Abstract
Leber hereditary optic neuropathy (LHON) is the most extensively studied mitochondrial disease, with the majority of the cases being caused by one of three primary mitochondrial DNA (mtDNA) mutations. Incomplete disease penetrance and gender bias are two features of LHON and indicate involvement of additional genetic or environmental factors in the pathogenesis of the disorder. Haplogroups J, K, and H have been shown to influence the clinical expression of LHON in subjects harboring primary mutations in European families. However, whether mtDNA haplogroups would affect the penetrance of LHON in East Asian families has not been evaluated yet. By studying the penetrance of LHON in 1859 individuals from 182 Chinese families (including one from Cambodia) with the m.11778G→A mutation, we found that haplogroup M7b1′2 significantly increases the risk of visual loss, whereas M8a has a protective effect. Analyses of the complete mtDNA sequences from LHON families with m.11778G→A narrow the association of disease expression to m.12811T→C (Y159H) in the NADH dehydrogenase 5 gene (MT-ND5) in haplogroup M7b1′2 and suggest that the specific combination of amino acid changes (A20T-T53I) in the ATP synthase 6 protein (MT-ATP6) caused by m.8584G→A and m.8684C→T might account for the beneficial background effect of M8a. Protein secondary-structure prediction for the MT-ATP6 with the two M8a-specific amino acid changes further supported our inferences. These findings will assist in further understanding the pathogenesis of LHON and guide future genetic counseling in East Asian patients with m.11778G→A.
Main Text
Leber hereditary optic neuropathy (LHON, MIM 535000) is a common cause of acute or subacute visual loss in young adults, predominately affecting males.1–4 The prevalence of LHON in western Europe is about one in 25,000–50,000 individuals.1,5,6 Genetic defects in the mitochondrial DNA (mtDNA) genome play a key role in the development of LHON, in which the three primary mtDNA mutations (m.11778G→A [R340H] in NADH dehydrogenase 4 gene [MT-ND4, MIM 516003], m.14484T→C [M64V] in MT-ND6 [MIM 516006], and m.3460G→A [A52T] in MT-ND1 [MIM 516000]) contribute to about 95% of LHON cases.1,4,6 However, the phenotypic expression of these primary mutations is very complex. Only about one-third of individuals harboring one of these three mutations eventually develop LHON, and the penetrance varies among different families.1,7,8 Therefore, identification of other factors affecting LHON penetrance would be of value in elucidating the pathophysiology of retinal neuron loss, as well as in searching for clues that might relieve visual loss or prevent the onset of LHON. Many factors, such as mtDNA background, heteroplasmy of mtDNA mutation, nuclear gene(s), and environmental factors, have been shown to play active roles in the phenotypic expression of LHON.1,4,9–20
Most recently, Hudson et al.7 provided clear evidence that the expression of LHON primary mutations was influenced by the mtDNA haplogroup background in European families. The risk of visual failure is higher when m.11778G→A or m.14484T→C mutations are present in haplogroup J and when m.3460G→A is present in haplogroup K, whereas haplogroup H reduces the disease manifestation in families with m.11778G→A.7 The cause of the association of mtDNA background effect (subclades J1 and J2b) with LHON expression in families with m.11778G→A or m.14484T→C has been narrowed to two specific combinations of amino acid changes (L236I-F19L and L236I-D171N-V356M) in the cytochrome b gene (MT-CYB; MIM 516020).21 Because the distribution patterns of the three primary mutations7,8 and the matrilineal genetic structures22,23 differ remarkably in populations from Europe and East Asia, it is indispensable to disclose the potential haplogroup effects on LHON expression in East Asians.
One hundred and seventy-five families with LHON and the m.11778G→A mutation were identified across China from our routine clinical diagnosis of 1369 unrelated subjects suspected of having LHON (including those families that were described in our previous studies8,24–26). The presence of the primary mutation m.11778G→A was verified by direct sequencing or allele-specific PCR in all families in the present study. In all cases, we detected no heteroplasmy for the m.11778G→A mutation. The clinical diagnoses were determined at the Zhongshan Ophthalmic Center or by local ophthalmologists. Unaffected individuals were defined as having no vision impairment. Informed consent, conforming to the tenets of the Declaration of Helsinki and following the guidance of sample collection of Human Genetic Disease (863 program) by the ministry of Public Health of China, was obtained from participants prior to this study, which was approved by the institutional review boards of the Zhongshan Ophthalmic Center and the Kunming Institute of Zoology. We followed the available approach to assign each mtDNA to its respective haplogroup, as previously described.23,24,27 In brief, each sample was analyzed for a 1.4 kb fragment (region 16024-850) that covers the entire mtDNA control-region sequence and was classified on the basis of the recognition of the haplogroup motif and its matching or near-matching with reported Chinese mtDNAs.23,27 Coding-region mutation motifs (e.g., m.5178C→A [MT-ND2: L237M; MIM 516001] for haplogroup D, recognized by −5176AluI) were screened to further solidify the inferred haplogroup status for some lineages. In addition, LHON penetrance information of ten reported Chinese families with m.11778G→A25,26,28–32 was also included for analysis in this study. Note that there were some errors in seven of those reported complete mtDNA sequences28–32 and that we classified those samples following a well-described strategy.33,34 The complete mtDNA sequence was determined in probands from seven M7b families, three F families, three M8a families, and two G families via the same strategy and amplification and sequencing conditions as described in our recent study.25 Sequence variations were scored relative to the revised Cambridge reference sequence (rCRS).35 The classification tree of the complete mtDNA sequences was drawn via the same procedure as described in our previous studies and others.33,36–38
We did not consider the age of subjects as a risk factor for the disease penetrance and included all subjects, irrespective of age, in order to avoid an ascertainment bias in elevating the penetrance value in the pedigree.7 The following family members were excluded from the analysis: (1) the first generation, (2) spouses of the matrilineal members, and (3) children of the male member in each family. In total, we evaluated the penetrance of LHON in 1859 individuals carrying the m.11778G→A mutation, from 182 Chinese families that were located in South and North China (including one family from Cambodia). Among these subjects, 50.6% were male and 49.4% were female. Binary logistic regression was used for determining the effects of the variables (sex and haplogroup) on the phenotypic expression of LHON with the use of SPSS 13.0 (SPSS, Chicago, IL). In regard to sample size and statistical power, we only considered the haplogroups shared by at least four families as variables in the statistical analysis. The remaining haplogroups were aggregated together into one variable. In total, 14 variables, including haplogroups M7b1′2, M7c, M8a, C, M10, D4, D5, A, B4, B5, G, Y, F, and others (lumping together Z, M9, M12, N9a, R11, and U), were separately introduced into the regression equation, with the independent variable sex and the dependent variable visual failure. Other potential variables, such as heteroplasmy of the primary mutation and pedigree generation, were neglected in our study. A p value less than 0.05 was regarded as statistically significant.
