Abstract
Breast cancer is a heterogeneous disease and risk factors could be differentially associated with the development of distinct tumors subtypes that manifest different biological behavior and progression. In support of this view, there is growing evidence that known breast cancer risk factors vary by hormone receptor status and perhaps other pathological characteristics of disease. Recent work from large consortial studies has lead to the discovery of novel breast cancer susceptibility loci in genic (CASP8, FGFR2, TNRC9, MAP3K1, LSP1) and non-genic regions (8q24, 2q35, 5p12) of the genome, and has demonstrated substantial heterogeneity by tumor characteristics. In particular, susceptibility loci in FGFR2, TNRC9, 8q24, 2q35, 5p12 have stronger associations for estrogen receptor positive disease (ER +) than estrogen receptor negative disease (ER −). These findings suggest that common genetic variants can influence the pathological subtype of breast cancer, and provide further support for the hypothesis that ER+ and ER− disease result from different etiologic pathways. Current studies had limited power to detect susceptibility loci for less common tumor subtypes, such as ER − disease including triple negative and basal-like tumors. Ongoing work targeting uncommon subtypes is likely to identify additional tumor specific susceptibility loci in the near future. Characterization of etiologic heterogeneity of breast cancer may lead to improvements in the understanding of the biological mechanisms for breast cancer, and ultimately result in improvements in prevention, early detection and treatment.
Introduction
The presentation of breast cancer can vary greatly with respect to clinical features, pathological characteristics and biological behaviors 1. Age-specific incidence rates for breast cancer using data from the Surveillance Epidemiology and End Results (SEER) demonstrate different patterns for estrogen receptor positive (ER+) and estrogen receptor negative (ER−) tumors2, as well as for different histopathologies 3. Variance in rate patterns at the population level could reflect underlying heterogeneity of environmental and/or genetic factors for tumor subtypes. In support of this view, epidemiological studies are providing growing evidence that breast cancer risk factors vary by hormone receptor status,4–6. Specifically, many environmental risk factors for breast cancer, i.e age at menarche, nulliparity, late age at first birth and post-menopausal obesity, are more strongly related to hormone receptor positive than negative tumors 4−6.
Expression profiling studies indicate that expression of hormone receptors is associated with consistent “molecular portraits” in tumor tissues that are established early in tumorigenesis and represent critical determinants of tumor biology 7–9. Accordingly, risk factors could be linked not only to the development of tumors but also to their biology and progression. Therefore, determining whether risk factor associations for breast cancer differ by morphological and molecular characteristics of the tumors represents a critical etiologic question.
Recent discoveries have lead to the identification of at least eight novel common breast cancer susceptibility loci derived primarily from genome-wide association studies (GWAS), and it is expected that further GWAS will confirm additional loci in the near future. The first wave of GWAS have been conducted primarily in Caucasian populations with a predominance of ER+ disease, and are not well powered to identify susceptibility factors that might more common in other ethnicities or that are specific to less common tumor subtypes. In this article we discuss the evidence for heterogeneity of genetic associations with breast cancer risk by tumor subtypes, in particular those defined by the expression of ER in the tumors.
Genetic susceptibility to breast cancer
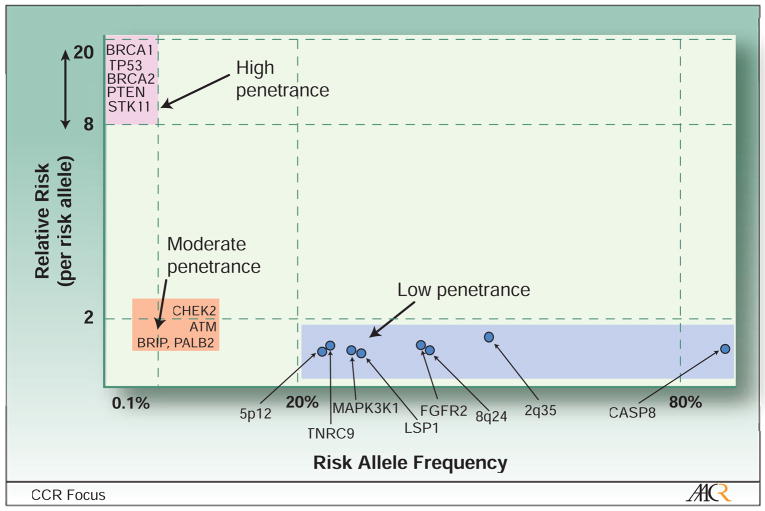
Although environmental factors, primarily hormonal and reproductive factors such as pregnancy history, age at menarche and age at menopause, are major contributors to breast cancer risk10, there is also strong evidence for a genetic component. First-degree relatives of breast cancer patients have approximately a 2-fold increase in risk for developing breast cancer and most of the excess risk is likely to be caused by genetic factors rather than shared environment11,12. Different approaches have lead to the identification of susceptibly loci that contribute to explain the excess familial risk. The loci identified range from those rare in the population associated with very high risk (relative risk of carriers versus non-carriers of 8 to 10) to common susceptibility loci associated with small increases in risk (relative risk less than 2) (Figure 1).
Figure 1. Breast cancer susceptibility loci according to the approximate magnitude of their associated relative risk (per risk allele) and frequency of the risk allele.
This figure shows that most low penetrance variants in susceptibly loci discovered to date fall into the lower right corner (risk allele frequencies over 20% and relative risk per risk allele under 1.3), and that common risk alleles associated with higher relative risk (upper right corners) are unlikely to exist. While additional high penetrance mutations in susceptibly loci are unlikely; moderate and low penetrance variants are likely to be discovered in the near future as the genetic coverage of whole genome scans improves for uncommon variants and the size of studies increases.
High-penetrance mutations
Family linkage studies have identified rare mutations in genes associated with very high risks (high-penetrance mutations) that are responsible for inherited breast cancer syndromes. These include mutations in BRCA1 and BRCA2 causing breast and ovarian cancer syndromes, PTEN in Cowden syndrome, TP53 in Li-Fraumeni syndrome and STK11/LKB1 in Peutz-Jegher syndrome 13,14. In spite of the high risks, because these variants are rare in the population, they account for a relatively small percentage of the familial risk, estimated to 20–25% 14,15. Tumors in BRCA1 carriers have different morphological and immunohistochemical characteristics than those tumors occurring in BRCA2 mutation carriers or in non-carrier tumors 16–21. Specifically, breast tumors in BRCA1 mutation carriers tend to be triple negative (i.e. ER−, PR−, HER2−), high grade and with lymphatic node invasion. These tumors are primarily of infiltrating ductal histology, and they also show a higher percentage of medullary tumors than in non-carriers. The high percentage of ER− tumors is not explained by differences in the age at onset or other tumor characteristics 17,22, suggesting that BRCA1 tumors might originate from ER−cells. More recently, profiling studies have shown that most triple negative tumors among BRCA1 carriers show expression patterns corresponding to the “basal-like” gene cluster7,21,23,24. These studies also noted similarities in the expression and immunohistochemical profiles of BRCA1 and basal-like non-hereditary tumors, suggesting similar origins of these tumors 7,21,23,24 These observations underscore the etiologic heterogeneity of breast cancer subtypes by pathological characteristics.
