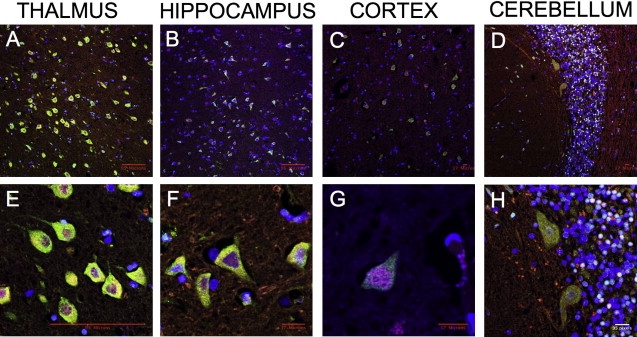
Expression of PRICKLE1 in Adult Human Brain
Low-power (A–D) and high-power (E–H) confocal images from immunostaining of adult human thalamus (A, E), hippocampus (B, F), cerebral cortex (C, G), and cerebellum (D, H). PRICKLE1 staining is in green, NeuN is in red, and nuclear staining is in blue. Scale markers are represented on the bottom right corner of each image.
To evaluate the functional effect of this mutation on PRICKLE1-partner protein interactions, we tested whether mutant PRICKLE1 can coimmunoprecipitate REST (Figure 4). Although, as previously described, wild-type PRICKLE1 binds REST directly,12,13 mutant PRICKLE1 fails to bind REST in vitro. Furthermore, because REST nuclear localization is altered in brains from patients with Huntington's disease with known mutations in Huntingtin,21 we tested whether mutant PRICKLE1 affected the subcellular localization of REST. Indeed, in HeLa cells, immunolocalization shows that whereas overexpression of wild-type PRICKLE1 results in cytoplasmic retention of REST, overexpressed mutant PRICKLE1 fails to retain REST in the cytoplasm (representative cells are shown in Figure 5A). To quantify the effect of mutant PRICKLE1 on REST nuclear localization, we counted cells based on REST expression in the cytoplasm, nucleus, or both cytoplasm and nucleus, in cells cotransfected with REST+PRICKLE1 versus REST + mutant PRICKLE1 and found significantly increased nuclear REST in cells cotransfected with mutant PRICKLE1 (Figure 5B). Therefore, the identified PRICKLE1 mutation disrupts both normal REST-binding and REST subcellular localization.
Figure 4.
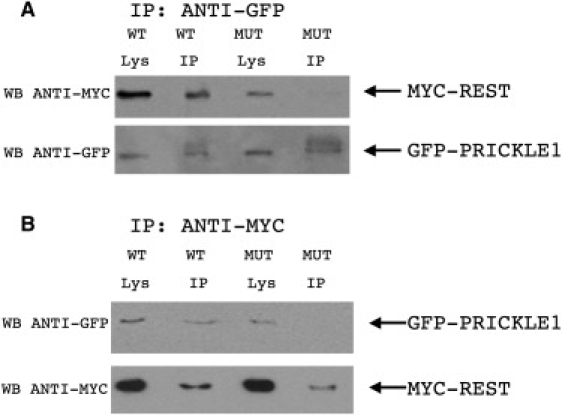
R104Q Mutant PRICKLE1 Has Impaired NRSF/REST Binding
Coimmunoprecipitation of REST with wild-type (WT) or R104Q mutant (MUT) encoding PRICKLE1 demonstrates decreased REST binding for MUT PRICKLE1. MYC-REST and either GFP-tagged WT or R104Q MUT PRICKLE1 plasmids were transfected into HeLa cells. Cell lysates were prepared in RIPA buffer and immunoprecipitated with agarose-conjugated anti-GFP antibody (A) or anti-MYC antibody (B). Immunoprecipitates (IP) were subjected to SDS-PAGE followed by western blotting (WB) with MYC or GFP antibodies. WB antibody noted to left of gels. The input (1/5) of immunoprecipitation is shown in the “Lys” lanes. Arrows to the right of the gels note the position of MYC-REST and GFP-PRICKLE1.
Figure 5.