Table 1 and Table S1 (available online) list the distribution frequencies of mtDNA haplogroups in our cohort of families. The overall pattern was similar to the general profile that was observed in 41 families in our recent study,24 with haplogroups D, B, and M7 being the prevalent haplogroups. Only four of the 182 families (2.2%) with m.11778G→A belonged to haplogroup F (including family WZ4 reported by Qian et al.29); this frequency was significantly lower than the expected frequency, which ranges from 6%–27% in the regional Chinese populations across China.23,37,39 However, the overall LHON penetrance in these four families ranged from 25%–75% and was not at a lower rate of penetrance compared to that in those families with other haplogroup status (Table 2 and Table S2). We also failed to observe a significant haplogroup effect of F on the penetrance of LHON (p = 0.530; odds ratio [OR] = 1.302; 95% confidence interval [CI] = 0.571–2.966) in these F families (Table 3). The exact reason for such a low frequency of F in LHON lineages24 with no effect on penetrance remains unclear. Population stratification might account for this pattern. When we pooled the published regional Han Chinese23,37,39 as one population, to mimic the heterogeneous nature of the LHON population, haplogroup F was still significantly lower in the LHON population compared to the aggregated sample (Fisher's exact test, two tailed p = 1 × 10−6). Conversely, the frequencies of haplogroups D4 and M7c were significantly higher (p < 0.05) in the LHON patients compared to the pooled Han Chinese, but none of them affected the LHON penetrance (Tables 1 and 3). Analysis of the complete mtDNA genomes of the four haplogroup F families with LHON and m.11778G→A failed to provide any useful information (Figure 1 and Table 4), because these mtDNA samples belonged to different subbranches (two F1a, one F1b, and one F2a).
Table 1.
Haplogroup Frequency for the 1859 Subjects, from 182 Pedigrees, with the Primary LHON Mutation m.11778G→A
| Haplogroup | No. of Subjects (%)a | No. of Families (%)a | Pooled Han Chinese (%)b | p Valuec |
|---|---|---|---|---|
| D4 | 432 (23.24) | 43 (23.63) | 56 (13.73) | 0.004 |
| D5 | 161 (8.66) | 15 (8.24) | 27 (6.62) | 0.491 |
| B4 | 197 (10.60) | 19 (10.44) | 55 (13.48) | 0.347 |
| B5 | 115 (6.19) | 10 (5.49) | 17 (4.17) | 0.523 |
| M7b1′2 | 136 (7.32) | 15 (8.24) | 24 (5.88) | 0.370 |
| M7c | 122 (6.56) | 12 (6.59) | 11 (2.70) | 0.036 |
| G | 139 (7.48) | 12 (6.59) | 14 (3.43) | 0.126 |
| M8a | 112 (6.02) | 10 (5.49) | 18 (4.41) | 0.675 |
| M10 | 113 (6.08) | 8 (4.40) | 8 (1.96) | 0.104 |
| A | 75 (4.03) | 10 (5.49) | 25 (6.13) | 0.852 |
| Y | 60 (3.23) | 6 (3.30) | 6 (1.47) | 0.203 |
| C | 46 (2.47) | 7 (3.85) | 11 (2.70) | 0.605 |
| F | 27 (1.45) | 4 (2.20) | 68 (16.67) | 1×10−6 |
| N9a | 34 (1.83) | 3 (1.65) | 15 (3.68) | 0.209 |
| M12 | 35 (1.88) | 2 (1.10) | 2 (0.49) | 0.591 |
| R11 | 9 (0.48) | 2 (1.10) | 4 (0.98) | 1.000 |
| M9a | 8 (0.43) | 1 (0.55) | 10 (2.45) | 0.186 |
| Z | 9 (0.48) | 1 (0.55) | 8 (1.96) | 0.287 |
| Other | 29 (1.56) | 2 (1.10) | 29 (7.11) | 0.001 |
| Total | 1859 | 182 | 408 | – |
The reported families24–26,28–32 were also included.
Pooled Han Chinese individuals from Yunnan, Hubei, Xinjiang, Liaoning, Shandong, and Guangdong Provinces reported by Yao et al.23,39 and Kivisild et al.37
Fisher's exact test (two-tailed) was performed on the basis of the number of lineages in the pooled Han Chinese and LHON samples.
Table 2.
Haplogroup Distribution of Affected and Unaffected Individuals in 182 Families with the Primary Mutation m.11778G→A
| Sex and Haplogroup | No. of Individuals with G11778A |
|
|---|---|---|
| Affected | Unaffected | |
| Male | ||
| M7b1′2 | 41 | 27 |
| M7c | 25 | 42 |
| M8a | 15 | 39 |
| C | 20 | 9 |
| M10 | 30 | 33 |
| D4 | 91 | 129 |
| D5 | 37 | 34 |
| A | 24 | 16 |
| B4 | 44 | 57 |
| B5 | 21 | 32 |
| G | 41 | 26 |
| Y | 14 | 20 |
| F | 7 | 5 |
| Other | 26 | 35 |
| Total | 436 | 504 |
| Female | ||
| M7b1′2 | 15 | 53 |
| M7c | 9 | 46 |
| M8a | 7 | 51 |
| C | 3 | 14 |
| M10 | 10 | 40 |
| D4 | 43 | 169 |
| D5 | 23 | 67 |
| A | 7 | 28 |
| B4 | 20 | 76 |
| B5 | 10 | 52 |
| G | 21 | 51 |
| Y | 1 | 25 |
| F | 3 | 12 |
| Other | 11 | 52 |
| Total | 183 | 736 |
The previously reported Chinese families with LHON and m.11778G→A25,26,28–32 were included. For detailed information, refer to Tables S1 and S2.
Table 3.
