Abstract
The enamel matrix proteins (amelogenin, enamelin and ameloblastin) are degraded by matrix metalloproteinase-20 and kallikrein-4 during enamel development and mature enamel is virtually protein free. The precise mechanism of removal and degradation of the enamel protein cleavage products from the matrix, however, remains poorly understood. It has been proposed that receptor-mediated endocytosis allows for the cleaved proteins to be removed from the matrix during enamel formation and then transported to the lysosome for further degradation. This study aims to identify lysosomal proteases that are present in maturation-stage enamel organ. RNA from first molars of 11-day-old mice was collected and expression was initially assessed by RT-PCR and then quantified by qPCR. The pattern of expression of selected proteases was assessed by immunohistochemical staining of demineralized mouse incisors. With the exception of cathepsin G, all lysosomal proteases assessed were expressed in maturation-stage enamel organ. Identified proteases included cathepsins B, D, F, H, K, L, O, S and Z. Tripeptidyl peptidases I and II as well as dipeptidyl peptidases I, II, III and IV were also found to be expressed. Immunohistochemical staining confirmed that the maturation-stage ameloblasts express cathepsins L and S and tripeptidyl peptidase II. Our results suggest that the ameloblasts are enriched by a large number of lysosomal proteases at maturation that are likely involved in the degradation of the organic matrix.
Key Words: Enamel, Proteases, Ameloblasts, Lysosome, Cathepsin
Introduction
Dental enamel is unique because it starts as a soft, protein-rich substance and ends as a hard, almost protein-free mineral. Matrix metalloproteinase-20 and kallikrein-4 are known to degrade enamel matrix proteins during development, however, little is known about how these cleavage products are removed from mature enamel. Maturation-stage ameloblasts have been shown to have resorptive capabilities [Nanci et al., 1996] and it has been proposed that once inside the cell, the cleaved proteins are then transported to the lysosome for further degradation.
Several studies have found that there is an increase in lysosome number in ameloblasts as enamel matures [Nanci et al., 1987; Salama, 1990]. Lysosomes are enriched in a variety of enzymes capable of degrading proteins, lipids and carbohydrates from the extracellular environment. These include a family of cysteine proteases known as cathepsins (Cat), which are often ubiquitously expressed. Within the lysosome, proteins can also be cleaved by a variety of aminopeptil, dipeptil (DPP) and tripeptidyl (TPP) peptidases. Once proteins are fully degraded into single amino acids, these free amino acids can be recycled for protein synthesis.
Cat B, D and L as well as DPP II have previously been identified in ameloblasts [Andujar et al., 1989; Al Kawas et al., 1996; Smid et al., 2001; Nishikawa, 2005]. This study sought to identify previously unidentified lysosomal proteases present during enamel maturation.
Materials and Methods
Reverse Transcriptase Polymerase Chain Reaction
TotalRNA was extracted from 11-day-old (maturation stage) mouse enamel organ with TRIzol reagent, and cDNA was transcribed using the SuperScript III First-Strand Synthesis system (Invitrogen). Primer sets were designed by analysis of annealing sites by DNAStar software. In order to ensure a large sample size as well as a clearly defined developmental stage, molars were selected for extraction of enamel organ RNA.
Real-Time PCR Analysis of Gene Expression in Enamel Organ
Mouse molars were harvested at 11 days postnatally and the dental papilla was carefully removed. The enamel organ mRNA was subjected to qPCR analysis by iQ SYBR green (Bio-Rad). Gene-specific primers were designed using DNAStar. Data are representative of 6 individual mice with duplicate measurements.
Immunohistochemistry of Mouse Incisors
Immunohistochemical analysis of demineralized, paraffin-embedded and sectioned mouse incisors was performed. Sections were incubated in blocking agent followed by overnight incubation in specific antisera. Staining was visualized by incubation in peroxidase-conjugated antibody [Vectastain Elite ABC Kit (Goat IgG)] and Sigma Fast 3,3′-diaminobenzidine substrate. Sections were counterstained with 0.1% Fast Green and examined by light microscopy. Antibodies used were Cat S (ab-18822, 20 μg/ml; Abcam), Cat L (sc-6500, 1:500; Santa Cruz Biotechnology) or TPP II (sc-15148, 1:1,000; Santa Cruz Biotechnology).
Results
Lysosomal protease expression was detected by RT-PCR of maturation-stage (day 11) enamel organ (fig. 1) and quantified by real-time PCR (table 1). As indicated by the cycle threshold numbers, Cat B, D, K and L are the most abundant lysosomal enzymes at maturation (17–18 cycles). TPP I, DPP I and III as well as Cat F, O and Z are midrange (19–21 cycles), while Cat H and S, TPP II as well as DPP II and IV are in lower abundance (22–25 cycles). Cat G was not detectable.