Moderate-penetrance variants
A combination of family-based and population-based approaches has identified relatively uncommon variants associated with modest increases in risk (moderate-penetrance variants) (Figure 1). These include the 1100delC protein-truncating variant in CHEK225–27, and rare variants in ATM causing ataxiatelangiectasia28, in the BRIP1 gene encoding a BRCA1 -interacting protein29, and the PALB2 gene encoding a BRCA2-interacting protein30. Because of the modest increases in risk and relatively low frequency of this class of genetic variants, their contribution to familial risk is estimated to be less than 3% 31. Current studies have been too small to be able to evaluate differences in the association between these susceptibility loci and tumor subtypes, and additional studies are needed to address this question.
Low-penetrance variants
Current modeling suggests that the majority of the unexplained fraction of familial risk is likely to be explained by a polygenic model implying a combination of many individual variants with weak associations with risk 32–34. Most of these variants are likely to be common (minor allele frequency>0.05) genetic susceptibility loci associated with small increases in risk (low-penetrance variants), which are best studied in population-based studies. This class of genetic variants will be the focus of the reminder of this review.
Discovery of common, low-penetrance variants in susceptibility loci
The approaches to studying common genetic susceptibility factors have evolved very quickly over the last several years, owing to the completion of the sequencing of the human genome35 and the mapping of haplotypes of a large subset of the most common genetic variation, namely, the single nucleotide polymorphism (SNP)36,37. Rapid advances in annotating genomes in populations coupled with efficient developments in genotyping technologies together and substantial reductions in genotyping costs now enable determination of hundreds of thousands of SNPs simultaneously. Genetic variants are determined for each individual from a source of genomic DNA (usually lymphocytes or buccal cells) using different genotyping technologies such as TaqMan assays for single SNPs or multiplexed genotyping platforms to determine several SNPs simultaneously. The technical advances have enabled investigators to move beyond evaluating a few candidate variants in key genes, to conduct more comprehensive as well as exploratory evaluation of common genetic variation in candidate pathways to cancer, and perform GWAS.
1. Candidate gene approach
Over the last 15 years, many studies have used candidate gene approaches to identify common genetic susceptibility loci for breast cancer and other diseases but with somewhat disappointing results. Because of the central role of hormones and reproductive history in the development of breast cancer, genes involved in hormone biosynthesis, metabolism and bioavailability have been prime candidates for study. DNA double strand break repair and related cell cycle checkpoints are also important candidate pathways because of the links between these processes and high to moderate penetrance breast cancer genes. Other etiologic pathways evaluated include those related to known or suspected risk factors for breast cancer such as carcinogen metabolism, alcohol metabolism, and obesity; and genes involved in key carcinogenic processes such as inflammation, immunity, DNA repair, apoptosis, cell signaling, methylation, regulation of telomere length, as well as genes with frequent somatic alterations in breast tumors.
Most findings reported as ‘positive’ from individual studies evaluating candidate genes end up failing to replicate across studies and thus are considered to be false positive findings 38–44. The lack of replication, assuming absence of biases, can be explained by two main factors, namely low statistical power of individual studies to detect weak associations with risk, and low prior probability of a disease association for a given variant. The latter is true even for genes considered to be strong candidates because of limited knowledge of carcinogenic processes and the functional implications of genetic variants. Therefore, large efforts aimed at replicating findings from individual studies using stringent criteria (e.g. very low P-values) are required to identify variants conclusively associated with risk 45. An example of such effort is the Breast and Prostate Cancer Cohort Consortium (BPC3)a that is pooling data from 10 prospective cohort studies to evaluate common variation in candidate genes in the steroid metabolism and IGF pathways in relation to the risk of breast and prostate cancers 46.
The Breast Cancer Association Consortium (BCAC)b, attempted to replicate previously reported associations for 15 candidate gene variants that had been evaluated in at least three independent studies with at least 10,000 subjects in total, but none of these were confirmed to be associated with risk 47,48. The best finding that has been successfully replicated is for a coding variant (D302N) in the caspase 8 (CASP8) gene 48. The N allele was found in 13% of women of Caucasian origin, and was associated with an estimated 12% reduction in breast cancer risk (the relative risk measured by the per-allele odds ratio (OR) was 0.88 (95% confidence interval (CI), 0.84–0.92, P-value for trend=10−7) (see Table 1 for more details). This corresponds to an OR of 1.14 (=1/0.88) for carriers of the common D allele compared to the N allele, which is a substantially weaker association with risk compared to previously identified high and moderate penetrance loci such BRCA1, BRCA2, CHEK2 and others (Figure 1). This finding was based on a pooled analysis of 16,432 cases and 17,106 controls from 14 studies participating in BCAC. This work illustrated the value of large consortia in the replication of findings from initial studies, particularly when the strength of the association is weak. Caspase-8 is a cysteine protease that plays an important role in the initiation of apoptosis or programmed cell death in response to DNA damage 49. The functional implications of the D302N variant are unknown and therefore it is possible that other variants in linkage disequilibrium are the causative variants. The D302N variant is very rare in Asian populations, however a 6-bp deletion polymorphism (−652 6N del) in the promoter of CASP8 has been associated with the risk of multiple cancers, including breast cancer in a Chinese population50. This variant abolished an Spl transcription binding site and is associated with decreased mRNA expression in lymphocytes, and with lower caspase-8 activity after activation-induced cell death in T-lymphocytes 50. Subsequent studies have not been able to confirm the association with breast cancer risk in Caucasian populations 51–54, nor in populations of Asian and African origin in the USA51. A relatively small study in Italy suggested that this variant might be associated with age at diagnosis in familial breast cancer cases 53. Ongoing studies are further evaluating genetic variants in CASP8 and related genes and these efforts might result in the identification of additional variants associated with breast cancer risk. BCAC also reported a weak association with a coding variant (L10P) in the transforming growth factor-b (TGFB1) gene coding of a cytokine involved in the regulation of normal mammary gland development and function 48. This association was present only in progesterone receptor negative (PR−) tumors and remains to be confirmed.
Table 1.