Effect of Gender and mtDNA Haplogroups on Phenotypic Manifestation of the Primary Mutation m.11778G→A
| Variable | All 182 Families |
140 Familiesa |
||||
|---|---|---|---|---|---|---|
| p Value | Odds Ratio | 95% CI | p Value | Odds Ratio | 95% CI | |
| Sex | 2.613 × 10−32 | 3.479 | 2.830–4.277 | 5.576 × 10−27 | 3.324 | 2.671–4.138 |
| M7b1′2 | 0.032 | 1.503 | 1.035–2.183 | 0.015 | 1.631 | 1.100–2.416 |
| M7c | 0.101 | 0.702 | 0.460–1.071 | 0.123 | 0.706 | 0.454–1.099 |
| M8a | 0.002 | 0.460 | 0.282–0.751 | 0.001 | 0.391 | 0.224–0.682 |
| C | 0.050 | 1.845 | 1.000–3.404 | 0.154 | 1.655 | 0.828–3.308 |
| M10 | 0.861 | 1.038 | 0.685–1.571 | 0.451 | 1.173 | 0.775–1.778 |
| D4 | 0.213 | 0.858 | 0.675–1.092 | 0.151 | 0.827 | 0.638–1.071 |
| D5 | 0.092 | 1.353 | 0.952–1.923 | 0.076 | 1.394 | 0.965–2.013 |
| A | 0.157 | 1.426 | 0.872–2.331 | 0.154 | 1.467 | 0.867–2.481 |
| B4 | 0.744 | 0.947 | 0.682–1.314 | 0.911 | 0.981 | 0.697–1.381 |
| B5 | 0.204 | 0.752 | 0.484–1.167 | 0.165 | 0.714 | 0.443–1.149 |
| G | 0.001 | 1.827 | 1.266–2.637 | 0.003 | 1.825 | 1.230–2.709 |
| Y | 0.091 | 0.590 | 0.320–1.088 | 0.097 | 0.580 | 0.305–1.104 |
| Fb | 0.530 | 1.302 | 0.571–2.966 | – | – | – |
| Other | 0.429 | 0.847 | 0.560–1.280 | 0.705 | 0.928 | 0.629–1.368 |
Statistical testing was performed on the basis of the original data in Table S2. The visual failure was regarded as the dependent variable in the binary logistic-regression model. The haplogroups (present in at least four families) were separately introduced into the regression equation with the independent variable sex and the dependent variable visual failure.
42 small pedigrees were excluded.
When the small pedigrees were not considered, haplogroup F was excluded because of the small number of families.
Figure 1.
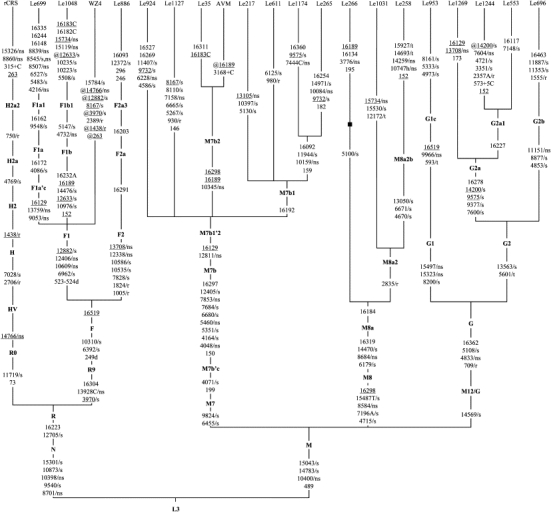
Classification Tree of 20 Complete mtDNAs with m.11778G→A and the Revised Cambridge Reference Sequence
Haplogroup names are inserted along the branches that determine the locations of the corresponding ancestral haplotypes, following the most recent update of the East Asian mtDNA phylogeny.36 Suffixes “C” and “A” refer to transversions, and “+5C” signifies an insertion of five cytosines. Deletion and heteroplasmy of a mutation are indicated by suffixes “d” and “h,” respectively. Back mutations are highlighted by the prefix “@,” and recurrent mutations are underlined. The synonymous and nonsynonymous coding-region variants in the samples are further denoted by “/s” and “/ns,” respectively. Nucleotide variations that are located in the tRNA genes and the rRNA genes are marked with “/t” and “/r,” respectively. Length mutations of the C-tract in region 303–309 and the m.11778G→A mutation in the 20 mtDNAs were omitted from the tree. Sequence WZ4 is taken from Qian et al.29 but obscured by several errors (33). Families Le1244, Le1269, and Le696 are taken from our recent studies.25,26 The Japanese LHON patient with intracranial arteriovenous malformation (AVM) is taken from Fujitake et al.40
Table 4.
Private Nonsynonymous mtDNA Sequence Variations in Chinese Families with LHON and m.11778G →A
| Family | Haplogroup | Nucleotide Variant (Amino Acid Change)a | Gene | Reported (Population Context)b | Reported (Disease Context)b | Conservationc |
|---|---|---|---|---|---|---|
| Le699 | F1a1 | G8839A (A105T) | MT-ATP6 | yes | yes | yes |
| G8545A (A7T) | MT-ATP6 | yes | no | no | ||
| A8507G (N48D) | MT-ATP8 | yes | no | no | ||
| T4216C (Y304H) | MT-ND1 | yes | yes | no | ||
| Le1048 | F1b1 | G15119A (A125T) | MT-CYB | yes | no | yes |
| G15734A (A330T) | MT-CYB | yes | yes | yes | ||
| Le924 | M7b1′2 | C6228T (L109F) | MT-CO1 | no | no | no |
| Le1127 | M7b1′2 | A7158G (I419V) | MT-CO1 | yes | yes | no |
| Le217 | M7b1 | A13105G (I257V) | MT-ND5 | yes | yes | no |
| Le1174 | M7b1 | G7444C (X514T) | MT-CO1 | no | no | – |
| C10159T (S34F) | MT-ND3 | no | no | no | ||
| Le265 | M7b1 | T10084C (I9T) | MT-ND3 | yes | no | no |
| C10159T (S34F) | MT-ND3 | no | no | no | ||
| Le266 | M8a | G3776A (S157N) | MT-ND1 | no | no | no |
| Le1031 | M8a2 | G15734A (A330T) | MT-CYB | yes | yes | yes |
| Le258 | M8a2b | T10747A (L93Q)d | MT-ND4L | no | no | yes |
| G14259A (P139S) | MT-ND6 | yes | no | no | ||
| Le953 | G1c | G9966A (V254I) | MT-CO3 | yes | yes | yes |
| Le1269 | G2a | G13708A (A458T) | MT-ND5 | yes | yes | no |
| Le1244 | G2a1 | G7604A (V7M) | MT-CO2 | yes | no | no |
The nucleotide variants were listed in a format for web-based searches, e.g. mutation G8839A should be presented as 8839G→A and m.8839G→A according to the “traditional” and “approved” formats for mtDNA-mutation nomenclature56, respectively.
