Main Text
To the Editor: Recently, Li et al.1 reported on the frequency of the derived allele ADH1B∗47His in Eastern and Western Asia.
The data were based on meta-analysis of the published results for 131 populations and the results of genotyping of samples from 37 additional populations, performed by the authors. The authors made the suggestion that there had been separate and independent increases in the frequency of the ADH1B∗47His allele in Eastern and Western Asia. Their worldwide-frequency-distribution diagram for this allele also includes the previous reports that there is a regional elevation in the ADH1B∗47His allele for Eastern Europeans (Russians). The authors acknowledged that Central Asian population data that would support their conclusion about this distribution are, as yet, absent.
The allele-frequency data derived from the two previous studies2,3 were essential for the hypothesis that a local maxima for distribution of ADH1B∗47His might exist on the Russian Plain and in Southwest Asia.1 We noted, however, that the frequencies of the ADH1B∗47His allele presented in these studies for Russians2 and Iranian populations3 are significantly distinct from the allele frequency for the same or neighboring population groups reported in other studies.4–7 There are several fairly obvious factors that could produce these differences: first, genotyping errors might occur, depending on methods that vary between the different studies; second, small (nonrepresentative) numbers of genotyped samples were included; third, genetic drift might occur in a local community enrolled in genotyping within the ethnic group of interest; fourth, there might have been recent undetected gene flow from a distant geographic region with a different frequency of the allele. There are reasonable concerns that errors in genotype data for some populations might affect the meta-analysis and the resulting conclusions regarding the evolution of this functionally significant polymorphism.
In this Letter, we elucidated the allele-frequency data for Russians and Southwest Asian populations and added data on Central Asian and Siberian populations to provide a complete description of the Eurasian distribution of the ADH1B∗47His allele. We have typed the ADH1B Arg47His polymorphism for 3408 individuals in 46 additional populations by PCR-RFLP with MslI using primers and protocols described eslewhere4 and have developed a refined geographic map that includes 172 populations from Africa and Eurasia (Figure 1, Table 1).
Figure 1.
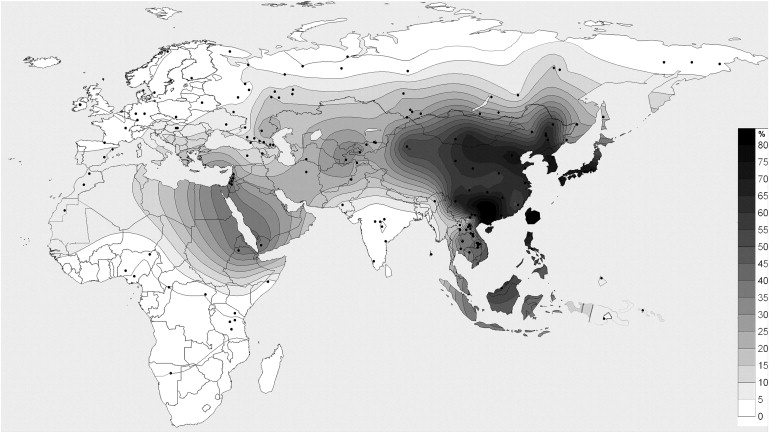
The Geographic Distribution of the ADH1B∗47His Allele
The frequency data and geographic coordinates for populations under study (Table 1), added to the frequency data and geographic coordinates for 126 populations published by Li et al.1 and ALFRED. Surface Mapping System program Surfer 8.00 (Golden Software) was used.
Table 1.