Established breast cancer susceptibility loci
| rs number | Gene | Chromosome | MAF | Heterozygote OR (95% CI) | Rare homozygote OR (95% CI) | Trend test P*** | Study |
|---|---|---|---|---|---|---|---|
| Candidate gene | |||||||
| rs1045485 | CASP8 | 2q | 0.13 | 0.89 (0.85–0.94) | 0.74 (0.62–0.87) | 1.1×10−7 | Cox etal. 2007 48 |
| GWAS | |||||||
| rs2981582* | FGFR2 | 10q | 0.38 | 1.23 (1.18–1.28) | 1.63 (1.53–1.72 | 2.0×l0−76 | Eastonetal. 2007 59 |
| rs1219648* | 1.20 (1.07–1.42) | ) 1.64 (1.42–1.90) | 1.1×l0−10 | Hunter etal 2007 60 | |||
| rs10941679 | 5p12 | 0.24 | 1.19 (1.13–1.26)** | Not reported | 2.9×10−11 | Stacey etal. 2008 62 | |
| rs3803662 | TNRC9 | 16q | 0.25 | 1.23 (1.18–1.29) | 1.39 (1.26–1.45) | 10−36 | Eastonetal. 2007 59 |
| 0.27 | 1.27 (1.19–1.36) | 1.64 (1.45–1.85) | 5×10−19 | Stacey etal. 2007 61 | |||
| rs13387042 | 2q34 | 0.50 | 1.11 (1.03–1.20) | 1.44 (1.30–1.58) | 1.3×10−13 | Stacey etal. 2007 61 | |
| rs13281615 | 8q24 | 0.40 | 1.06(1.01–1.11) | 1.18 (1.10–1.25) | 5×10−12 | Eastonetal. 2007 59 | |
| rs889312 | MAP3K1 | 5p | 0.28 | 1.13 (1.09–1.18) | 1.27 (1.19–1.36) | 7×10−20 | Eastonetal. 2007 59 |
| rs3817198 | LSP1 | 11p | 0.30 | 1.06 (1.02–1.11) | 1.17 (1.08–1.25) | 3×10−9 | Eastonetal. 2007 59 |
The two reported SNPs have an r2 of 1.0.
Per-allele OR calculated under a log additive model
P value for trend test under a log additive model
Numbers are from published data cited in references shown in the study column.
CASP8: caspase 8; FGFR2 fibroblast growth factor receptor; LSP1 lymphocyte-specific protein 1; MAP3K1 mitogen-activated protein kinase 1; TNRC9: trinucleotide repeat containing 9; MAF: minor allele frequency among controls
Studies of candidate genes in breast cancer have evaluated a few hundred ‘favorite’ genes, which is a small percentage of the more than 25,000 genes across the genome. In some respects, GWAS seek to identify regions or genes that should then be considered as candidate genes in subsequent studies. Previously, most candidate gene studies did not perform comprehensive evaluations of genetic variation in these genes. Therefore, it is possible that additional associations can be found using this approach in large collaborative efforts. The candidate gene approach is likely to be particularly important for the discovery of relatively uncommon variants in genes with high probability of being related to disease since these are unlikely to be captured by GWAS approaches described below.
2. Agnostic approach: genome-wide association studies (GWAS)
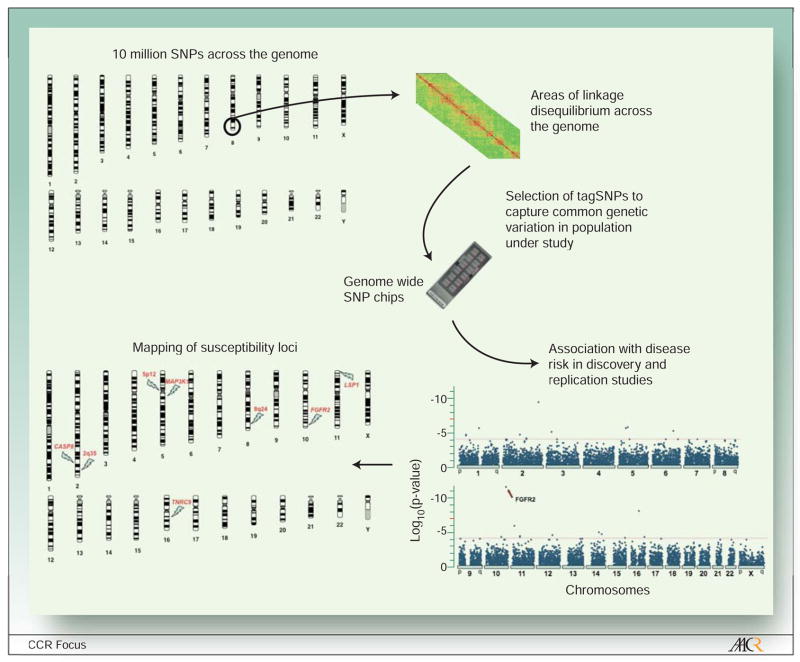
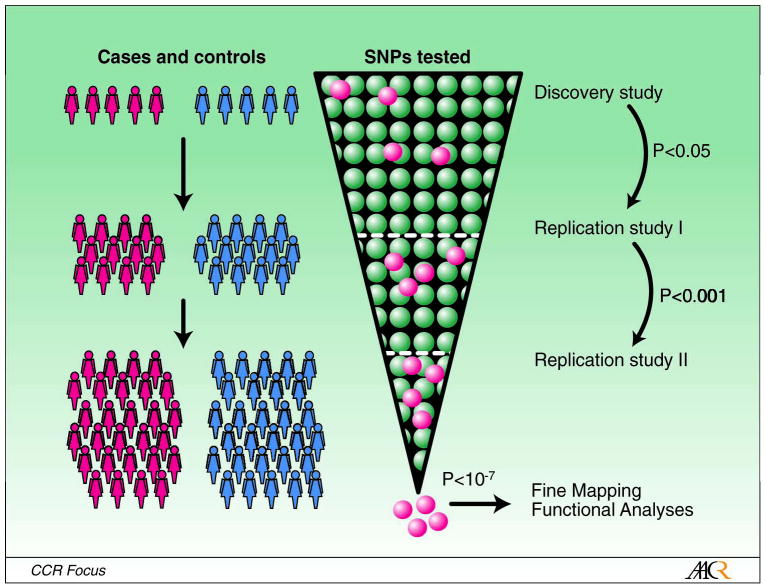
Although the genotyping costs have decreased dramatically in the last few years, it is still be too expensive to genotype all known SNPs (over 10 million SNPs have been identified by the Human Genome Project with a minor allele frequency greater than 1% in at least one population) or sequence the entire human genome. The GWAS strategy selects subsets of genetic markers that serve as surrogates for untested markers, taking advantage of the linkage disequilibrium (LD), defined as correlation among genetic variants located close together 55–57 (Figure 2). The current generation of SNP panels used to conduct genome wide scans includes up to one million SNPs selected using data from the International HapMap Projectc. These SNP panels should be able to capture the majority of common genetic variation in human populations- although the extent of coverage can vary by population genetics history58. Performing whole genome scans in large numbers of individuals can be very be very costly, therefore most GWAS use multi-stage designs in order to reduce genotyping costs56. In these designs, a relatively small proportion of samples from cases and controls are scanned in a discovery study (Figure 3). In subsequent stages, only those markers showing the most significant associations with disease risk are genotyped in additional samples from cases and controls. Because of the large number of makers being tested, very stringent statistical criteria (e.g P value<10−7) are needed to confirm findings in final stages, thus minimizing the number of false positive findings.
Figure 2. Mapping of disease susceptibility loci using genome wide scans.