Fig. 1.
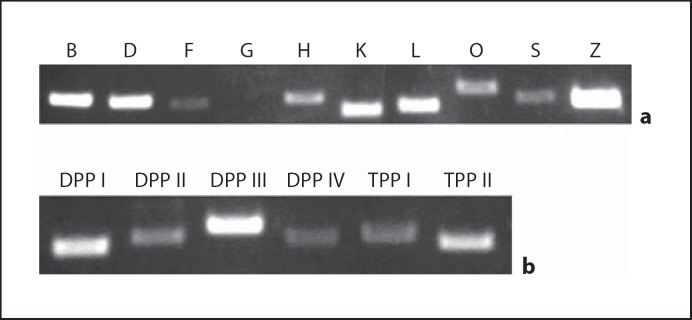
Lysosomal protease expression in mature mouse enamel by RT-PCR. Total RNA from 11-day-old mouse first molars was isolated, reverse transcribed and amplified following established protocols. a Cat B, D, F, G, H, K, L, O, S and Z. b DPP I, II, III and IV as well as TPP I and II. All cathepsins examined were found to be expressed in mature enamel with the exception of Cat G.
Table 1.
qPCR of lysosomal proteases at maturation stage
| Cycles | |
|---|---|
| Cat B | 17.36 ± 0.94 |
| Cat D | 17.49 ± 0.22 |
| Cat F | 20.98 ± 1.24 |
| Cat G | n/d |
| Cat H | 23.20 ± 0.39 |
| Cat K | 17.57 ± 0.76 |
| Cat L | 18.70 ± 0.39 |
| Cat O | 20.49 ± 0.30 |
| Cat S | 23.04 ± 0.45 |
| Cat Z | 20.97 ± 0.33 |
| TPP I | 19.41 ± 0.30 |
| TPP II | 23.24 ± 0.66 |
| DPP I (Cat C) | 20.36 ± 0.34 |
| DPP II | 22.95 ± 0.28 |
| DPP III | 19.39 ± 0.32 |
| DPP IV | 25.73 ± 0.40 |
Mouse molars were harvested from 11-day-old mice and subjected to qPCR analysis using gene-specific primers. The number of cycles to reach a predetermined cycle threshold is given. Data are representative of 6 individual mice with measurements in duplicate. n/d = Not detected.
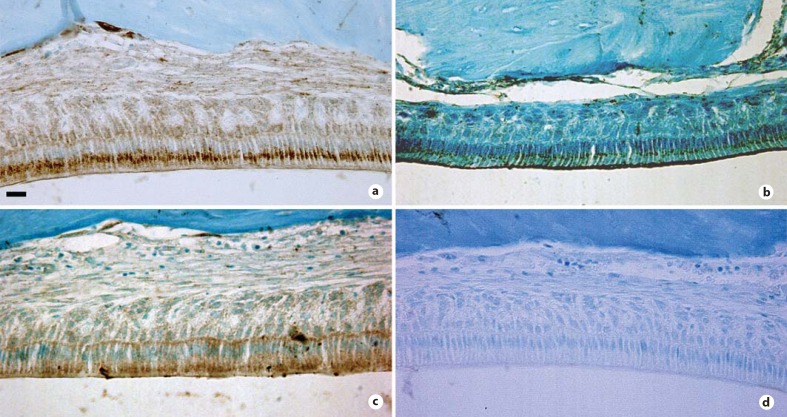
Immunohistochemical analysis of mouse incisors was performed using selected protease antibodies to identify the cells responsible for the protease expression. Figure 2 shows that maturation-stage ameloblasts expressed the proteases.
Fig. 2.
Immunohistochemical analysis of lysosomal protease expression in maturation stage ameloblasts. Immunohistochemistry of demineralized, paraffin sections of mouse incisors. Sections were counterstained with 0.1% Fast Green. The maturation stage of enamel development is depicted. Mature ameloblasts are shown to express Cat L (a), Cat S (b), TPP II (c) and no primary antibody control (d). Scale bar = 50 μm.
Discussion
With the exception of Cat G, all lysosomal enzymes examined were present at maturation. It is not surprising that Cat G was not identified in enamel as its expression appears to be restricted to neutrophils, neutrophil precursors in bone marrow and tissues that contain them (such as spleen) [Salveson, 2004].