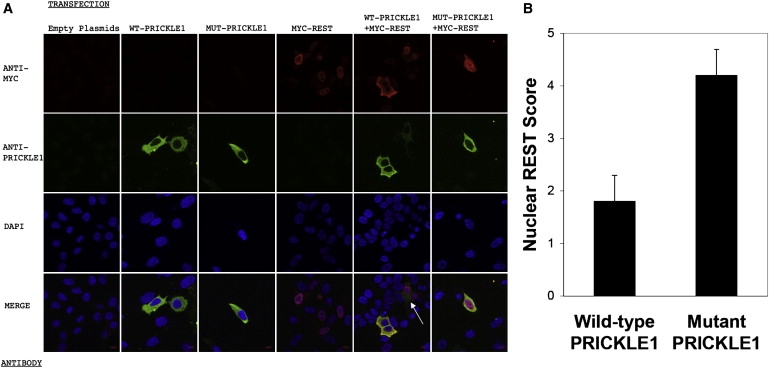
Subcellular Localization of Recombinant MYC-REST and WT or MUT PRICKLE1
(A) HeLa cells were transfected with MYC-REST and WT PRICKLE1 or MUT PRICKLE1, as noted on the top of the images in (A). Antibody staining with ANTI-MYC antibody (red), ANTI-PRICKLE1 antibody (green), DAPI nuclear staining (blue), and MERGED confocal images are noted on the left. Scale bar represents 9 microns. All confocal images were captured with identical exposure settings. The white arrow in the merged WT-PRICKLE1+MYC-REST marks a cell with relatively less WT PRICKLE1 expressed versus the two other WT PRICKLE1-expressing cells in the same panel. Note that increased REST in the nucleus is inversely proportional to WT PRICKLE1 expression, whereas the nuclear REST signal is strong even in the presence of a strong MUT PRICKLE1 signal (bottom right).
(B) Quantification of nuclear REST in PRICKLE1 versus mutant PRICKLE1 cotransfections. Results are the mean (±SD) of 100 cells for each group.
To investigate whether the mutant Prickle1 protein is functionally different from the wild-type in vivo, we injected zebrafish embryos with RNA encoding either wild-type or mutant zebrafish prickle1. As previously demonstrated, overexpressing wild-type prickle1 alters gastrulation, resulting in a reduced anterior-posterior length (Figures 6A and 6B) and lateral expansion of somites (Figures 6C and 6D). Here, we further demonstrated that, compared to wild-type Prickle1, expressing similar levels of R104Q mutant Prickle1 showed a significantly less pronounced phenotype (Figure 6E). Thus in a zebrafish overexpression assay, the identified PRICKLE1 mutation alters the in vivo function of Prickle1.
Figure 6.
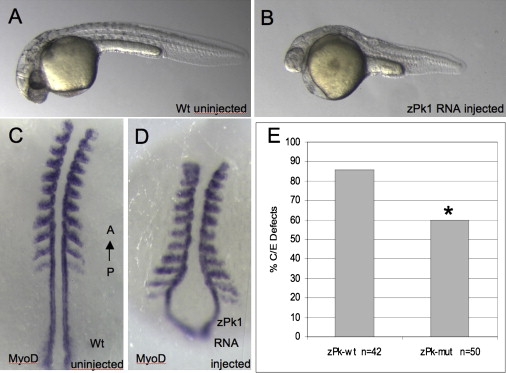
Mutant prickle1 Shows Decreased Activity In Vivo in Zebrafish
(A–D) Equivalent amounts of RNA encoding wild-type zebrafish prickle1 (pk-wt) or zebrafish prickle1 encoding the human R104Q homologous amino acid (pk-mut) were injected into zebrafish embryos and assayed for convergence extension defects by morphology (A) wild-type compared to (B) pk-overexpression, anterior to the left and with molecular markers to axial tissues in (C) wild-type and (D) pk-overexpressing embryos, anterior to the top.
(E) Decreased gastrulation defects are observed in mutant prickle1 expressing embryos compared to wild-type prickle expressing embryos. The mutant Prickle1 showed significantly decreased overexpression phenotype, with a p value of 0.01 by the Fisher's exact test.
Discussion
The finding of a shared haplotype and identical PRICKLE1 mutation in three separately ascertained families of the same ethnic group with PME suggests a founder effect. The variation reported in the original three reports is explained by the differences in ages of the subjects ascertained in the earlier studies. Longitudinal assessment showed a uniform phenotype summarized in Table 1. The genealogical data and smaller ancestral haplotype found in family C suggests that this family separated earlier from the common ancestral line.