The search was performed on Aug 18, 2008, with the strategy described in Bandelt et al.41 followed (e.g. both “G8839A mtDNA” and “8839G→A mtDNA” were queried).
The conservation analysis was performed by a comparison of human mtDNA (GenBank accession no. J01415) to eight different vertebrate species, including zebrafish (NC_002333), frog (AB043889), blue whale (NC_001601), mouse (AY466499), cattle (AY526085), horse (EF597513), dog (DQ480502), and gorilla (NC_001645).
This site is heterogeneous for T and A.
Consistent with a previous report for European LHON patients,7 sex is also the strongest predictor for visual loss in Chinese families (p = 2.613 × 10−32; OR = 3.479; 95% CI = 2.830–4.277), with a 3.5-fold increased risk of visual failure for males compared with females. Haplogroups M7b1′2 and G increased the risk of visual failure 1.5-fold (p = 0.032; OR = 1.503; 95% CI = 1.035–2.183) and 1.8-fold (p = 0.001, OR = 1.827, 95% CI = 1.266–2.637), respectively. Haplogroup M8a was found to be associated with a reduced risk (p = 0.002; OR = 0.460; 95% CI = 0.282–0.751). Because some of the pedigrees studied here were relatively small, we then excluded 42 pedigrees (each having five maternally related individuals at most) in order to eliminate the potential bias in scoring the affected and unaffected individuals in these small pedigrees. Analysis for the residual 1710 subjects from 140 families then yielded similar results, with an increased risk for haplogroups M7b1′2 (p = 0.015, OR = 1.631, 95% CI = 1.100–2.416) and G (p = 0.003, OR = 1.825, 95% CI = 1.230–2.709) and a reduced risk for haplogroup M8a (p = 0.001, OR = 0.391, 95% CI = 0.224–0.682) (Table 3). To minimize the probability of type II errors in the above test, we performed logistic regression by introducing all 14 variables in the regression equation, with the independent variable sex and the dependent variable visual failure. The increased risk for haplogroups M7b1′2 and G and the decreased risk for haplogroup M8a in phenotypic manifestation of m.11778G→A was further confirmed (considering all 182 families: M7b1′2, p = 0.012, OR = 1.630, 95% CI = 1.116–2.382; G, p = 4.59 × 10−4, OR = 1.949, 95% CI = 1.342–2.832; M8a, p = 0.013, OR = 0.535, 95% CI = 0.326–0.878; excluding 42 small pedigrees: M7b1′2, p = 0.008, OR = 1.714, 95% CI = 1.150–2.554; G, p = 0.002, OR = 1.903, 95% CI = 1.275–2.840; M8a, p = 0.005, OR = 0.445, 95% CI = 0.254–0.779).
The association between an increased risk of visual loss and haplogroup G is unexpected, because different families belonging to this haplogroup presented strikingly different penetrance patterns.25,26 In particular, one reported family (Le696)26 had a very high penetrance (78.6%) and harbored two pathogenic mutations, m.1555A→G (MT-RNR1; MIM 561000) and m.11778G→A, which might have enhanced the phenotypic expression and caused a bias in estimation of the haplogroup background effect. Indeed, when we excluded this family, together with four small pedigrees from the analysis, haplogroup G did not significantly increase the penetrance of LHON (p = 0.051, OR = 1.526, 95% CI = 0.998–2.335). Therefore, the effect of haplogroup G on the clinical expression of LHON should be treated with caution and further verified in a future study with more pedigrees. Analysis of the five LHON families with G status shows that these mtDNA samples can be grouped into subhaplogroups G1c, G2a, and G2b and thus share only one nonsynonymous haplogroup-specific variant, viz. m.4833A→G (T122A) in the MT-ND2 gene (Figure 1), which might account for the predisposing effect of haplogroup G in LHON penetrance.
To further define the effect of haplogroup M7b1′2 on LHON penetrance, we narrowed the potential association to specific mtDNA mutations by analyzing the entire mtDNA genomes of eight M7b1′2 families (including one previously reported Japanese LHON proband with intracranial arteriovenous malformation40) (Figure 1). All probands shared a string of nonsynonymous mutations (m.4048G→A [D248N] in MT-ND1, m.5460G→A [A331T] in MT-ND2, and m.7853G→A [V90I] in MT-CO2 [cytochrome c oxidase II; MIM 516040]) and synonymous variants (m.4164A→G in MT-ND1, m.5351A→G in MT-ND2, m.6680T→C in MT-CO1 [MIM 516030], m.7684T→C in MT-CO2) that are characteristic of haplogroup M7b, as well as m.12811T→C (Y159H) in MT-ND5 (MIM 516005), which defines haplogroup M7b1′2. At the twig level, we identified five nonsynonymous mutations in families Le924 (m.6228C→T [MT-CO1: L109F]), Le1127 (m.7158A→G [MT-CO1: I419V]), Le1174 (m.7444G→C [MT-CO1: X514F]; m.10159C→T [MT-ND3: S34F; MIM 516002]), and Le217 (m.13105A→G [MT-ND5: I257V]). With the exception of m.6228C→T, m.7444G→C, and m.10159C→T, all variants can be found in reported mtDNA samples via standard database and web-based searches.41 Some of the variants are evolutionarily conserved and have been reported in disease context (Table 4). None of these mtDNA variants has been reported to be associated with LHON, except for m.12811T→C, which was considered to be a secondary mutation for LHON expression42 and was present in two out of 35 Finnish LHON probands.5 In addition, m.12811T→C could be found in three out of 63 Dutch LHON patients,43 one case individual with cancer,44 and one patient with cerebral autosomal-dominant arteriopathy with subcortical infarcts and leukoencephalopathy (MIM 125310),45 according to extensive database searches. In the non-European context, m.12811T→C was regarded as a haplogroup-specific polymorphism in East Asians (M7b1′2)36 and Native Americans (A2h).46 The potentially synergistic effect of m.11778G→A and m.12811T→C might be the reason for an increased penetrance on the haplogroup M7b1′2 background.