The Human Genome Project has identified over 10 million single nucleotide polymorphisms (SNPs), which are the most common form of genetic variation in the genome. GWAS take advantage of the correlation (i.e. linkage disequilibrium) between neighboring SNPs in the same chromosome (characterized by the HapMap Project) to select a subset of SNPs (called tagSNPs) that capture most common genetic variation across the genome. Genome wide SNP chips are used to genotype a large number of tagSNPs (500,000 to 1M) on DNA samples from participants in case-control studies to evaluate their association with risk of disease (see Figure 3 for a description of a multistage GWAS design). This strategy is used to map SNPs to areas of the genome likely to include disease-causing variants.
Figure 3. Multi-stage design for genome wide association studies (GWAS).
In a multi-stage design, a large number of single nucleotide polymorphisms (SNPs) selected to capture most common genetic variation across the genome (genome wide scan chip) are tested in a relatively small number of cases and controls in a “discovery study”. The SNPs showing the most significant associations with disease risk in the discovery study (e.g. P value from an association test <0.05) are re-tested in subsequent replication studies including large independent sets of cases and controls. In the example shown in the figure, SNPs with P values <0.001 in a first replication study are re-tested in a second replication study. SNPs showing strong evidence for an association with disease risk based on data from the three phases (e.g. P value from an association test <10–7) are selected as markers for chromosomal regions likely to contain disease causing variants. Very large studies and stringent statistical criteria are necessary to have sufficient power to detect associations while minimizing the probability of false positive findings. The selected markers in GWAS are further evaluated in fine mapping studies to identify causal variants, and functional studies to understand the biological mechanism of the observed associations with disease.
Red and blue individuals represent cases of breast cancer and controls, respectively, being tested in different stages of the design. The green and red dots in the inverted cone represent SNPs being tested in each stage. The red dots are markers for disease susceptibly alleles.
Over the last year, we have witnessed an explosion of new discoveries of susceptibility loci for a wide range of diseases derived from GWASd. Three large-scale GWAS in breast cancer59–62 have discovered seven novel genetic susceptibility loci (FGFR2, TNRC9, MAP3K1, LSP1, 8q24, 2q35, 5p12). Each of these variants shows independent associations with risk of breast cancer, although statistical gene-gene interactions resulting in larger joint effects than expected by their individual relative risks could exist. None of these variants were in coding regions of genes and four variants were in non-genic regions ((8q24, 2q35, 5p12), and the magnitude of the association with breast cancer risk was small (Table 1).
The strongest association was for a SNP in the second intron of the fibroblast growth factor receptor 2, FGFR2, (rs number, rs2981582; Table 1). The high-risk allele was present in 38% of Caucasian populations and women with two copies of this allele (rare homozygote genotype) had a relative risk (measured by the odds ratio) of 1.63 compared to women with two copies of the low-risk allele (common homozygote genotype; Table 1). Although this association was found by an agnostic approach, the FGF signaling pathway has been shown to be important in mammary tumorogenesis by mouse models 63 and FGFR2, in particular, encodes a transmembrane tyrosine kinase that has been involved in mammary gland development and breast carcinogenesis 64,65. FGFR2 is overexpressed or amplified in up to 10% of breast tumors65–68, and its expression is associated with ER+ tumors 69, suggesting a hormone-dependent action of this gene.
Fine mapping studies of the FGFR2 region around the initial SNP discovered in GWAS identified a haplotype of eight strongly linked SNPs that could be the as-yet unidentified causative SNPs70. Gene expression studies have shown increasing FGFR2 expression levels associated with the rare homozygote genotype, and functional studies identified the OCT1/RUNX2 binding site as the main determinant of the increased expression levels 70. This work illustrates how initial discoveries of genetic markers associated with risk in GWAS can lead to additional work aimed at studying the biologic mechanisms underlying the observed associations.
The rs13281615 variant lies in a non-genic region of chromosome 8q24, more than 350 kb from the protooncogene c-MYC (V-myc myelocytomatosis viral oncogene homolog (avian)). This variant was found in 40% of Caucasians and those with the homozygous variant genotype had a relative risk of 1.18 compared to the homozygous common genotype (Table 1). This chromosomal region of 8q24 is of great interest because multiple independent variants in this non-genic region have been associated with the risk of prostate71–79, colorectal73,77,80–82, and ovarian77 cancer risk. Genetic markers in this region are located in five distinct haplotype blocks, one associated only with breast cancer risk, three blocks associated with prostate cancer risk and one block associated with prostate, colorectal and ovarian cancer risk 73,77. The underlying mechanism(s) that explain the striking associations is not well understood. As mentioned above, MYC is the closest gene in this region and functional studies suggest that it might be implicated in prostate cancer by down-regulating the prostate tumor suppressor KLF6 gene 83. Ongoing and future association and functional studies in this region are likely to lead to a better understanding of mechanisms of carcinogenesis.
Little is know about the function of the other loci identified in GWAS. The mechanisms of association with the other two SNPs in non-genic regions, 2q35 and 5p12 are unknown. The closest gene to rs10941679 in 5p12 is MRPS30 (also known as programmed cell death protein 9, PDCD9) which is implicated in apoptosis 84, is associated with ER+ tumors 85 and with tumor characteristics associated with good prognosis 86. In addition, the fibroblast growth factor 10 (FGF10), a breast cancer oncogene and FGFR2 ligand, is also in the vicinity of this region. The identification of SNPs in non-genic regions associated with the risk of breast cancer and other common disorders through GWAS highlight the role of intra-genic regions in the regulation of transcription, and suggest that polymorphisms in non-coding RNAs might also play a role in these diseases 87.
The rs889312 variant is in a LD block that contains the mitogen-activated protein kinase 3 K1(MAP3K1) and two hypothetical genes (MGC33648 and mesoderm induction early response 1, family member 3 or MIER3). MAP3K1 forms part of the MAPK cell-signaling pathway implicated in cellular response to mitogens, however MAP3K1 has not been previously linked to breast carcinogenesis. The rs383662 variant is in an area of LD that contains the trinucleotide repeat containing 9 (TNRC9, also known as TOX high mobility group box family member 3, TOX3) and a hypothetical gene LOC643714. The function of TNRC9 is unknown although it contains a putative high mobility group suggesting that it might function as a transcription factor. In addition, TNRC9 expression has been associated with the presence of bone metastasis derived from breast tumors 88. rs38017198 lies in intron 10 of lymphocyte-specific protein 1 (LSP1) gene (also known as WP43) that encodes an F-actin bundling cytoskeletal protein that is expressed in haemotopoietic and endothelial cells. LSP1 has been implicated in malignant lymphoma and Hodgkin disease 89, and other variants in this gene have been associated with risk of developing non-Hodgkin lymphoma 90.
Since GWAS encompass dense data sets, it is critical for other bona fide investigators to access the data through a registered system to explore new hypotheses and also use the resources to confirm possible associations, as has been done by the Cancer Genetic Markers of Susceptibility initiative of the USA National Cancer Institute (NCI)e ,60.