The identification of a PRICKLE1 mutation causing this PME-ataxia syndrome raises several issues regarding the biology of PRICKLE1 and the pathophysiology of neurological disease in these affected individuals. PRICKLE1 is part of the noncanonical or planar cell polarity (WNT/PCP) signaling pathway. In vivo studies in developing flies, frogs, and fish clearly demonstrated that Prickle1 is important for planar polarity signaling. Recently, mice lacking Prickle1 were shown to die early in gestation, confirming an essential role for Prickle1 in development (H. Tao et al., 2008, Jap. Soc. Dev. Biol., abstract). At least some PRICKLE1 functions seem to be present in protein with the R104Q mutation because the mutation does not affect PRICKLE1 protein expression nor subcellular localization (Figure 5) and it still retains some wild-type function in our zebrafish overexpression system (Figure 6).
Our in vitro studies suggest that PRICKLE1 normally binds and translocates REST to the cytoplasm, thereby preventing REST from silencing target genes. The R104Q PRICKLE1 mutation lies within a known protein binding domain and thus disrupts REST binding (Figure 4), blocking the normal transport of REST out of the nucleus (Figure 5). These results suggest that tissues expressing mutant PRICKLE1 contain constitutively active REST which inappropriately downregulates REST target genes. This is significant because in addition to silencing neuronal genes in nonneuronal cells and neuronal precursors, REST also regulates target genes in mature neurons.22 REST targets include ion channels and neurotransmitters, and the PME-ataxia syndrome may occur when brain regions expressing mutant PRICKLE1 misexpress these target genes. Although Prickle function was implicated in the control of cell division and morphogenesis during zebrafish neurulation23 and REST activity was recently described in fish and frogs,24 a role for the PRICKLE/REST interaction during neurogenesis has not yet been studied.
PRICKLE1 has also been implicated in the canonical WNT/β-catenin pathway.25 Mutations in EPM2A, which cause Lafora disease—a severe form of PME with neurodegeneration—can affect the stability of the β-catenin complex in the canonical WNT/β-catenin pathway by dephosphorylating GSK3-β.26 Although the involvement of PRICKLE1 and the influence of Epm2A on β-CATENIN may be unrelated, it is possible that a common pathway connects the effects of PRICKLE1 mutations to Lafora disease and WNT signaling. It is notable that many affected individuals with PME respond to medications such as valproic acid, which also affects the WNT/β-catenin signaling pathway through inhibition of GSK3-β.27,28 On the other hand, valproic acid was also shown to alter inositol levels, which would interfere with calcium regulation.29–31
It is not yet clear how a mutation in human PRICKLE1 leads to the PME syndrome, which is pathophysiologically characterized by increased cortical hyperexcitability with involvement of the cerebellum and probably other deep gray matter nuclei as well.
Future analysis of the effects of specific point mutations of Prickle1 in animal models will further elucidate the molecular mechanisms underlying this PME-ataxia syndrome. Understanding the molecular and cellular basis of the disease may lead to improved diagnostic and therapeutic approaches for afflicted individuals.
Supplemental Data
Supplemental Data include one figure and two tables and can be found with this article online at http://www.ajhg.org/.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
PHRAP, http://www.phroap.org
Acknowledgments
We thank the members of the families for their participation. We thank Chantal Allamargot and Kathy Walters for their assistance with immunostaining and confocal microscopy at the University of Iowa, and Kaye Suyama for her work with the antibodies at Stanford. A.G.B. is supported by NIH/NINDS grant K08NS48174. M.P.S. is an investigator of the Howard Hughes Medical Institute. D.C.S. and H.L.G. were supported by NIH CA112369. M.S. was supported by NIH/NCRR P20RR020171 and NIH MH067123. A.B. was supported by a grant from the Epilepsy Foundation. We thank Jeffrey Murray for access to the CEPH-HGD panel and for commentary on the manuscript. The authors have no disclosures and no conflicts of interest. A.G.B. wrote the manuscript, performed all immunohistochemical studies in HeLa cells and half of the tissue immunostaining, and oversaw all aspects of the resequencing, control genotyping, and coimmunoprecipitations. H.E. and A.D. clinically evaluated part of pedigree B. S.B. clinically evaluated all the pedigrees and integrated the clinical data, together with Z.A., S.K., A.K., M.N., S.W., A.Z., and R.S. H.E. and S.B. designed the mapping strategies. R.W., A.B., and S.C. performed the fine mapping, resequencing, and genotyping studies. A.B. compiled Table S1. S.C. compiled Table S2. A.R.B. performed the coimmunoprecipitations. P.F. and J.M. assisted in the design and analysis of he genotyping assays. M.N. ascertained and prepared the specimens for human tissue staining. S.W. prepared the Prickle1 antibody at the University of Iowa. P.G. assisted in the development of the cell-culture experiments. D.C.S. oversaw all zebrafish injections and analyses performed by H.L.G. M.S. provided the myc-REST construct and guidance in cell transfections. S.M. provided a mouse prickle1-GFP expression vector that was used in pilot studies in preparation for the use of a human Prickle1-EGFP construct. J.A., M.P.S., D.A., and E.K.V. produced the Prickle1 antibodies and performed the immunostaining at Stanford University.
