Abstract
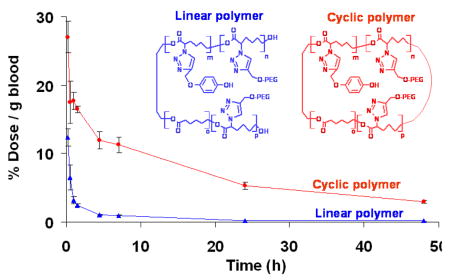
The ability of a polymer to reptate through a nanopore has an influence on its circulatory half-life and biodistribution since many physiological barriers contain nanopores. A cyclic polymer lacks chain ends, therefore, cyclic polymers with a MW above the renal threshold for elimination should circulate longer than their linear polymer counterparts when injected into animals. As predicted, radiolabeled cyclic polymers with MW above the renal threshold have a longer blood circulation time in mice compared to linear polymers of comparable MW.
Long circulation time of water-soluble polymers is essential for the successful delivery of drugs to solid tumors. The circulation time of such polymers depends upon molecular weight and polymer architecture.1–4 This is because physiological barriers in the kidneys, have a nanoporous structure which retards the permeation of soluble polymers but allows the passage and elimination from the body of low molecular weight substances.5,6 Linear polymers traverse a nanopore by the end-on motion of the polymer chain and since only one polymer segment needs to enter the pore for a linear polymer to traverse it, linear polymers cross nanopores more easily than star polymers.1,5 Cyclic polymers lack chain ends, so two chain segments would need to enter the pore for the cyclic polymer to transit. Therefore, we predicted that cyclic polymers would behave differently in vivo compared to linear polymers of the same molecular weight.
Linear polymer precursor and cyclic copolymer of α-cholo-ε-caprolactone (ClCL) and ε-caprolactone (ε-CL) were synthesized following published procedures.7–9 Briefly, the cyclic random copolymer of ε-CL and ClCL were polymerized using cyclic tin-based catalyst, 2,2-dibutyl-2-stanna-1,3-dioxepane. ClCL was introduced as a copolymer to allow further modification of the polymer chain. To create the cyclic polymers, α-(1-acryloxyethyl)-ε-caprolactone was added to the polymerization medium to enable cyclization via intramolecular photocrosslinking (S1).
A comparison of the linear polymer precursor and the cyclic polymer on GPC (Figure S2–4) shows a shift of the elution time toward a smaller molecular weight (MW) for the cyclic polymer indicating the formation of intramolecular cyclization. The number average MW of the linear polymer precursor and cyclic polymer measured by GPC were 11,900 (PDI = 1.46) and 9,300 (PDI = 1.38), respectively(S2). The larger MW value of the linear polymers is expected because they have larger hydrodynamic volumes than cyclic polymers of the same mass.4,10
An alkyne phenol moiety was quantitatively introduced on the azide containing polyester (P(N3CL-co-CL)) backbone using very mild conditions to avoid hydrolytic degradation. The polyester backbone was further modified by cycloaddition of an alkyne, ω-methoxyl-PEG to increase water solubility (S5,6). The grafting density of PEG chains on the linear and cyclic polyester is stoichiometrically controlled using “click chemistry”.7 The MW of the linear and cyclic polymers can be tuned from less than to greater than the elimination thresholds in the kidney by the addition of PEG chains of 1.1, 2 or 5 kD.11
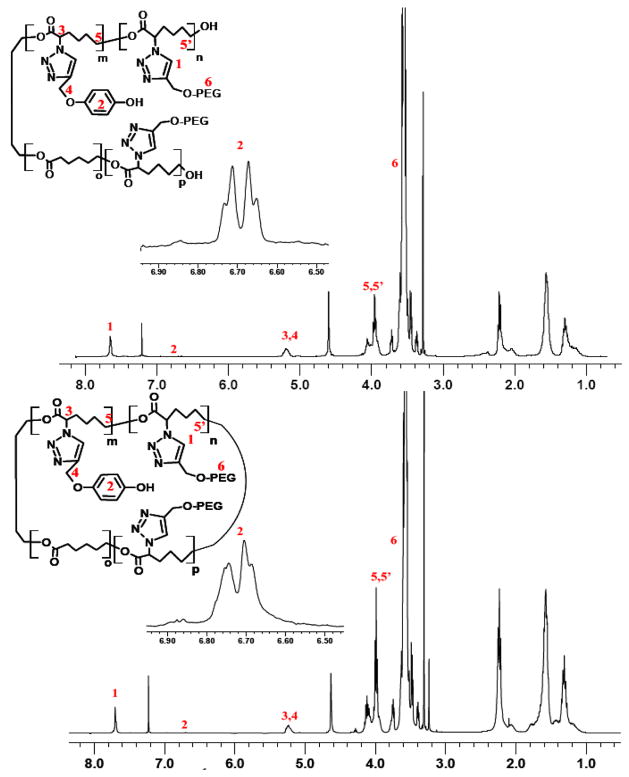
The grafting of PEG is confirmed by a characteristic chemical shift of -CH2O- resonance at 3.6 ppm and the triazole proton at 7.7 ppm by 1HNMR. By calculating the integral intensity of the chemical shift at 3.6 ppm (-CH2CH2O-) and 4 ppm (-CH2CH2CH2O-) of PEG and ε-caprolactone, respectively, about twenty-one 1.1 and 2 kD PEG chains and sixteen 5kD PEG chains were grafted onto poly(N3CL-co-CL) (72 units). The number average MW of grafted polymer was about 32, 50 and 90 kD for PEG 1.1, 2 and 5 kD conjugation, respectively (Table 1). We adjusted the amount of propargyl phenol in the reactions so that on average, a polyester chain contained one phenol moiety.