Intriguingly, three of the five M7b1′2 lineages that harbored nonsynonymous mutations at the twig level also had private amino acid changes in the MT-CO1 gene (Figure 1 and Table 4); this suggests that the decreased activity of cytochrome c oxidase and the partial dysfunction of complex IV might be related to the onset of LHON.9,47–49 For instance, m.7444G→A in the MT-CO1 gene causes a change of the termination codon of MT-CO1 to lysine and was claimed to be associated with LHON,47 although this variant was prevalent in haplogroup V and should be categorized as a polymorphism.33 Note that a recent study showed that pathogenic mutations are also common in the general population.50 The previously unpublished variant m.7444G→C in Le1174 causes a similar problem as that of m.7444G→A and results in a change of the termination codon to threonine. Whether the change of the mitochondrial respiratory-chain complexes I and IV activities caused by the above mutations in MT-CO1, MT-CO2, MT-ND1, MT-ND2, MT-ND3, and MT-ND5 in M7b1′2 lineages would account for an increased risk for LHON awaits further experimental study. It is worth noting that in a recent study by Kazuno et al.,51 the four cybrid lines containing mtDNA with haplogroup status G1a1 (two), M7b2, and M7a1a generally had a lower cytosolic calcium response to histamine and a higher-level mitochondrial matrix pH compared to those cybrids containing mtDNA belonging to haplogroups N9a, A, B4, etc. This result suggests that potential alterations in mitochondrial pH and calcium concentration caused by the haplogroup background effects of M7b1′2 and G might be one of the mechanisms for the increased risk of LHON penetrance.
A protective effect of mtDNA haplogroup background has been reported for several diseases; e.g., haplogroup N9a confers resistance to type 2 diabetes in Asians52 and to metabolic syndrome in Japanese women,53 and haplogroup H reduces the risk of visual failure in European families with m.11778G→A.7 In this study, we found that M8a enacted a protective effect on the disease expression in Chinese LHON families, and this protective effect became even more pronounced when we discarded small pedigrees. Analysis of three complete M8a mtDNA sequences from these families showed that each mtDNA had at least one private nonsynonymous nucleotide change at the twig level (Figure 1 and Table 4). Family Le258 had a previously unpublished heterogeneous mutation at site 10747, and this site was conserved in vertebrates. Intriguingly, the haplogroup-specific nonsynonymous-variant pair m.8584G→A and m.8684C→T, causing a combination of amino acid changes A20T and T53I, is located in the ATP synthase 6 gene (MT-ATP6; MIM 516060). We performed protein secondary-structure modeling for the MT-ATP6 protein harboring the two M8a-specific amino acid changes in comparison to mutants containing a well-known pathogenic mutation at site 8993 (m.8993T→G or m.8993T→C), a rare LHON mutation m.9101T→C,54,55 as well as the wild-type (rCRS) by using the TMpred program. As shown in Figure 2, MT-ATP6 is a largely hydrophobic protein and contains two hydrophilic loops. Both m.8993T→C and m.8993T→G mutants alter the hydrophobicity, but m.8993T→G has a stronger effect, whereas m.9101T→C decreases the hydrophobicity close to the C-terminal end. The amino acid change A20T of M8a decreases the hydrophobicity, but this change is balanced by a reduction of hydrophilicity in the adjacent region, caused by T53I. It thus seems that the two specific amino acid changes are the cause of the protective effect of M8a and that they enhance the activity of the mitochondrial ATP synthase complex. Experimental data will be essential for confirming this speculation.
Figure 2.
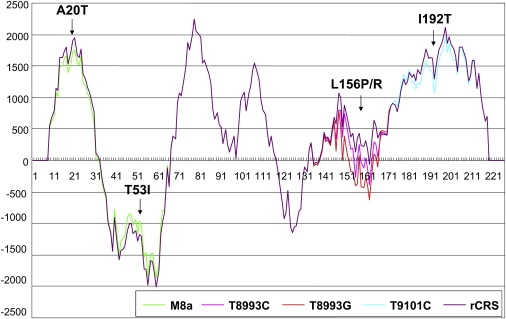
A Hydrophobicity Chart for the MT-ATP6 Protein Predicted by the TMpred Program
The hydrophobicity of the MT-ATP6 protein harboring the two haplogroup M8a specific amino acid changes (A20T-T53I) is compared to the wild-type MT-ATP6 (rCRS) and the known pathogenic mutants caused by m.8993T→C (L156P) and m.8993T→G (L156R), as well as, a rare LHON mutation m.9101T→C (I192T).
In summary, by studying 1859 individuals in 182 Chinese families with LHON and m.11778G→A, we found that haplogroup M7b1′2, as well as, possibly, haplogroup G, significantly increased the risk of visual failure in Chinese individuals with m.11778G→A, whereas M8a might have a protective effect on the penetrance of LHON. Sex is the most significant factor for influencing the clinical expression of LHON (3.48-fold) in Chinese families but this influence is lower than that in European LHON families (5.41-fold). Haplogroup F is present at a much lower frequency in these affected families than in the general Han Chinese when we only counted the matrilines, whereas haplogroups D4 and M7c are present at a significantly higher frequency in the affected families. However, none of these haplogroups showed any effect on the penetrance of LHON. Similarly, frequencies of haplogroups M7b1′2, G, and M8a are not significantly increased in LHON pedigrees despite their apparent background effect on the penetrance. The exact reason for this apparent inconsistency remains unclear and this pattern is in contrast to the European study,7 in which an internal consistency of the haplogroup association was observed, namely, haplogroup J is present at an increased frequency in LHON families with m.11778G→A and m.14484T→C, and subdivisions of this haplogroup have increased penetrance. Analysis of the complete mtDNA sequences of LHON probands with M7b1′2 and G status did not identify the two specific combinations of cytochrome b amino acid changes that are responsible for the background effect of haplogroups J1c and J2b in the penetrance of LHON in western European patients,21 suggesting different mtDNA mutation spectra and mechanisms in the penetrance of LHON in the East and the West. The increased risk of LHON penetrance of haplogroup M7b1′2 may be due to the coexistence of m.11778G→A and m.12811T→C, whereas the effect of haplogroup G as a risk factor in the disease expression may be related to the nonsynonymous mutation m.4833A→G in the MT-ND2 gene. The haplogroup-specific combination of two amino acid changes A20T and T53I in the MT-ATP6 protein may be the cause for a beneficial background effect of M8a. The identification of haplogroup background in LHON expression in Chinese families will undoubtedly help to understand the pathogenesis of LHON and guide future genetic counseling.