Heterogeneity of genetic associations by tumor ER status
Recent work from large consortial studies has demonstrated that the associations between novel breast cancer susceptibility loci and breast cancer described above could vary by clinically important tumor characteristics. Most notably, associations with most of the susceptibility loci identified to date are evidently stronger for ER+ than ER− disease, although some of these differences are small and not statistically significant (Table 2). Susceptibility loci significantly modified by ER status include FGFR2, TNRC9, 8q24, 2q35, 5p12 61,62,91. The strongest evidence is for a variant in FGRF2 that was primarily associated with ER+ disease in two separate reports 62,91. This finding is consistent with the involvement of FGFR2 in estrogen-related breast carcinogenesis 68,92–94, and with higher levels of FGFR2 expression in ER+ than ER− cell lines and tumors 69,95. The variant in FGFR2 was also associated with low tumor grade independent of ER status 91. Interestingly, the variant in the 5p12 region, which is close to the FGFR2 ligand FGF10, also shows strong evidence for an association primarily with ER+ tumors 62.
Table 2.
Established breast cancer susceptibility loci by estrogen receptor status
| Per-allele OR (95% CI) | ||||||
|---|---|---|---|---|---|---|
| Gene/region | rs number | MAF | ER+ tumors | ER− tumors | HeterogeneityP-value | Study |
| FGFR2 | rs2981582 | 0.38 | 1.31 (1.27–1.36) | 1.08 (1.03–1.14) | 10−13 | Garcia-Closas et al. 2008 91 |
| 0.41 | 1.29 (1.22–1.38) | 0.99 (0.88–1.10) | 2.9×10−5 | Stacey et al. 2008 62 | ||
| 5p12 | rs10941679 | 0.24 | 1.27 (1.19–1.35) | 1.05 (0.92–1.18) | 0.004 | Stacey et al. 2008 62 |
| TNRC9 | rs3803662 | 0.27 | 1.23 (1.19–1.27) | 1.14 (1.09–1.24) | 0.015 | Garcia-Closas et al. 2008 91 |
| 1.32 (1.22–1.42) | 1.07 (0.94–1.23) | 0.0098 | Stacey et al. 2007 61 | |||
| 2q35 | rs13387042 | 0.54 | 1.22 (1.14–1.31) | 1.06 (0.94–1.19) | 0.036 | Stacey et al. 2007 61 |
| 8q24 | rs13281615 | S0.41 | 1.13 (1.10–1.17) | 1.03 (0.96–1.08) | 0.001 | Garcia-Closas et al. 2008 91 |
| MAP3K1 | rs889312 | 0.28 | 1.12 (1.09–1.16) | 1.07(1.01–1.13) | 0.11 | Garcia-Closas et al. 2008 91 |
| LSP1 | rs3817198 | 0.30 | 1.07 (1.04–1.11) | 1.04 (0.99–1.10) | 0.31 | Garcia-Closas et al. 2008 91 |
| CASP8 | rs1045485 | 0.13 | 0.89 (0.82–0.96)* | 0.95 (0.84–1.07)* | 0.24 | Cox etal. 2007 48 |
Per allele OR not shown in original paper. ORs are for heterozygous vs homozygous common genotype
MAF: minor allele frequency among controls
Numbers are from published data cited in references shown in the study column
CASP8: caspase 8; FGFR2 fibroblast growth factor receptor; LSP1 lymphocyte-specific protein 1; MAP3K1 mitogen-activated protein kinase 1; TNRC9: trinucleotide repeat containing 9
The variant in TNRC9 was found to be associated more strongly with ER+ disease by Stacey at al. based on analyses of 2,128 ER+ and 589 ER− tumors 61. A much larger study including 12,974 ER+ and 3,765 ER− cases from BCAC found an association with both tumor subtypes, although the association was slightly weaker for ER− tumors 91. Based on this report, the TNRC9 variant shows the strongest association with ER− tumors among all loci identified to date. Finally, the variant in the 8q24 region also showed a significantly stronger association for ER+ than ER−disease in the BCAC analyses 91.
The above susceptibility loci also showed associations with progesterone receptor (PG) status, however, these did not appear to be independent of ER. It is also possible that the observed associations between susceptibility loci and ER status reflect stronger underlying associations with other correlated tumor markers, or with particular marker profiles such as molecular subtypes previously defined by expression profiling.7,9. For instance, triple negative tumors (ER−, PR−, HER−), which include a large percentage of basal-like tumors, are a clinically distinct subgroup of breast cancer and could have different etiology21. Limited epidemiological data supports differences in non-genetic risk factors for triple negative tumors compared to “luminal A” tumors (ER+, PER+ HER−) 21,96,97. In addition, initial analyses using data from studies participating in BCAC support the notion additional heterogeneity on genetic risk factors can be detected within hormone receptor positive or hormone receptor negative tumors according to the expression of HER2 and basal markers (EGFR and basal cytokeratins) 98.
The Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA)f; has recently evaluated whether variants in FGFR2 (rs2981582), TNRC9 (rs3803662) and MAP3K1 (rs889312) are associated with the risk of breast cancer in over 10,000 BRCA1 and BRCA2 mutation carriers from 23 studies 99. The minor alleles in SNPs in FGFR2 and MAP3K1 were associated with increases in risk among BRCA2 mutation carriers similar to those previously reported in non-carriers; however these SNPs were not associated with an increased risk in BRCA1 carriers. On the other hand, the SNP in TNRC9 was associated with risk of both BRCA1 and BRCA2 related tumors. Over 90% of BRCA1 related tumors are ER−, in contrast with BRCA2 tumors that tend to be ER+, similarly to tumors in non-carriers 100. Thus, these findings further support the observed heterogeneity of these genetic associations by ER status of tumors in non-carriers reported by the BCAC 91. Unfortunately, the CIMBA report did not have detailed information on the ER status of the tumors and thus could not evaluate if ER status could explain the observed differences by carrier status.
An important limitation of studies of etiologic heterogeneity is the very large sample size required to study less common subtypes and the use of non-standardized tissue collection/processing protocols and immunohistochemical assays performed at different study centers. Tissue microarray (TMA) blocks consist of recipient paraffin blocks that contain small tissue cores removed from targets of many individual donor paraffin blocks, thus offering an attractive method for obtaining standardized, rapid, and cost-effective immunohistochemical characterization of many tumors 101. This technique is facilitating the evaluation of etiologic heterogeneity in large epidemiological studies. In addition, improvements in automated image analysis technologies 102 and web-based systems allowing access to pathological images from multiple centers might facilitate the daunting task pathologists face when scoring thousands of tissue cores, while providing more standardized and quantitative measures.