Populations Sampled in This Study
| Country | Population | Latitude | Longitude | N | Number of Genotypes |
ADH1B∗47His Frequency (%) | ||
|---|---|---|---|---|---|---|---|---|
| Arg/Arg | Arg/His | His/His | ||||||
| Russia | Russians (Kostroma)a | 57.8 N | 40.9 E | 118 | 111 | 7 | 0 | 3.0 |
| Russia | Russians (Kursk) | 51.7 N | 36.2 E | 86 | 73 | 13 | 0 | 7.6 |
| Russia | Russians (Rostov district) | 47.3 N | 39.8 E | 96 | 88 | 8 | 0 | 4.2 |
| Russia | Russians (Moscow) | 55.8 N | 37.6 E | 104 | 100 | 4 | 0 | 1.9 |
| Russia | Russians (Bashkortostan) | 54.7 N | 56.0 E | 99 | 94 | 5 | 0 | 2.5 |
| Russia | Russians (Siberia)b | 84.9 N | 56.4 E | 487 | 429 | 58 | 0 | 6.0 |
| Russia | Russians (Chukotka) | 64.7 N | 177.5 E | 29 | 25 | 4 | 0 | 6.9 |
| Ukraine | Ukrainiansa | 48.0 N | 34.0 E | 109 | 91 | 18 | 0 | 8.3 |
| Belarus | Byelorussians | 54.0 N | 27.0 E | 126 | 119 | 7 | 0 | 2.8 |
| Russia | Kola Saami | 68.0 N | 35.0 E | 62 | 57 | 5 | 0 | 4.0 |
| Russia | Komi-zyrians | 61.4 N | 50.8 E | 49 | 44 | 5 | 0 | 5.1 |
| Russia | Maris | 57.0 N | 48.0 E | 98 | 77 | 20 | 1 | 11.2 |
| Russia | Udmurts (Purga) | 56.3 N | 53.0 E | 68 | 51 | 17 | 0 | 12.5 |
| Russia | Udmurts (Igra) | 57.9 N | 53.3 E | 94 | 75 | 16 | 3 | 11.7 |
| Russia | Tatars | 55.6 N | 49.3 E | 21 | 16 | 5 | 0 | 11.9 |
| Armenia | Armenians | 40.1 N | 44.5 E | 39 | 32 | 6 | 1 | 10.3 |
| Russia | Ingush | 43.2 N | 44.8 E | 47 | 33 | 13 | 1 | 16.0 |
| Russia | Darginians | 42.4 N | 47.5 E | 49 | 36 | 13 | 0 | 13.3 |
| Russia | Avars | 42.5 N | 46.7 E | 50 | 33 | 15 | 2 | 19.0 |
| Georgia | Abkhaz | 43.0 N | 41.0 E | 50 | 41 | 8 | 1 | 10.0 |
| Russia | Balkars | 43.5 N | 43.6 E | 40 | 30 | 9 | 1 | 13.7 |
| Russia | Cherkess | 44.2 N | 42.1 E | 53 | 40 | 12 | 1 | 13.2 |
| Russia | Kalmyks | 46.3 N | 44.2 E | 59 | 30 | 27 | 2 | 26.3 |
| Iran | Iraniansa | 35.0 N | 57.0 E | 41 | 24 | 14 | 3 | 24.4 |
| Turkmenistan | Turkmen | 38.8 N | 57.0 E | 54 | 35 | 16 | 3 | 20.4 |
| Uzbekistan | Uzbeks | 42.8 N | 74.6 E | 25 | 13 | 11 | 1 | 26.0 |
| Kirghizia | Kirghizs | 40.5 N | 72.6 E | 102 | 41 | 54 | 7 | 33.3 |
| Tadjikistan | Tadjiks | 38.6 N | 68.8 E | 60 | 23 | 32 | 5 | 35.0 |
| Tadjikistan | Pamir mountain dwellersa | 37.8 N | 71.6 E | 30 | 18 | 11 | 1 | 21.7 |
| Kazakhstan | Kazakhs | 43.3 N | 76.6 E | 35 | 22 | 12 | 1 | 20.0 |
| Kazakhstan | Uyghurs | 43.0 N | 77.0 E | 29 | 19 | 9 | 1 | 19.0 |
| Russia | Altaians Northern | 51.9 N | 85.6 E | 96 | 57 | 34 | 5 | 22.9 |
| Russia | Altaians Southern | 50.8 N | 85.5 E | 65 | 40 | 24 | 1 | 20.0 |
| Russia | Tuvinians | 50.9 N | 90.1 E | 51 | 29 | 18 | 4 | 25.5 |
| Russia | Buryats (Ulan Ude) | 51.8 N | 107.6 E | 118 | 65 | 43 | 10 | 26.7 |
| Russia | Buryats (Kurumkan) | 54.2 N | 110.2 E | 61 | 38 | 22 | 1 | 19.7 |
| Russia | Buryats (Aginskoe) | 51.1 N | 114.3 E | 65 | 34 | 29 | 2 | 25.4 |
| Russia | Khants | 63.7 N | 67.1 E | 145 | 143 | 2 | 0 | 0.7 |
| Russia | Kets | 62.5 N | 86.2 E | 51 | 49 | 2 | 0 | 2.0 |
| Russia | Chukchi | 64.9 N | 176.0 E | 45 | 43 | 2 | 0 | 2.2 |
| Russia | Evenks (Eastern) | 56.0 N | 118.0 E | 72 | 60 | 11 | 1 | 9.0 |
| Russia | Yakuts | 62.9 N | 130.2 E | 53 | 44 | 8 | 1 | 9.4 |
| Russia | Siberian Tatars | 56.4 N | 84.5 E | 75 | 48 | 25 | 2 | 19.3 |
| Russia | Nivkhs | 50.1 N | 142.5 E | 31 | 20 | 11 | 0 | 17.7 |
| Russia | Udege | 46.8 N | 134.2 E | 58 | 31 | 23 | 4 | 26.7 |
| Russia | Nanais | 44.0 N | 132.0 E | 13 | 8 | 3 | 2 | 26.9 |
As indicated by Li et al.,1 “the Moscow Russian sample appears anomalous with a fairly high frequency (41%) of ADH1B∗47His” resulting in a local maximum in the central part of the Russian Plain on the allele-frequency map (Figure 2 in Li et al.1). In addition, in another study,8 the frequency of the allele for Russians in Siberia was also found to be relatively high (∼20%), which is significantly different from other European populations. To estimate the frequency among Russians more extensively, we have carried out further genotyping in Muscovites and in other Russian populations from different geographic regions (Table 1). The frequency of the ADH1B∗47His allele in Russians across the country (including both European and Asian parts of Russia) varies between 1.9% and 7.6%, with a mean frequency of 4.9% in the total group of 1019 Russian individuals. These data agree with other data on ADH1B∗47His allele frequency for Northern Russian populations of Archangelsk (5%)9 and Vologda (6%)4, and are similar to the frequencies that we estimated for the closest relative Slavic groups (Ukrainians and Byelorussians; see Table 1). The higher frequencies of the allele reported previously for Russians2,8 are, therefore, most likely the result of genotyping error. In support of such an explanation, a significant deviation in Hardy-Weinberg equilibrium is observed in at least one of these studies (p < 0.001).8