Acknowledgments
The authors thank the patients for donating DNA samples, Jin-Xin Zhang (The Department of Medical Statistics and Epidemiology, School of Public Health, Sun Yat-Sen University) for statistical assistance, and the reviewers for critical comments on the early version of the manuscript. This study was supported by grants from the Chinese Academy of Sciences (Y.-G.Y. and Y.-P.Z.) and the National Science Fund for Distinguished Young Scholars (30725044 to Q.Z.). Y.-G.Y. was supported by the “Century Program” (or Hundreds-Talent Program) of the Chinese Academy of Sciences. Y.J. is a Ph.D. student of Q.Z.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
GenBank, http://www.ncbi.nlm.nih.gov/Genbank/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
Accession Numbers
The mtDNA sequences reported herein have been submitted to GenBank under accession numbers FJ198229–FJ198385 and FJ198214–FJ198228.
References
- 1.Man P.Y.W., Turnbull D.M., Chinnery P.F. Leber hereditary optic neuropathy. J. Med. Genet. 2002;39:162–169. doi: 10.1136/jmg.39.3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carelli V., Ross-Cisneros F.N., Sadun A.A. Mitochondrial dysfunction as a cause of optic neuropathies. Prog. Retin. Eye Res. 2004;23:53–89. doi: 10.1016/j.preteyeres.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Carelli V., Rugolo M., Sgarbi G., Ghelli A., Zanna C., Baracca A., Lenaz G., Napoli E., Martinuzzi A., Solaini G. Bioenergetics shapes cellular death pathways in Leber's hereditary optic neuropathy: a model of mitochondrial neurodegeneration. Biochim. Biophys. Acta. 2004;1658:172–179. doi: 10.1016/j.bbabio.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Yen M.-Y., Wang A.-G., Wei Y.-H. Leber's hereditary optic neuropathy: a multifactorial disease. Prog. Retin. Eye Res. 2006;25:381–396. doi: 10.1016/j.preteyeres.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Puomila A., Hamalainen P., Kivioja S., Savontaus M.L., Koivumaki S., Huoponen K., Nikoskelainen E. Epidemiology and penetrance of Leber hereditary optic neuropathy in Finland. Eur. J. Hum. Genet. 2007;15:1079–1089. doi: 10.1038/sj.ejhg.5201828. [DOI] [PubMed] [Google Scholar]
- 6.Man P.Y.W., Griffiths P.G., Brown D.T., Howell N., Turnbull D.M., Chinnery P.F. The epidemiology of Leber hereditary optic neuropathy in the North East of England. Am. J. Hum. Genet. 2003;72:333–339. doi: 10.1086/346066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hudson G., Carelli V., Spruijt L., Gerards M., Mowbray C., Achilli A., Pyle A., Elson J., Howell N., La Morgia C. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am. J. Hum. Genet. 2007;81:228–233. doi: 10.1086/519394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia X., Li S., Xiao X., Guo X., Zhang Q. Molecular epidemiology of mtDNA mutations in 903 Chinese families suspected with Leber hereditary optic neuropathy. J. Hum. Genet. 2006;51:851–856. doi: 10.1007/s10038-006-0032-2. [DOI] [PubMed] [Google Scholar]
- 9.Smith K.H., Johns D.R., Heher K.L., Miller N.R. Heteroplasmy in Leber's hereditary optic neuropathy. Arch. Ophthalmol. 1993;111:1486–1490. doi: 10.1001/archopht.1993.01090110052022. [DOI] [PubMed] [Google Scholar]
- 10.Hudson G., Keers S., Man P.Y.W., Griffiths P., Huoponen K., Savontaus M.L., Nikoskelainen E., Zeviani M., Carrara F., Horvath R. Identification of an X-chromosomal locus and haplotype modulating the phenotype of a mitochondrial DNA disorder. Am. J. Hum. Genet. 2005;77:1086–1091. doi: 10.1086/498176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Man P.Y., Brown D.T., Wehnert M.S., Zeviani M., Carrara F., Turnbull D.M., Chinnery P.F. NDUFA-1 is not a nuclear modifier gene in Leber hereditary optic neuropathy. Neurology. 2002;58:1861–1862. doi: 10.1212/wnl.58.12.1861. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa K., Funayama T., Ohde H., Inagaki Y., Mashima Y. Genetic variants of TP53 and EPHX1 in Leber's hereditary optic neuropathy and their relationship to age at onset. Jpn. J. Ophthalmol. 2005;49:121–126. doi: 10.1007/s10384-004-0166-8. [DOI] [PubMed] [Google Scholar]
- 13.Danielson S.R., Carelli V., Tan G., Martinuzzi A., Schapira A.H., Savontaus M.L., Cortopassi G.A. Isolation of transcriptomal changes attributable to LHON mutations and the cybridization process. Brain. 2005;128:1026–1037. doi: 10.1093/brain/awh447. [DOI] [PubMed] [Google Scholar]
- 14.Torroni A., Petrozzi M., D'Urbano L., Sellitto D., Zeviani M., Carrara F., Carducci C., Leuzzi V., Carelli V., Barboni P. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am. J. Hum. Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- 15.Brown M.D., Sun F., Wallace D.C. Clustering of Caucasian Leber hereditary optic neuropathy patients containing the 11778 or 14484 mutations on an mtDNA lineage. Am. J. Hum. Genet. 1997;60:381–387. [PMC free article] [PubMed] [Google Scholar]
- 16.Lamminen T., Huoponen K., Sistonen P., Juvonen V., Lahermo P., Aula P., Nikoskelainen E., Savontaus M.L. mtDNA haplotype analysis in Finnish families with leber hereditary optic neuroretinopathy. Eur. J. Hum. Genet. 1997;5:271–279. [PubMed] [Google Scholar]
- 17.Hofmann S., Jaksch M., Bezold R., Mertens S., Aholt S., Paprotta A., Gerbitz K.D. Population genetics and disease susceptibility: characterization of central European haplogroups by mtDNA gene mutations, correlation with D loop variants and association with disease. Hum. Mol. Genet. 1997;6:1835–1846. doi: 10.1093/hmg/6.11.1835. [DOI] [PubMed] [Google Scholar]