Current GWAS have been conducted in breast cancer cases unselected by ER status. Because ER+ tumors are more common that ER− tumors, these studies had better statistical power to detect SNPs preferentially associated with the more common subtype rather than less common groups, such as E− tumors or specific ER− subtypes (e.g. triple negative or basal-like disease). Ongoing GWAS including large numbers of ER− tumors will be better suited to identify ER− specific susceptibility loci. Understanding the etiology of basal-like tumors is of particular importance since these tumors tend to occur early in life, are harder to detect by screening mammography and are associated with poor prognosis 103.
The observed differences in genetic associations by tumor subtypes support for the notion that ER− and ER+ tumors result from different etiologic pathways (some of which might be shared), rather than different stages of tumor evolution within a common carcinogenic pathway104, Although the magnitude of the observed differences is small, and by themselves these findings are unlikely to have any immediate clinical implications, the observed differences provide clues to the biological mechanisms that underpin tumor heterogeneity.
Concluding remarks
In the last year, at least eight novel susceptibility loci have been discovered primarily by GWAS. Theses recent discoveries are shading light to important mechanisms in breast carcinogenesis, and provide support for the presence of etiologic heterogeneity across breast cancer subtypes. Under a polygenic model of disease susceptibility, seven of the eight low-penetrance genes were estimated to explain about 5% of the genetic risk for breast cancer31, with an additional small contribution expected from the eighth SNP in 5p12 not included in that report. Given that high-penetrance genes explain 20–25% and moderate-penetrance genes explain less than 3% of the genetic risk, a large number of additional variants are likely to exist31. Each of the unknown susceptibility loci is expected to be weakly associated with risk and thus very large studies are required. tudies evaluating different ethnic groups and tumor subtypes should increase the power to find variants that are more common in these population subgroups. Finally, the study of strong candidate genes and pathways such as those already implicated in breast cancer risk, as well as the study of gene-gene and gene-environment interactions might enhance the ability to identify and characterize additional variants. However, evaluation of interactions as a means of genetic discovery can increase the chances of false positive findings dramatically and thus will require even larger replication studies to be confirmed.
Because of the weak associations with risk, low-penetrance loci are unlikely to have utility for individualized prevention strategies 31. The value of these studies is to identify noel loci in the genome that could shed light on new mechanisms of disease, and thus become targets for therapy or prevention. Although the currently identified loci provide low discriminatory accuracy to distinguish between low and high risk groups in the population105, many additional loci are yet to be discovered, and the combination of loci might prove useful in population screening strategies 31,g
Current studies have focused on SNPs as markers of genetic susceptibility, and other less common forms of genetic variation such as copy number variants might also contribute to susceptibility to breast and other cancers. In addition, integrated biomarkers that reflect multiple genetic, epigenetic and environmental challenges such as DNA repair capacity, telomere length or methylation patterns 106 are also promising markers to identify susceptible groups in the population. Characterization of common genetic variation in breast cancer cases can also lead to the discovery of variants that influence response to therapy and survival that could prove useful in the clinical management of patients 107.
In conclusion, the discovery of disease susceptibility loci and characterization of underlying etiologic heterogeneity may lead to improvements in the understanding of the biological mechanisms for breast cancer, and ultimately result in improvements in prevention, early detection and treatment.
Footnotes
References
- 1.Dowsett M, Dunbier A. Emerging biomarkers and new understanding of traditional markers in personalized therapy for breast cancer. Clin Cancer Res. 2008;14 doi: 10.1158/1078-0432.CCR-08-0974. in press. [DOI] [PubMed] [Google Scholar]
- 2.Anderson WF, Jatoi I, Devesa SS. Distinct breast cancer incidence and prognostic patterns in the NCI’s SEER program: suggesting a possible link between etiology and outcome. Breast Cancer Res Treat. 2005;90:127–37. doi: 10.1007/s10549-004-3777-3. [DOI] [PubMed] [Google Scholar]
- 3.Anderson WF, Chu KC, Chang S, Sherman ME. Comparison of age-specific incidence rate patterns for different histopathologic types of breast carcinoma. Cancer Epidemiol Biomarkers Prev. 2004;13:1128–35. [PubMed] [Google Scholar]
- 4.Ma H, Bernstein L, Pike MC, Ursin G. Reproductive factors and breast cancer risk according to joint estrogen and progesterone receptor status: a meta-analysis of epidemiological studies. Breast Cancer Res. 2006;8:R43. doi: 10.1186/bcr1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reeves GK, Beral V, Green J, Gathani T, Bull D. Hormonal therapy for menopause and breast-cancer risk by histological type: a cohort study and meta-analysis. Lancet Oncol. 2006;7:910–8. doi: 10.1016/S1470-2045(06)70911-1. [DOI] [PubMed] [Google Scholar]
- 6.Althuis MD, Fergenbaum JH, Garcia-Closas M, Brinton LA, Madigan MP, Sherman ME. Etiology of hormone receptor-defined breast cancer: a systematic review of the literature. Cancer Epidemiol Biomarkers Prev. 2004;13:1558–68. [PubMed] [Google Scholar]
- 7.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 8.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colditz GA, Baer HJ, Tamimi RM. Breast cancer. In: Schottenfeld D, Fraumeni JF, editors. Cancer Epidemiology and Prevention. New York: Oxford University Press; 2006. pp. 995–1012. [Google Scholar]
- 11.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. The New England journal of medicine. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 12.Peto J, Mack TM. High constant incidence in twins and other relatives of women with breast cancer. Nat Genet. 2000;26:411–4. doi: 10.1038/82533. [DOI] [PubMed] [Google Scholar]
- 13.Antoniou AC, Easton DF. Models of genetic susceptibility to breast cancer. Oncogene. 2006;25:5898–905. doi: 10.1038/sj.onc.1209879. [DOI] [PubMed] [Google Scholar]
- 14.Easton DF. How many more breast cancer predisposition genes are there? Breast Cancer Res. 1999;l:14–7. doi: 10.1186/bcr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson D, Easton D. The genetic epidemiology of breast cancer genes. Journal of mammary gland biology and neoplasia. 2004;9:221–36. doi: 10.1023/B:JOMG.0000048770.90334.3b. [DOI] [PubMed] [Google Scholar]
- 16.Lakhani SR, Jacquemier J, Sloane JP, et al. Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. Journal of the National Cancer Institute. 1998;90:1138–45. doi: 10.1093/jnci/90.15.1138. [DOI] [PubMed] [Google Scholar]
- 17.Lakhani SR, Van De Vijver MJ, Jacquemier J, et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20:2310–8. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Breast Cancer Linkage Consortium. Pathology of familial breast cancer: differences between breast cancers in carriers of BRCA1 or BRCA2 mutations and sporadic cases. Lancet. 1997;349:1505–10. [PubMed] [Google Scholar]