References
- 1.Berkovic S.F., Mazarib A., Walid S., Neufeld M.Y., Manelis J., Nevo Y., Korczyn A.D., Yin J., Xiong L., Pandolfo M. A new clinical and molecular form of Unverricht-Lundborg disease localized by homozygosity mapping. Brain. 2005;128:652–658. doi: 10.1093/brain/awh377. [DOI] [PubMed] [Google Scholar]
- 2.El-Shanti H., Daoud A., Sadoon A.A., Leal S.M., Chen S., Lee K., Spiegel R. A distinct autosomal recessive ataxia maps to chromosome 12 in an inbred family from Jordan. Brain Dev. 2006;28:353–357. doi: 10.1016/j.braindev.2005.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Straussberg R., Basel-Vanagaite L., Kivity S., Dabby R., Cirak S., Nurnberg P., Voit T., Mahajnah M., Inbar D., Saifi G.M. An autosomal recessive cerebellar ataxia syndrome with upward gaze palsy, neuropathy, and seizures. Neurology. 2005;64:142–144. doi: 10.1212/01.WNL.0000148600.60470.E6. [DOI] [PubMed] [Google Scholar]
- 4.Ciani L., Salinas P.C. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat. Rev. Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- 5.Veeman M.T., Axelrod J.D., Moon R.T. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev. Cell. 2003;5:367–377. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 6.Carreira-Barbosa F., Concha M.L., Takeuchi M., Ueno N., Wilson S.W., Tada M. Prickle 1 regulates cell movements during gastrulation and neuronal migration in zebrafish. Development. 2003;130:4037–4046. doi: 10.1242/dev.00567. [DOI] [PubMed] [Google Scholar]
- 7.Veeman M.T., Slusarski D.C., Kaykas A., Louie S.H., Moon R.T. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi M., Nakabayashi J., Sakaguchi T., Yamamoto T.S., Takahashi H., Takeda H., Ueno N. The prickle-related gene in vertebrates is essential for gastrulation cell movements. Curr. Biol. 2003;13:674–679. doi: 10.1016/s0960-9822(03)00245-8. [DOI] [PubMed] [Google Scholar]
- 9.Bastock R., Strutt H., Strutt D. Strabismus is asymmetrically localised and binds to Prickle and Dishevelled during Drosophila planar polarity patterning. Development. 2003;130:3007–3014. doi: 10.1242/dev.00526. [DOI] [PubMed] [Google Scholar]
- 10.Bellaiche Y., Beaudoin-Massiani O., Stuttem I., Schweisguth F. The planar cell polarity protein Strabismus promotes Pins anterior localization during asymmetric division of sensory organ precursor cells in Drosophila. Development. 2004;131:469–478. doi: 10.1242/dev.00928. [DOI] [PubMed] [Google Scholar]
- 11.Tree D.R., Shulman J.M., Rousset R., Scott M.P., Gubb D., Axelrod J.D. Prickle mediates feedback amplification to generate asymmetric planar cell polarity signaling. Cell. 2002;109:371–381. doi: 10.1016/s0092-8674(02)00715-8. [DOI] [PubMed] [Google Scholar]
- 12.Shimojo M., Hersh L.B. REST/NRSF-interacting LIM domain protein, a putative nuclear translocation receptor. Mol. Cell. Biol. 2003;23:9025–9031. doi: 10.1128/MCB.23.24.9025-9031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimojo M., Hersh L.B. Characterization of the REST/NRSF-interacting LIM domain protein (RILP): localization and interaction with REST/NRSF. J. Neurochem. 2006;96:1130–1138. doi: 10.1111/j.1471-4159.2005.03608.x. [DOI] [PubMed] [Google Scholar]
- 14.Galigniana M.D., Radanyi C., Renoir J.M., Housley P.R., Pratt W.B. Evidence that the peptidylprolyl isomerase domain of the hsp90-binding immunophilin FKBP52 is involved in both dynein interaction and glucocorticoid receptor movement to the nucleus. J. Biol. Chem. 2001;276:14884–14889. doi: 10.1074/jbc.M010809200. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez-Alegre P., Paulson H.L. Aberrant cellular behavior of mutant torsinA implicates nuclear envelope dysfunction in DYT1 dystonia. J. Neurosci. 2004;24:2593–2601. doi: 10.1523/JNEUROSCI.4461-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cann H.M., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 17.Crompton L.A., Du Roure C., Rodriguez T.A. Early embryonic expression patterns of the mouse Flamingo and Prickle orthologues. Dev. Dyn. 2007;236:3137–3143. doi: 10.1002/dvdy.21338. [DOI] [PubMed] [Google Scholar]