A second study3 used by Li et al. for their comparative allele-frequency estimation in Western Asia shows that the frequency of ADH1B∗47His in Iran is also relatively high (46% in Turks from Iran, 68% in Persian Zoroastrians from Iran, and 51% in Turkmen from Northeastern Iran bordering Turkmenistan) compared with the main part of other populations from the same geographic area. Again, potential errors in genotyping may also be suspected, given that an excess of heterozygotes and significant deviation from Hardy-Weinberg equilibrium was observed in this study.3 The methods used in these studies, such as PCR-RFLP with MaeIII2,8 or amplified product-length polymorphism assay,3 could potentially contribute to genotyping errors. Because ADH1B, ADH1A, and ADH1C are highly homologous genes, the genotyping requires highly specific PCR primers for the ADH1B gene. We previously genotyped a limited number of individuals from Iran and found a significantly lower frequency of the allele (24%),5 a value that is close to the allele frequency in another population in the region (Druze, 27%).4 We have also tested samples from Southern Turkmen native to a region bordering Northern Iran and found a similar frequency (20.4%). Given the ADH1B∗47His frequency in Turks that was reported earlier (12.5%),7 the general frequency of the allele in Western Asia is at least 2- to 3-fold lower than that in the data from the literature used by Li et al.1 for the estimation of geographic distribution. The Samaritans are another population group from Southwest Asia who have a very high frequency of the allele and were also employed in their analysis.1,4 They are a genetic isolate with an apparent bottleneck in their recent evolutionary history, which results in marked genetic differences between Samaritans and other populations in the region.10,11 In other Southwest Asian groups, Yemenites and Sephardic Jews, the frequency of the allele reached a maximum of 41%,4 with an average frequency ∼30% in this region. The mean is much lower than that used for genogeography reconstruction.1
To estimate the detailed geographic distribution of the ADH1B∗47His allele, we genotyped 23 populations across Asia, including Central Asian populations (Table 1). The overall allele frequencies in Southwestern Asia were relatively close to those in Central Asia (∼19%–32%). Thus, the discontinuity between West and East Asia seems to be less pronounced than previously suggested1 (Figure 1).
As was shown, Southeast Asia has the highest recorded allele frequency (70% and higher), whereas South Asia as a whole has a relatively low frequency of the ADH1B∗47His allele (∼10% or lower).1,4,7,12 According to our data, the Southwest Asian local maximum reaches 30% frequency and is connected with the Southeast Asian maximum via the Asian steppe belt, where the average allele frequency is ∼20%–30%. The frequency from the steppe region toward the North and West reduces gradually in the range of ∼10%–16% in populations across the Caucasus and Volga-Ural regions. The only exception is the Kalmyk population (26.3%), with ancestor roots from Mongol-Oirat tribes, who migrated to this region from Central Asia approximately 300 years ago. This frequency is similar to those in other related Mongoloid groups (Altaians and Buryats) (Table 1) and might reflect a relatively high frequency of the allele in ancestor Mongoloid groups in Central Asia.
This wider distribution might be explained by the migration processes in the Asian steppes. More extensive data must now be collected for ADH-locus haplotype analysis in different ethnic populations to elucidate whether the geographic frequency of the ADH1B∗47His allele is indeed increased separately; i.e., to see whether it is under direct differential selection pressure in different ethnic groups or whether the observed pattern reflects migration processes in Asia and nearby European regions.