- 19.Karp SE, Tonin PN, Begin LR, et al. Influence of BRCA1 mutations on nuclear grade and estrogen receptor status of breast carcinoma in Ashkenazi Jewish women. Cancer. 1997;80:435–41. doi: 10.1002/(sici)1097-0142(19970801)80:3<435::aid-cncr11>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 20.Loman N, Johannsson O, Bendahl PO, Borg A, Ferno M, Olsson H. Steroid receptors in hereditary breast carcinomas associated with BRCA1 or BRCA2 mutations or unknown susceptibility genes. Cancer. 1998;83:310–9. [PubMed] [Google Scholar]
- 21.Schneider BP, Winer EP, Foulkes WD, et al. Triple negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14 doi: 10.1158/1078-0432.CCR-08-1208. in press. [DOI] [PubMed] [Google Scholar]
- 22.Foulkes WD, Metcalfe K, Sun P, et al. Estrogen receptor status in BRCA1- and BRCA2-related breast cancer: the influence of age, grade, and histological type. Clin Cancer Res. 2004;10:2029–34. doi: 10.1158/1078-0432.ccr-03-1061. [DOI] [PubMed] [Google Scholar]
- 23.Turner NC, Reis-Filho JS. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25:5846–53. doi: 10.1038/sj.onc.1209876. [DOI] [PubMed] [Google Scholar]
- 24.Foulkes WD, Stefansson IM, Chappuis PO, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. Journal of the National Cancer Institute. 2003;95:1482–5. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 25.CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9.065 controls from 10 studies. American journal of human genetics. 2004;74:1175–82. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–9. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 27.Vahteristo P, Bartkova J, Eerola H, et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. American journal of human genetics. 2002;71:432–8. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renwick A, Thompson D, Seal S, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38(8):873–5. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 29.Seal S, Thompson D, Renwick A, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–41. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 30.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. The New England journal of medicine. 2008;358:2796–803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 32.Ponder BA, Antoniou A, Dunning A, Easton DF, Pharoah PD. Polygenic inherited predisposition to breast cancer. Cold Spring Harbor symposia on quantitative biology. 2005;70:35–41. doi: 10.1101/sqb.2005.70.029. [DOI] [PubMed] [Google Scholar]
- 33.Nathanson KL, Wooster R, Weber BL. Breast cancer genetics: what we know and what we need. Nature medicine. 2001;7:552–6. doi: 10.1038/87876. [DOI] [PubMed] [Google Scholar]
- 34.Mack TM, Hamilton AS, Press MF, Diep A, Rappaport EB. Heritable breast cancer in twins. British journal of cancer. 2002;87:294–300. doi: 10.1038/sj.bjc.6600429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 36.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 39.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–9. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 40.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–82. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 41.Ioannidis JP. Common genetic variants for breast cancer: 32 largely refuted candidates and larger prospects. Journal of the National Cancer Institute. 2006;98:1350–3. doi: 10.1093/jnci/djj392. [DOI] [PubMed] [Google Scholar]
- 42.Ioannidis JP, Trikalinos TA, Khoury MJ. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. American journal of epidemiology. 2006;164:609–14. doi: 10.1093/aje/kwj259. [DOI] [PubMed] [Google Scholar]
- 43.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–72. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 45.Chanock SJ, Manolio T, Boehnke M, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–60. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 46.Hunter DJ, Riboli E, Haiman CA, et al. A candidate gene approach to searching for low-penetrance breast and prostate cancer genes. Nat Rev Cancer. 2005;5:977–85. doi: 10.1038/nrc1754. [DOI] [PubMed] [Google Scholar]
- 47.Commonly studied single-nucleotide polymorphisms and breast cancer: results from the Breast Cancer Association Consortium. Journal of the National Cancer Institute. 2006;98:1382–96. doi: 10.1093/jnci/djj374. [DOI] [PubMed] [Google Scholar]
- 48.Cox A, Dunning AM, Garcia-Closas M, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–8. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 49.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–6. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 50.Sun T, Gao Y, Tan W, et al. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet. 2007;39:605–13. doi: 10.1038/ng2030. [DOI] [PubMed] [Google Scholar]
- 51.Haiman CA, Garcia RR, Kolonel LN, Henderson BE, Wu AH, Le Marchand L. A promoter polymorphism in the CASP8 gene is not associated with cancer risk. Nat Genet. 2008;40:259–60. 60–1. doi: 10.1038/ng0308-259. [DOI] [PubMed] [Google Scholar]
- 52.Frank B, Rigas SH, Bermejo JL, et al. The CASP8 -652 6N del promoter polymorphism and breast cancer risk: a multicenter study. Breast cancer research and treatment. 2008;111:139–44. doi: 10.1007/s10549-007-9752-z. [DOI] [PubMed] [Google Scholar]
- 53.De Vecchi G, Verderio P, Pizzamiglio S, et al. Evidences for association of the CASP8 -652 6N del promoter polymorphism with age at diagnosis in familial breast cancer cases. Breast cancer research and treatment. 2008 doi: 10.1007/s10549-008-9963-y. [DOI] [PubMed] [Google Scholar]
- 54.Cybulski C, Wokolorczyk D, Gliniewicz B, et al. A six-nucleotide deletion in the CASP8 promoter is not associated with a susceptibility to breast and prostate cancers in the Polish population. Breast cancer research and treatment. 2007 doi: 10.1007/s10549-007-9864-5. [DOI] [PubMed] [Google Scholar]
- 55.Johnson GC, Esposito L, Barratt BJ, et al. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–7. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 56.Kraft P, Cox DG. Study designs for genome-wide association studies. In: Rao D, Gu C, editors. Genetic dissection of complex traits. San Diego: Academic Press; 2008. [Google Scholar]
- 57.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 58.Barrett JC, Garden LR. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38:659–62. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- 59.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 62.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–6. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 63.Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 2005;16:179–86. doi: 10.1016/j.cytogfr.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 64.Dickson C, Spencer-Dene B, Dillon C, Fantl V. Tyrosine kinase signalling in breast cancer: fibroblast growth factors and their receptors. Breast Cancer Res. 2000;2:191–6. doi: 10.1186/bcr53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh M. Cancer genomics and genetics of FGFR2 (Review) International journal of oncology. 2008;33:233–7. [PubMed] [Google Scholar]
- 66.Heiskanen M, Kononen J, Barlund M, et al. CGH, cDNA and tissue microarray analyses implicate FGFR2 amplification in a small subset of breast tumors. Anal Cell Pathol. 2001;22:229–34. doi: 10.1155/2001/981218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adnane J, Gaudray P, Dionne CA, et al. BEK and FLG, two receptors to members of the FGF family, are amplified in subsets of human breast cancers. Oncogene. 1991;6:659–63. [PubMed] [Google Scholar]