- 18.Tsao K., Aitken P.A., Johns D.R. Smoking as an aetiological factor in a pedigree with Leber's hereditary optic neuropathy. Br. J. Ophthalmol. 1999;83:577–581. doi: 10.1136/bjo.83.5.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadun A.A., Carelli V., Salomao S.R., Berezovsky A., Quiros P.A., Sadun F., DeNegri A.M., Andrade R., Moraes M., Passos A. Extensive investigation of a large Brazilian pedigree of 11778/haplogroup J Leber hereditary optic neuropathy. Am. J. Ophthalmol. 2003;136:231–238. doi: 10.1016/s0002-9394(03)00099-0. [DOI] [PubMed] [Google Scholar]
- 20.Kerrison J.B., Miller N.R., Hsu F., Beaty T.H., Maumenee I.H., Smith K.H., Savino P.J., Stone E.M., Newman N.J. A case-control study of tobacco and alcohol consumption in Leber hereditary optic neuropathy. Am. J. Ophthalmol. 2000;130:803–812. doi: 10.1016/s0002-9394(00)00603-6. [DOI] [PubMed] [Google Scholar]
- 21.Carelli V., Achilli A., Valentino M.L., Rengo C., Semino O., Pala M., Olivieri A., Mattiazzi M., Pallotti F., Carrara F. Haplogroup effects and recombination of mitochondrial DNA: novel clues from the analysis of Leber hereditary optic neuropathy pedigrees. Am. J. Hum. Genet. 2006;78:564–574. doi: 10.1086/501236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards M., Macaulay V., Hickey E., Vega E., Sykes B., Guida V., Rengo C., Sellitto D., Cruciani F., Kivisild T. Tracing European founder lineages in the Near Eastern mtDNA pool. Am. J. Hum. Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Y.-G., Kong Q.-P., Bandelt H.-J., Kivisild T., Zhang Y.-P. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am. J. Hum. Genet. 2002;70:635–651. doi: 10.1086/338999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji Y., Jia X., Zhang Q., Yao Y.-G. mtDNA haplogroup distribution in Chinese patients with Leber's hereditary optic neuropathy and G11778A mutation. Biochem. Biophys. Res. Commun. 2007;364:238–242. doi: 10.1016/j.bbrc.2007.09.111. [DOI] [PubMed] [Google Scholar]
- 25.Wang H.-W., Jia X., Ji Y., Kong Q.-P., Zhang Q., Yao Y.-G., Zhang Y.-P. Strikingly different penetrance of LHON in two Chinese families with primary mutation G11778A is independent of mtDNA haplogroup background and secondary mutation G13708A. Mutat. Res. 2008;643:48–53. doi: 10.1016/j.mrfmmm.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Zhang A.-M., Jia X., Yao Y.-G., Zhang Q. Co-occurrence of A1555G and G11778A in a Chinese family with high penetrance of Leber's hereditary optic neuropathy. Biochem. Biophys. Res. Commun. 2008;376:221–224. doi: 10.1016/j.bbrc.2008.08.128. [DOI] [PubMed] [Google Scholar]
- 27.Yao Y.-G., Kong Q.-P., Wang C.-Y., Zhu C.-L., Zhang Y.-P. Different matrilineal contributions to genetic structure of ethnic groups in the Silk Road region in China. Mol. Biol. Evol. 2004;21:2265–2280. doi: 10.1093/molbev/msh238. [DOI] [PubMed] [Google Scholar]
- 28.Li R., Qu J., Zhou X., Tong Y., Hu Y., Qian Y., Lu F., Mo J.-Q., West C.-E., Guan M.-X. The mitochondrial tRNA(Thr) A15951G mutation may influence the phenotypic expression of the LHON-associated ND4 G11778A mutation in a Chinese family. Gene. 2006;376:79–86. doi: 10.1016/j.gene.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Qian Y., Zhou X., Hu Y., Tong Y., Li R., Lu F., Yang H., Mo J.-Q., Qu J., Guan M.-X. Clinical evaluation and mitochondrial DNA sequence analysis in three Chinese families with Leber's hereditary optic neuropathy. Biochem. Biophys. Res. Commun. 2005;332:614–621. doi: 10.1016/j.bbrc.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Qu J., Li R., Tong Y., Hu Y., Zhou X., Qian Y., Lu F., Guan M.-X. Only male matrilineal relatives with Leber's hereditary optic neuropathy in a large Chinese family carrying the mitochondrial DNA G11778A mutation. Biochem. Biophys. Res. Commun. 2005;328:1139–1145. doi: 10.1016/j.bbrc.2005.01.062. [DOI] [PubMed] [Google Scholar]
- 31.Qu J., Li R., Zhou X., Tong Y., Lu F., Qian Y., Hu Y., Mo J.Q., West C.E., Guan M.-X. The novel A4435G mutation in the mitochondrial tRNAMet may modulate the phenotypic expression of the LHON-associated ND4 G11778A mutation. Invest. Ophthalmol. Vis. Sci. 2006;47:475–483. doi: 10.1167/iovs.05-0665. [DOI] [PubMed] [Google Scholar]
- 32.Qu J., Li R., Zhou X., Tong Y., Yang L., Chen J., Zhao F., Lu C., Qian Y., Lu F. Cosegregation of the ND4 G11696A mutation with the LHON-associated ND4 G11778A mutation in a four generation Chinese family. Mitochondrion. 2007;7:140–146. doi: 10.1016/j.mito.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao Y.-G., Salas A., Bravi C.M., Bandelt H.-J. A reappraisal of complete mtDNA variation in East Asian families with hearing impairment. Hum. Genet. 2006;119:505–515. doi: 10.1007/s00439-006-0154-9. [DOI] [PubMed] [Google Scholar]
- 34.Bandelt H.-J., Achilli A., Kong Q.-P., Salas A., Lutz-Bonengel S., Sun C., Zhang Y.-P., Torroni A., Yao Y.-G. Low “penetrance” of phylogenetic knowledge in mitochondrial disease studies. Biochem. Biophys. Res. Commun. 2005;333:122–130. doi: 10.1016/j.bbrc.2005.04.055. [DOI] [PubMed] [Google Scholar]
- 35.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 36.Kong Q.-P., Bandelt H.-J., Sun C., Yao Y.-G., Salas A., Achilli A., Wang C.-Y., Zhong L., Zhu C.-L., Wu S.-F. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum. Mol. Genet. 2006;15:2076–2086. doi: 10.1093/hmg/ddl130. [DOI] [PubMed] [Google Scholar]