The highest levels of expression, as assessed by qPCR, indicate that Cat B, D, K and L are most abundant at maturation stage. Cat B, D and L have previously been detected in ameloblasts [Andujar et al., 1989; Al Kawas et al., 1996; Smid et al., 2001; Nishikawa, 2005], however, identification of Cat K in enamel organ is novel. Cat K is a cysteine protease predominantly expressed in osteoclasts with high expression also seen in ovary, small intestine and colon, and at lower levels in heart, skeletal muscle, placenta, lung, prostate, testes, spleen, thymus, kidney, pancreas and liver [Brömme and Okamoto, 1995]. Discovery of Cat K expression in enamel organ is not surprising, given its wide tissue distribution and the fact that the protease has the capacity to cleave near proline residues [Xia et al., 1999].
The most abundant protein in enamel, amelogenin, is proline rich like collagen. The ability of the lysosome to degrade high concentrations of amelogenin would likely require a large amount of proteases capable of digesting proline peptide bonds. DPP II is a protease with a preference for digestion of proline peptide bonds [McDonald and Okhkubo, 2004], whereas Cat Z (also known as Cat X) is one of the few enzymes capable of cleaving C-terminal proline bonds [Klemenčič et al., 2000]. DPP IV is an exopeptidase specific for cleavage of N-terminal dipeptides where the C-terminal amino acid of the removed dipeptide is a proline. Interestingly, this protease, which is generally enriched in tissues with polarized epithelial cells (such as kidney, intestine and liver) [Misumi and Ikehara, 2004], was found to be the lowest in abundance in mature enamel organ.
DPP I and III, TPP I and II as well as Cat F, H and O were also present at maturation at a significant level. DPP I is an aminopeptidase that sequentially removes 2 N-terminal amino acid residues from folded proteins. DPP I is interesting because in addition to its lysosomal activity, it is also involved in the activation of the proenzymes of chymotrypsin-like serine proteases [Turk et al., 2004]. Loss-of-function mutations in the DPP I gene result in early-onset periodontitis and palmoplantar keratosis, characteristic of Haim-Munk and Papillon-Lefevre syndromes [Hart et al., 1999; Toomes et al., 1999]. This may be the result of incomplete processing of an unidentified protease [Nuckolls and Slavkin, 1999]. It is possible that DPP I is secreted from ameloblasts and could act as the yet unidentified activator of the serine protease kallikrein-4.
In conclusion, our results have revealed that ameloblasts are enriched in a variety of lysosomal enzymes at maturation when protein is actively being removed from enamel.
Acknowledgement
This work was supported by National Institute of Dental and Craniofacial Research grant DE016276 (J.D.B.).
Abbreviations used in this paper
- DPP
dipeptidyl peptidase
- TPP
tripeptidyl peptidase
References
- Al Kawas S., Amizuka N., Bergeron J.J., Warshawsky H. Immunolocalization of the cation-independent mannose 6-phosphate receptor and cathepsin B in the enamel organ and alveolar bone of the rat incisor. Calcif Tissue Int. 1996;59:59–192. doi: 10.1007/s002239900108. [DOI] [PubMed] [Google Scholar]
- Andujar M.B., Hartmann D.J., Caillot G., Ville G., Magloire H. Immunolocalization of cathepsin D in dental tissues. Matrix. 1989;9:9–397. doi: 10.1016/s0934-8832(89)80045-9. [DOI] [PubMed] [Google Scholar]
- Brömme D., Okamoto K. Human cathepsin O2, a novel cysteine protease highly expressed in osteoclastoma and ovary molecular cloning, sequencing and tissue distribution. Biol Chem Hoppe Seyler. 1995;376:376–379. doi: 10.1515/bchm3.1995.376.6.379. [DOI] [PubMed] [Google Scholar]
- Brömme D. In: Handbook of Proteolytic Enzymes. ed 2. Cathepsin F., Barrett A.J., Rawlings N.D., Woessner J.F., editors. Vol. 2. London: Elsevier Academic Press; 2004. pp. 1087–1088. [Google Scholar]