- 68.Hishikawa Y, Tamaru N, Ejima K, Hayashi T, Koji T. Expression of keratinocyte growth factor and its receptor in human breast cancer: its inhibitory role in the induction of apoptosis possibly through the overexpression of Bcl-2. Arch Histol Cytol. 2004;67:455–64. doi: 10.1679/aohc.67.455. [DOI] [PubMed] [Google Scholar]
- 69.Luqmani YA, Graham M, Coombes RC. Expression of basic fibroblast growth factor, FGFR1 and FGFR2 in normal and malignant human breast, and comparison with other normal tissues. Br J Cancer. 1992;66:273–80. doi: 10.1038/bjc.1992.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyer KB, Maia AT, O’Reilly M, et al. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS biology. 2008;6:e108. doi: 10.1371/journal.pbio.0060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 72.Freedman ML, Haiman CA, Patterson N, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–73. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haiman CA, Le Marchand L, Yamamato J, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–6. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 75.Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meyer A, Schurmann P, Ghahremani M, et al. Association of chromosomal locus 8q24 and risk of prostate cancer: A hospital-based study of German patients treated with brachytherapy. Urologic oncology. 2008 doi: 10.1016/j.urolonc.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 77.Ghoussaini M, Song H, Koessler T, et al. Multiple Loci with different cancer specificities within the 8q24 gene desert. Journal of the National Cancer Institute. 2008;100:962–6. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beebe-Dimmer JL, Levin AM, Ray AM, et al. Chromosome 8q24 markers: risk of early-onset and familial prostate cancer. Int J Cancer. 2008;122:2876–9. doi: 10.1002/ijc.23471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schumacher FR, Feigelson HS, Cox DG, et al. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–6. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- 80.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q2421. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 81.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 82.Gruber SB, Moreno V, Rozek LS, et al. Genetic Variation in 8q24 Associated with Risk of Colorectal Cancer. Cancer Biol Ther. 2007:6. doi: 10.4161/cbt.6.7.4704. [DOI] [PubMed] [Google Scholar]
- 83.Sole X, Hernandez P, de Heredia ML, et al. Genetic and genomic analysis modeling of germline c-MYC overexpression and cancer susceptibility. BMC genomics. 2008;9:12. doi: 10.1186/1471-2164-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cavdar Koc E, Ranasinghe A, Burkhart W, et al. A new face on apoptosis: death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEES letters. 2001;492:166–70. doi: 10.1016/s0014-5793(01)02250-5. [DOI] [PubMed] [Google Scholar]
- 85.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 86.Yu K, Ganesan K, Miller LD, Tan P. A modular analysis of breast cancer reveals a novel low-grade molecular signature in estrogen receptor-positive tumors. Clin Cancer Res. 2006;12(11 Pt l):3288–96. doi: 10.1158/1078-0432.CCR-05-1530. [DOI] [PubMed] [Google Scholar]
- 87.Glinsky GV. Phenotype-defining functions of multiple non-coding RNA pathways. Cell cycle (Georgetown, Tex. 2008;7:1630–9. doi: 10.4161/cc.7.11.5976. [DOI] [PubMed] [Google Scholar]
- 88.Smid M, Wang Y, Klijn JG, et al. Genes associated with breast cancer metastatic to bone. J Clin Oncol. 2006;24:2261–7. doi: 10.1200/JCO.2005.03.8802. [DOI] [PubMed] [Google Scholar]
- 89.Marafioti T, Jabri L, Pulford K, Brousset P, Mason DY, Delsol G. Leucocyte-specific protein (LSP1) in malignant lymphoma and Hodgkin’s disease. British journal of haematology. 2003;120:671–8. doi: 10.1046/j.1365-2141.2003.04137.x. [DOI] [PubMed] [Google Scholar]
- 90.Cerhan JR, Ansell SM, Fredericksen ZS, et al. Genetic variation in 1253 immune and inflammation genes and risk of non-Hodgkin lymphoma. Blood. 2007;110:4455–63. doi: 10.1182/blood-2007-05-088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Garcia-Closas M, Hall P, Nevanlinna H, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS genetics. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y, Sugimoto Y, Kulp SK, Farrar WB, Brueggemeier RW, Lin YC. Estrogen-induced keratinocyte growth factor mRNA expression in normal and cancerous human breast cells. Oncol Rep. 1998;5:577–83. [PubMed] [Google Scholar]
- 93.Zhang Y, Kulp SK, Sugimoto Y, Farrar WB, Brueggemeier RW, Lin YC. Keratinocyte growth factor (KGF) induces aromatase activity in cultured MCF-7 human breast cancer cells. Anticancer Res. 1998;18:2541–6. [PubMed] [Google Scholar]
- 94.Tamaru N, Hishikawa Y, Ejima K, et al. Estrogen receptor-associated expression of keratinocyte growth factor and its possible role in the inhibition of apoptosis in human breast cancer. Lab Invest. 2004;84:1460–71. doi: 10.1038/labinvest.3700166. [DOI] [PubMed] [Google Scholar]
- 95.Zang XP, Pento JT. Keratinocyte growth factor-induced motility of breast cancer cells. Clin Exp Metastasis. 2000;18:573–80. doi: 10.1023/a:1011997317994. [DOI] [PubMed] [Google Scholar]
- 96.Millikan RC, Newman B, Tse CK, et al. Epidemiology of basal-like breast cancer. Breast cancer research and treatment. 2008;109:123–39. doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang XR, Sherman ME, Rimm DL, et al. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev. 2007;16:439–43. doi: 10.1158/1055-9965.EPI-06-0806. [DOI] [PubMed] [Google Scholar]
- 98.Garcia-Closas M, Couch F, Chang-Claude J, et al. Associations for breast cancer susceptibility loci vary by breast cancer subtypes: findings from the Breast Cancer Association Consortium. Proceedings of the 99th Annual Meeting of the American Association for Cancer Research; Philadelphia (PA): AACR. 2008. [Google Scholar]
- 99.Antoniou AC, Spurdle AB, Sinilnikova OM, et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. American journal of human genetics. 2008;82:937–48. doi: 10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lakhani SR, Reis-Filho JS, Fulford L, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11:5175–80. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 101.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nature medicine. 1998;4:844–7. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 102.Chung GG, Zerkowski MP, Ghosh S, Camp RL, Rimm DL. Quantitative analysis of estrogen receptor heterogeneity in breast cancer. Lab Invest. 2007;87:662–9. doi: 10.1038/labinvest.3700543. [DOI] [PubMed] [Google Scholar]
- 103.Da Silva L, Clarke C, Lakhani SR. Demystifying basal-like breast carcinomas. Journal of clinical pathology. 2007;60:1328–32. doi: 10.1136/jcp.2006.041731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Allred DC, Brown P, Medina D. The origins of estrogen receptor alpha-positive and estrogen receptor alpha-negative human breast cancer. Breast Cancer Res. 2004;6:240–5. doi: 10.1186/bcr938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. Journal of the National Cancer Institute. 2008;100:1037–41. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garcia-Closas M, Vermeulen R, Sherman ME, Moore LE, Smith MT, Rothman N. Application of biomarkers in cancer epidemiology. In: Fraumeni DSJF, editor. Cancer Epidemiology and Prevention. 3. New York: Oxford University Press; 2006. [Google Scholar]
- 107.Tan SH, Lee SC, Goh BC, Wong J. Pharmacogenetics in breast cancer therapy. Clin Cancer Res. 2008;14 doi: 10.1158/1078-0432.CCR-08-0993. in press. [DOI] [PubMed] [Google Scholar]