- 18.Katoh M. Identification and characterization of human PRICKLE1 and PRICKLE2 genes as well as mouse Prickle1 and Prickle2 genes homologous to Drosophila tissue polarity gene prickle. Int. J. Mol. Med. 2003;11:249–256. [PubMed] [Google Scholar]
- 19.Okuda H., Miyata S., Mori Y., Tohyama M. Mouse Prickle1 and Prickle2 are expressed in postmitotic neurons and promote neurite outgrowth. FEBS Lett. 2007;581:4754–4760. doi: 10.1016/j.febslet.2007.08.075. [DOI] [PubMed] [Google Scholar]
- 20.Tissir F., Goffinet A.M. Expression of planar cell polarity genes during development of the mouse CNS. Eur. J. Neurosci. 2006;23:597–607. doi: 10.1111/j.1460-9568.2006.04596.x. [DOI] [PubMed] [Google Scholar]
- 21.Zuccato C., Tartari M., Crotti A., Goffredo D., Valenza M., Conti L., Cataudella T., Leavitt B.R., Hayden M.R., Timmusk T. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 22.Palm K., Belluardo N., Metsis M., Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J. Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciruna B., Jenny A., Lee D., Mlodzik M., Schier A.F. Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature. 2006;439:220–224. doi: 10.1038/nature04375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olguin P., Oteiza P., Gamboa E., Gomez-Skarmeta J.L., Kukuljan M. RE-1 silencer of transcription/neural restrictive silencer factor modulates ectodermal patterning during Xenopus development. J. Neurosci. 2006;26:2820–2829. doi: 10.1523/JNEUROSCI.5037-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan D.W., Chan C.Y., Yam J.W., Ching Y.P., Ng I.O. Prickle-1 negatively regulates Wnt/beta-catenin pathway by promoting Dishevelled ubiquitination/degradation in liver cancer. Gastroenterology. 2006;131:1218–1227. doi: 10.1053/j.gastro.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y., Wang Y., Wu C., Zheng P. Dimerization of Laforin is required for its optimal phosphatase activity, regulation of GSK3beta phosphorylation, and Wnt signaling. J. Biol. Chem. 2006;281:34768–34774. doi: 10.1074/jbc.M607778200. [DOI] [PubMed] [Google Scholar]
- 27.Chen G., Huang L.D., Jiang Y.M., Manji H.K. The mood-stabilizing agent valproate inhibits the activity of glycogen synthase kinase-3. J. Neurochem. 1999;72:1327–1330. doi: 10.1046/j.1471-4159.2000.0721327.x. [DOI] [PubMed] [Google Scholar]
- 28.Wiltse J. Mode of action: inhibition of histone deacetylase, altering WNT-dependent gene expression, and regulation of beta-catenin–developmental effects of valproic acid. Crit. Rev. Toxicol. 2005;35:727–738. doi: 10.1080/10408440591007403. [DOI] [PubMed] [Google Scholar]
- 29.Galit S., Shirley M., Ora K., Belmaker R.H., Galila A. Effect of valproate derivatives on human brain myo-inositol-1-phosphate (MIP) synthase activity and amphetamine-induced rearing. Pharmacol. Rep. 2007;59:402–407. [PubMed] [Google Scholar]
- 30.Shimshoni J.A., Dalton E.C., Jenkins A., Eyal S., Ewan K., Williams R.S., Pessah N., Yagen B., Harwood A.J., Bialer M. The effects of central nervous system-active valproic acid constitutional isomers, cyclopropyl analogs, and amide derivatives on neuronal growth cone behavior. Mol. Pharmacol. 2007;71:884–892. doi: 10.1124/mol.106.030601. [DOI] [PubMed] [Google Scholar]
- 31.Tokuoka S.M., Saiardi A., Nurrish S.J. The mood stabilizer valproate inhibits both inositol- and diacylglycerol-signaling pathways in Caenorhabditis elegans. Mol. Biol. Cell. 2008;19:2241–2250. doi: 10.1091/mbc.E07-09-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