Web Resources
The URL for data presented herein is as follows:
Allele Frequency Database (ALFRED), http://alfred.med.yale.edu/alfred
Acknowledgments
The study was approved by Institutional Research Board of the Institute of General Genetics, with corresponding informed consent obtained from human subjects. This study was supported by a program of the Presidium of the Russian Academy of Sciences, “Biodiversity and Dynamics of Gene Pools,” and E.R. is supported, in part, by the National Institute of Neurological Disorders and Stroke, the National Institute of Diabetes and Digestive and Kidney Diseases, and the Russian Foundation for Basic Research. We thank G. Chambers for very helpful comments and discussion on the manuscript.
References
- 1.Li H., Mukherjee N., Soundararajan U., Tarnok Z., Barta C., Khaliq S., Mohyuddin A., Kajuna S.L.B., Mehdi S.Q. Geographically Separate Increases in the Frequency of the Derived ADH1B∗47His Allele in East and West Asia. Am. J. Hum. Genet. 2007;81:842–846. doi: 10.1086/521201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogurtsov P.P., Garmash I.V., Miandina G.I., Guschin A.E., Itkes A.V., Moiseev V.S. Alcohol dehydrogenase ADH2–1 and ADH2–2 allelic isoforms in the Russian population correlate with type of alcoholic disease. Addict. Biol. 2001;6:377–383. doi: 10.1080/13556210020077109. [DOI] [PubMed] [Google Scholar]
- 3.Sepehr A., Kamangar F., Abnet C.C., Fahimi S., Pourshams A., Poustchi H., Zeinali S., Sotoudeh M., Islami F., Nasrollahzadeh D. Genetic polymorphisms in three Iranian populations with different risks of esophageal cancer, an ecologic comparison. Cancer Lett. 2004;213:195–202. doi: 10.1016/j.canlet.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 4.Osier M.V., Pakstis A.J., Soodyall H., Comas D., Goldman D., Odunsi A., Okonofua F., Parnas J., Schulz L.O., Bertranpetit J. A global perspective on genetic variation at the ADH genes reveals unusual patterns of linkage disequilibrium and diversity. Am. J. Hum. Genet. 2002;71:84–99. doi: 10.1086/341290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borinskaya S.A., Gasemianrodsari F., Kalyina N.R., Sokolova M.V., Yankovsky N.K. Polymorphism of Alcohol Dehydrogenase Gene ADH1B in Eastern Slavic and Iranian-Speaking Populations. Genetika (Mosk.) 2005;41:1563–1566. [PubMed] [Google Scholar]
- 6.Marusin A.V., Stepanov V.A., Spiridonova M.G., Puzyrev V.P. Alcohol dehydrogenases ADH1B and ADH7 gene polymorphism in Russian population from the Siberian region. Mol. Biol. (Mosk.) 2004;38:625–631. [PubMed] [Google Scholar]
- 7.Goedde H.W., Agarwal D.P., Fritze G., Meier-Tackmann D., Singh S., Beckmann G., Bhatia K., Chen L.Z., Fang B., Lisker R. Distribution of ADH2 and ALDH2 genotypes in different populations. Hum. Genet. 1992;88:344–346. doi: 10.1007/BF00197271. [DOI] [PubMed] [Google Scholar]
- 8.Belkovets A., Kurilovich S., Avkenstyuk A., Agarwal D.P. Alcohol Drinking Habits and Genetic Polymorphism of Alcohol Metabolism Genes in West Siberia. International Journal of Human Genetics. 2001;1:165–171. [Google Scholar]
- 9.Han Y., Oota H., Osier M.V., Pakstis A.J., Speed W.C., Odunsi A., Okonofua F., Kajuna S.L., Karoma N.J., Kungulilo S. Considerable haplotype diversity within the 23kb encompassing the ADH7 gene. Alcohol Clin. Exp. Res. 2005;29:2091–2100. doi: 10.1097/01.alc.0000191769.92667.04. [DOI] [PubMed] [Google Scholar]
- 10.Bonné B. Genes and phenotypes in the Samaritan isolate. Am. J. Phys. Anthropol. 1966;24:1–20. doi: 10.1002/ajpa.1330240102. [DOI] [PubMed] [Google Scholar]
- 11.Shen P., Lavi T., Kivisild T., Chou V., Sengun D., Gefel D., Shpirer I., Woolf E. Reconstruction of patrilineages and matrilineages of Samaritans and other Israeli populations from Y-chromosome and mitochondrial DNA sequence variation. Hum. Mutat. 2004;24:248–260. doi: 10.1002/humu.20077. [DOI] [PubMed] [Google Scholar]
- 12.Rao V.R., Bhaskar L.V., Annapurna C., Reddy A.G., Thangaraj K., Rao A.P., Singh L. Single nucleotide polymorphisms in alcohol dehydrogenase genes among some Indian populations. Am. J. Hum. Biol. 2007;19:338–344. doi: 10.1002/ajhb.20589. [DOI] [PubMed] [Google Scholar]