Table 1.
Polymer characterization and pharmacokinetic data
| Polymer | Mn (kD) | PDI | Half life (h) |
AUC0→∞ (%ID.h/g) | |
|---|---|---|---|---|---|
| t1/2, α | t1/2, β | ||||
| Cyclic 90 | 93 | 1.33 | 1.00±0.30 | 20.9±4.4 | 587.1±13.6 |
| Cyclic 50 | 49 | 1.14 | 0.09±0.08 | 13.6±2.7 | 343.3±8.8 |
| Cyclic 32 | 33 | 1.21 | 0.27±0.02 | 2.7±1.3 | 11.7±1.4 |
| Linear 90 | 90 | 1.25 | 0.51±0.04 | 14.7±1.5 | 218.6±1.0 |
| Linear 50 | 52 | 1.21 | 0.26±0.03 | 4.4±1.3 | 22.7±1.0 |
| Linear 32 | 32 | 1.21 | 0.17±0.02 | 3.0±0.5 | 22.8±0.3 |
We determined the dependence of the blood circulation time and biodistribution as a function of the polymer architecture and molecular weight using polymers radiolabeled on the phenol group with 125I via the chloramine T method (S13).5 The polymer backbone degraded over the course of 10 days in phosphate buffered saline at 37°C (S9,11). The degradation rate was faster in human plasma (S10,11) but degradation did not interfere with the determination of the pharmacokinetic elimination rates (S15).
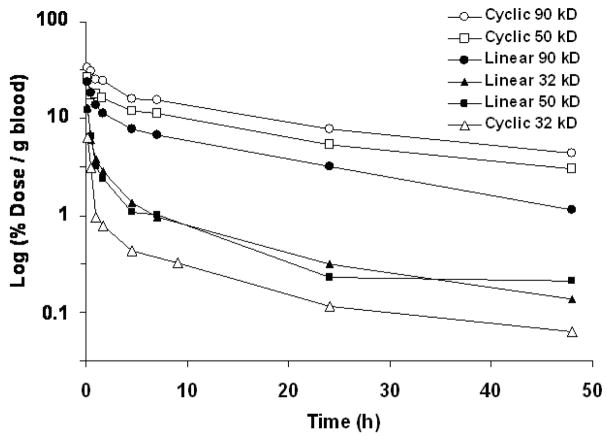
In Figure 2 we show a plot of polymer concentration in the blood with respect to time. Elimination half-lives (t1/2β) were calculated using the residuals method in a two-compartment model (Table 1). Cyclic polymers with molecular size greater than the renal filtration threshold (50 and 90 kD) have longer plasma circulation times than linear polymers of similar mass.11 This is because linear polymers can reptate through the nanopores in the glomeruli by an “end-on” motion more easily than cyclic polymers. Interestingly, the effect of polymer architecture was more profound in the 50 kD polymer (Rh determined from GPC is approximately 4.4 nm) than the 90kD polymer. A similar behavior has been reported for elongated polymers and more densely structured dendrimers.5,12 This behavior is manifested by a lower amount of cyclic polymer in the urine with the 50 and 90 kD but not for the 32 kD polymers. Radioactivity in the urine was both on polymer and lower molecular weight fragments (S14). The 32 kD polymers showed very rapid elimination. The 32 kD cyclic polymer was eliminated faster than the linear 32 kD polymer because the MW of both were below the nominal MW cutoff (30–40 kD) of the renal filtration of linear PEG11 but the cyclic polymer is a more compact molecule.
Figure 2.
Linear and cyclic polymer concentration in the blood with respect to time.