- 37.Kivisild T., Tolk H.-V., Parik J., Wang Y., Papiha S.S., Bandelt H.-J., Villems R. The emerging limbs and twigs of the East Asian mtDNA tree. Mol. Biol. Evol. 2002;19:1737–1751. doi: 10.1093/oxfordjournals.molbev.a003996. [DOI] [PubMed] [Google Scholar]
- 38.Derenko M., Malyarchuk B., Grzybowski T., Denisova G., Dambueva I., Perkova M., Dorzhu C., Luzina F., Lee H.K., Vanecek T. Phylogeographic analysis of mitochondrial DNA in northern Asian populations. Am. J. Hum. Genet. 2007;81:1025–1041. doi: 10.1086/522933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao Y.-G., Kong Q.-P., Man X.-Y., Bandelt H.-J., Zhang Y.-P. Reconstructing the evolutionary history of China: a caveat about inferences drawn from ancient DNA. Mol. Biol. Evol. 2003;20:214–219. doi: 10.1093/molbev/msg026. [DOI] [PubMed] [Google Scholar]
- 40.Fujitake J., Mizuta H., Fujii H., Ishikawa Y., Sasamoto K., Goto Y., Nonaka I., Tatsuoka Y. Leber's hereditary optic neuropathy with intracranial arteriovenous malformation: a case report. Acta Neurol. Belg. 2002;102:82–86. [PubMed] [Google Scholar]
- 41.Bandelt H.-J., Salas A., Taylor R.W., Yao Y.-G. The exaggerated status of “novel” and “pathogenic” mtDNA sequence variants due to inadequate database searches. Hum. Mutat. 2008 doi: 10.1002/humu.20846. in press. [DOI] [PubMed] [Google Scholar]
- 42.Huoponen K., Lamminen T., Juvonen V., Aula P., Nikoskelainen E., Savontaus M.L. The spectrum of mitochondrial DNA mutations in families with Leber hereditary optic neuroretinopathy. Hum. Genet. 1993;92:379–384. doi: 10.1007/BF01247339. [DOI] [PubMed] [Google Scholar]
- 43.Howell N., Oostra R.-J., Bolhuis P.A., Spruijt L., Clarke L.A., Mackey D.A., Preston G., Herrnstadt C. Sequence analysis of the mitochondrial genomes from Dutch pedigrees with Leber hereditary optic neuropathy. Am. J. Hum. Genet. 2003;72:1460–1469. doi: 10.1086/375537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gasparre G., Porcelli A.M., Bonora E., Pennisi L.F., Toller M., Iommarini L., Ghelli A., Moretti M., Betts C.M., Martinelli G.N. Disruptive mitochondrial DNA mutations in complex I subunits are markers of oncocytic phenotype in thyroid tumors. Proc. Natl. Acad. Sci. USA. 2007;104:9001–9006. doi: 10.1073/pnas.0703056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Annunen-Rasila J., Finnilä S., Mykkänen K., Moilanen J.S., Veijola J., Pöyhönen M., Viitanen M., Kalimo H., Majamaa K. Mitochondrial DNA sequence variation and mutation rate in patients with CADASIL. Neurogenetics. 2006;7:185–194. doi: 10.1007/s10048-006-0049-x. [DOI] [PubMed] [Google Scholar]
- 46.Achilli A., Perego U.A., Bravi C.M., Coble M.D., Kong Q.-P., Woodward S.R., Salas A., Torroni A., Bandelt H.-J. The phylogeny of the four pan-American mtDNA haplogroups: implications for evolutionary and disease studies. PLoS ONE. 2008;3:e1764. doi: 10.1371/journal.pone.0001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown M.D., Yang C.C., Trounce I., Torroni A., Lott M.T., Wallace D.C. A mitochondrial DNA variant, identified in Leber hereditary optic neuropathy patients, which extends the amino acid sequence of cytochrome c oxidase subunit I. Am. J. Hum. Genet. 1992;51:378–385. [PMC free article] [PubMed] [Google Scholar]
- 48.Mather M.W., Rottenberg H. Intrinsic uncoupling of cytochrome c oxidase may cause the maternally inherited mitochondrial diseases MELAS and LHON. FEBS Lett. 1998;433:93–97. doi: 10.1016/s0014-5793(98)00891-6. [DOI] [PubMed] [Google Scholar]
- 49.Brown M.D., Torroni A., Huoponen K., Chen Y.S., Lott M.T., Wallace D.C. Pathological significance of the mtDNA COX III mutation at nucleotide pair 9438 in Leber hereditary optic neuropathy. Am. J. Hum. Genet. 1994;55:410–412. [PMC free article] [PubMed] [Google Scholar]
- 50.Elliott H.R., Samuels D.C., Eden J.A., Relton C.L., Chinnery P.F. Pathogenic mitochondrial DNA mutations are common in the general population. Am. J. Hum. Genet. 2008;83:254–260. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kazuno A.A., Munakata K., Nagai T., Shimozono S., Tanaka M., Yoneda M., Kato N., Miyawaki A., Kato T. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2006;2:e128. doi: 10.1371/journal.pgen.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuku N., Park K.S., Yamada Y., Nishigaki Y., Cho Y.M., Matsuo H., Segawa T., Watanabe S., Kato K., Yokoi K. Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am. J. Hum. Genet. 2007;80:407–415. doi: 10.1086/512202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka M., Fuku N., Nishigaki Y., Matsuo H., Segawa T., Watanabe S., Kato K., Yokoi K., Ito M., Nozawa Y. Women with mitochondrial haplogroup N9a are protected against metabolic syndrome. Diabetes. 2007;56:518–521. doi: 10.2337/db06-1105. [DOI] [PubMed] [Google Scholar]
- 54.Lamminen T., Majander A., Juvonen V., Wikström M., Aula P., Nikoskelainen E., Savontous M.L. A mitochondrial mutation at nt 9101 in the ATP synthase 6 gene associated with deficient oxidative phosphorylation in a family with Leber hereditary optic neuroretinopathy. Am. J. Hum. Genet. 1995;56:1238–1240. [PMC free article] [PubMed] [Google Scholar]
- 55.Majander A., Lamminen T., Juvonen V., Aula P., Nikoskelainen E., Savontaus M.L., Wikström M. Mutations in subunit 6 of the F1F0-ATP synthase cause two entirely different diseases. FEBS Lett. 1997;412:351–354. doi: 10.1016/s0014-5793(97)00757-6. [DOI] [PubMed] [Google Scholar]
- 56.den Dunnen J.T., Antonarakis S.E. Nomenclature for the description of human sequence variations. Hum. Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