- Hart T.C., Hart P.S., Bowden D.W., Michalec M.D., Callison S.A., Walker S.J., Zhang Y., Firatli E. Mutations of the cathepsin C gene are responsible for Papillon-Lefevre syndrome. J Med Genet. 1999;36:36–881. [PMC free article] [PubMed] [Google Scholar]
- Kirschke H. Cathepsin H. In: Barrett A.J., Rawlings N.D., Woessner J.F., editors. Handbook of Proteolytic Enzymes. ed 2. Vol. 2. London: Elsevier Academic Press; 2004. pp. 1089–1092. [Google Scholar]
- Klemenčič I., Carmona A.K., Cezari M.H.S., Juliano M.A., Juliano L., Gunčar G., Turk D., Križaj I., Turk V., Turk B. Biochemical characterization of human cathepsin X revealed that the enzyme is an exopeptidase, acting as carboyxmonopeptidase or carboxydipeptidase. Eur J Biochem. 2000;267:267–5404. doi: 10.1046/j.1432-1327.2000.01592.x. [DOI] [PubMed] [Google Scholar]
- McDonald J.K., Okhkubo I. Dipeptidyl-peptidase II. In: Barrett A.J., Rawlings N.D., Woessner J.F., editors. Handbook of Proteolytic Enzymes. ed 2. Vol. 2. London: Elsevier Academic Press; 2004. pp. 1938–1943. [Google Scholar]
- Misumi Y., Ikehara Y. Dipeptidyl-peptidase IV. In: Barrett A.J., Rawlings N.D., Woessner J.F., editors. Handbook of Proteolytic Enzymes. ed 2. Vol. 2. London: Elsevier Academic Press; 2004. pp. 1905–1909. [Google Scholar]
- Nanci A., Slavkin H., Smith C.E. Immunocytochemical and radiographic evidence for secretion and intracellular degradation of enamel proteins by ameloblasts during maturation stage of amelogenesis in rat incisors. Anat Rec. 1987;217:217–107. doi: 10.1002/ar.1092170202. [DOI] [PubMed] [Google Scholar]
- Nanci A., Fortin M., Ghitescu L. Endocytic functions of ameloblasts and odontoblasts immunocytochemical and tracer studies on the uptake of plasma proteins. Anat Rec. 1996;245:245–219. doi: 10.1002/(SICI)1097-0185(199606)245:2<219::AID-AR9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Nishikawa S. Presence of anti-cystatin C-positive dendritic cells macrophages and localization of cysteine proteases apical bud of the enamel organ in the rat incisor. J Histochem Cytochem. 2005;53:53–643. doi: 10.1369/jhc.4A6533.2005. [DOI] [PubMed] [Google Scholar]
- Nuckolls G.H., Slavkin H.C. Paths of glorious proteases. Nat Genet. 1999;23:23–378. doi: 10.1038/70472. [DOI] [PubMed] [Google Scholar]
- Salama A.H., Bailey R.L., Eisenmann D.R., Zaki A.E. Quantitative cytochemistry of lysosomal structures in rat incisor maturation enamel organ. Arch Oral Biol. 1990;35:35–535. doi: 10.1016/0003-9969(90)90084-n. [DOI] [PubMed] [Google Scholar]
- Salveson G.S. Cathepsin G. In: Barrett A.J., Rawlings N.D., Woessner J.F., editors. Handbook of Proteolytic Enzymes. ed 2. Vol. 2. London: Elsevier Academic Press; 2004. pp. 1524–1526. [Google Scholar]
- Smid J.R., Young W.G., Monsour P.A. Dipeptidyl-peptidase II and cathepsin B activities in amelogenesis of the rat incisor. Eur J Oral Sci. 2001;109:109–260. doi: 10.1034/j.1600-0722.2001.00025.x. [DOI] [PubMed] [Google Scholar]
- Toomes C., James J., Wood A.J., Wu C.L., McCormick D., Lench N., Hewitt C., Moynihan L., Roberts E., Woods C.G., Markham A., Wong M., Widmer R., Ghaffer K.A., Pemberton M., Hussein I.R., Temtamy S.A., Davies R., Read A.P., Sloan P., Dixon M.J., Thakker N.S. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat Genet. 1999;23:23–421. doi: 10.1038/70525. [DOI] [PubMed] [Google Scholar]
- Turk B., Turk D., Dolenc I., Turk V. Dipeptidyl-peptidase I. In: Barrett A.J., Rawlings N.D., Woessner J.F., editors. Handbook of Proteolytic Enzymes. ed 2. Vol. 2. London: Elsevier Academic Press; 2004. pp. 1192–1196. [Google Scholar]
- Velasco G., López-Otín C. Cathepsin O. In: Barrett A.J., Rawlings N.D., Woessner J.F., editors. Handbook of Proteolytic Enzymes. ed 2. Vol. 2. London: Elsevier Academic Press; 2004. pp. 1102–1103. [Google Scholar]
- Xia L., Klib J., Wex H., Li Z., Lipyansky A., Breuil V., Stein L., Palmer J.T., Dempster D.W., Brömme D. Localization of rat cathepsin K in osteoclasts and resorption pits inhibition of bone resorption and cathepsin K-activity by peptidyl vinyl sulfones. Biol Chem. 1999;380:380–679. doi: 10.1515/BC.1999.084. [DOI] [PubMed] [Google Scholar]