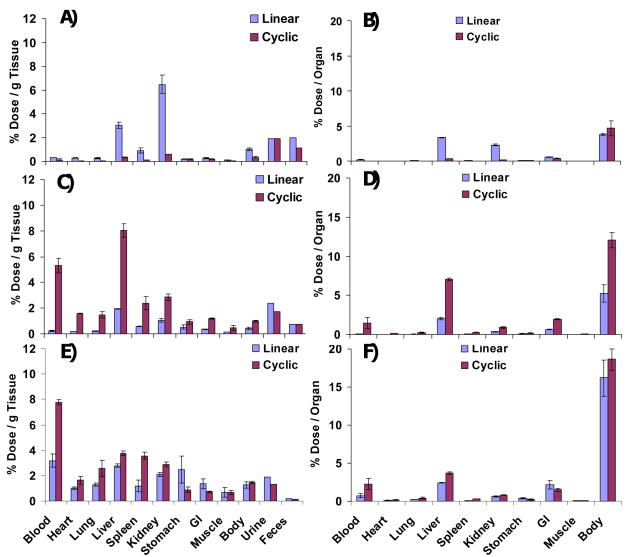
The accumulation of radioactivity in different organs at 24 h is displayed in Figure 3. First, as observed with other polymers, the greater the polymer molecular weight the greater the percent polymer retained in the body (S15,16). The 32 kD cyclic polymer had low accumulation in all organs due to its short circulation time and was secreted in urine 13 % more than the 32 kD linear polymer. The linear 32 kD although rapidly eliminated had a higher kidney accumulation than did any of the other polymers. The reason for this is not apparent. When the MW was increased to 50 kD, the cyclic polymer was secreted in urine 12% less than the linear polymer. As a result, the 50 kD cyclic polymer accumulated in organs more than 50 kD linear polymer. This tissue accumulation advantage of the cyclic polymer was also observed in the 90kD MW polymers. The comb polymers described herein exhibit blood retention values at 24 h that are greater than linear PEG11 and less than the dendritic bow-tie polymers1. The longer t1/2, β of the cyclic polymer compared to the linear polymer of the same MW may provide a window of opportunity for cyclic polymers as drug carriers or imaging agents; in the cyclized state the polymer would circulate releasing drug; when the chain was broken, the polymer would be more rapidly eliminated. This is not a particular advantage for degradable polymer backbones. It could be an advantage for cyclic non-degradable backbone, such as the methacrylate polymers widely used as drug carriers4,13.
Figure 3.
Biodistribution of linear and cyclic polymer A–B) 32 kD, C–D) 50 kD and E–F) 90 kD in mice at 24 h, plotted as % injected dose per gram of tissue versus time (left panel, urine and feces were calculated by dividing % injected dose with animal weight) and % injected dose per organ versus time (right panel). Error bars have been omitted from the feces and urine values because the respective excrements for each time point were not collected individually but were pooled together. The % recovery of linear and cyclic polymer with 32, 50 and 90 kD are 80.2, 68.3, 72.8, 76.9, 69.8 and 61.4, respectively.
In conclusion, we demonstrate polymer topology (cyclic vs linear) can play an important role in blood circulation time. This finding adds to the range of macromolecules whose architectures influence their behavior in circulation and may be useful for the creation of drug carriers with improved drug delivery attributes
Supplementary Material
Figure 1.
NMR (1H, 400 MHz) of A) Linear and B) Cyclic Phenol containing PEG(2 kD)-g-PCL(9 kD).
Acknowledgments
This work was supported by NIH EB002047. Supporting Information Available
References
- 1.Gillies ER, Dy E, Frechet JM, Szoka FC. Mol Pharm. 2005;2:129. doi: 10.1021/mp049886u. [DOI] [PubMed] [Google Scholar]
- 2.Lee CC, MacKay JA, Frechet JM, Szoka FC. Nat Biotechnol. 2005;23:1517. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 3.Geng Y, Dalhaimer P, Cai SS, Tsai R, Tewari M, Minko T, Discher DE. Nat Biotechnol. 2007;2:249. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grayson SM, Godbey WT. J Drug Target. 2008;16:329. doi: 10.1080/10611860801969616. [DOI] [PubMed] [Google Scholar]
- 5.Uzgiris E. Invest Radiol. 2004;39:131. doi: 10.1097/01.rli.0000107495.48025.97. [DOI] [PubMed] [Google Scholar]
- 6.Kricheldorf HR, Eggerstedt S. Macromol Chem and Phys. 1998;199:283. [Google Scholar]
- 7.Li H, Debuigne A, Jerome R, Lecomte P. Angew Chem Int Ed. 2006;45:2264. doi: 10.1002/anie.200503961. [DOI] [PubMed] [Google Scholar]
- 8.Bielawski CW, Benitez D, Grubbs RH. Science. 2002;297:2041. doi: 10.1126/science.1075401. [DOI] [PubMed] [Google Scholar]
- 9.Mehvar R, Robinson MA, Reynolds JM. J Pharm Sci. 1995;84:815. doi: 10.1002/jps.2600840706. [DOI] [PubMed] [Google Scholar]
- 10.Laurent BA, Grayson SM. J Am Chem Soc. 2006;128:4238. doi: 10.1021/ja0585836. [DOI] [PubMed] [Google Scholar]
- 11.Yamaoka T, Tabata Y, Ikada Y. J Pharm Sci. 1994;83:6. doi: 10.1002/jps.2600830432. [DOI] [PubMed] [Google Scholar]
- 12.Lim J, Guo Y, Rostollan CL, Stanfield J, Hsieh J, Sun X, Simanek EE. Mol Pharm. 2008;5:540. doi: 10.1021/mp8000292. [DOI] [PubMed] [Google Scholar]
- 13.Duncan R. Nature Reviews Cancer. 2006;6:688. doi: 10.1038/nrc1958. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.