Abstract
Translational Relevance
Previous reports suggested that abnormalities of INI1 could be detected in 70–75% of malignant rhabdoid tumors. The mechanism of inactivation in the other 25% remained unclear. The goal of this study was to perform a high-resolution genomic analysis of a large series of rhabdoid tumors with the expectation of identifying additional loci related to the initiation or progression of these malignancies. We also developed a comprehensive set of assays, including a new MLPA assay, to interrogate the INI1 locus in 22q11.2. Intragenic deletions could be detected using the Illumina 550K Beadchip, whereas single exon deletions could be detected using MLPA. The current study demonstrates that with a multi-platform approach, alterations at the INI1 locus can be detected in almost all cases. Thus, appropriate molecular genetic testing can be used as an aid in the diagnosis and for treatment planning for most patients.
Purpose
A high-resolution genomic profiling and comprehensive targeted analysis of INI1/SMARCB1 of a large series of pediatric rhabdoid tumors was performed. The aim was to identify regions of copy number change and loss of heterozygosity that might pinpoint additional loci involved in the development or progression of rhabdoid tumors, and define the spectrum of genomic alterations of INI1 in this malignancy.
Experimental Design
A multi-platform approach, utilizing Illumina single nucleotide polymorphism (SNP) based oligonucleotide arrays, multiplex ligation dependent probe amplification (MLPA), fluorescence in situ hybridization (FISH), and coding sequence analysis was used to characterize genome wide copy number changes, loss of heterozygosity, and genomic alterations of INI1/SMARCB1 in a series of pediatric rhabdoid tumors.
Results
The bi-allelic alterations of INI1 that led to inactivation were elucidated in 50 of 51 tumors. INI1 inactivation was demonstrated by a variety of mechanisms, including deletions, mutations, and loss of heterozygosity. The results from the array studies highlighted the complexity of rearrangements of chromosome 22, compared to the low frequency of alterations involving the other chromosomes.
Conclusions
The results from the genome wide SNP-array analysis suggest that INI1 is the primary tumor suppressor gene involved in the development of rhabdoid tumors with no second locus identified. In addition, we did not identify hot spots for the breakpoints in sporadic tumors with deletions of chromosome 22q11.2. By employing a multimodality approach, the wide spectrum of alterations of INI1 can be identified in the majority of patients, which increases the clinical utility of molecular diagnostic testing.
Keywords: INI1/SMARCB1, rhabdoid tumor, 22q11.2, SNP array, MLPA
Introduction
Malignant rhabdoid tumors (MRT) are rare, highly aggressive neoplasms found most commonly in infants and young children. Although they may present in any location in the body, they are predominantly found in the kidney and central nervous system (CNS). Patients may present with apparently sporadic tumors in one anatomic site or with multiple primary tumors arising in the brain, kidney and/or soft tissues. Due to their heterogeneous histologic features, diagnosis of these lesions can often be difficult. For example, central nervous system atypical teratoid/rhabdoid tumor (AT/RT) is often mis-classified as medulloblastoma, primitive neuroectodermal tumor (PNET), or choroid plexus carcinoma (CPC) (1).
The development of rhabdoid tumors was initially associated with alterations of chromosome 22 (2). Subsequent studies implicated the INI1/hSNF5/SMARCB1/BAF47 [MIM 601607] gene, located on chromosome band 22q11.2, as the gene responsible for the initiation of malignant rhabdoid tumors. Germline and somatic mutations and deletions of INI1 have been reported in renal and extra-renal rhabdoid tumors as well as AT/RT (3–5). INI1 has also been implicated in the development of epithelioid sarcoma (6), renal medullary carcinoma (7), and familial schwannomatosis (8), although it is not clear if each of these entities has a similar spectrum of mutations and deletions compared with rhabdoid tumors. In patients with malignant rhabdoid tumors, INI1 appears to function as a classic tumor suppressor gene, whereby germline mutations and deletions predispose to the development of these malignancies. Inactivation of both copies of the gene leads to loss of protein expression in the nucleus, which can be detected by immunohistochemistry. The immunohistochemistry assay for INI1 (BAF47) is currently used as an adjunct to histology in the differential diagnosis of rhabdoid tumors in both children and adults (1).
The INI1 gene codes for one of at least ten subunits of the SWI/SNF ATP- dependent chromatin-remodeling complex. INI1 is an invariant component of all SWI/SNF complexes, and is thus expressed in all normal cells at all stages of development. Studies in model organisms have shown that at least one copy of INI1 is required for normal development (9). The SWI/SNF complex appears to regulate transcriptional activity, resulting in both repression and activation of a wide variety of target genes (10, 11). INI1 appears to play a role in the Rb-cyclin D1 pathway, however, the specific function of INI1 with respect to development of human rhabdoid tumors has not yet been elucidated. The function of INI1 and the SWI/SNF complex and its role in the development of rhabdoid tumors has been recently reviewed (11, 12).
Using a combination of karyotype analysis, FISH and direct sequence analysis, deletions and mutations of the INI1 locus were previously detected in approximately 75% of patients (4). The underlying genetic basis of the remaining 25% of cases was unknown. The promoter region of the INI1 gene is not methylated in tumors (13), but alternative epigenetic mechanisms leading to loss of INI1 expression have not yet been explored. Copy number variations or intronic sequence alterations in the INI1 locus that are not detected by current screening methods could potentially account for a percentage of inactivating events in these tumors. Alternatively, there may be a second locus for rhabdoid tumors distinct from INI1 that has yet to be elucidated (14, 15).
Over the last several years, oligonucleotide-based microarrays have emerged as the platform of choice for genome-wide copy number and loss of heterozygosity analysis (16–20). In addition to the high resolution of these platforms, SNP-based oligonucleotide arrays allow for the detection of copy number neutral loss of heterozygosity (LOH) events that are prevalent in cancers (21, 22), which is particularly relevant for the loss of chromosome 22 observed in rhabdoid tumors (23). Although these arrays have an inter-marker distance of only a few kb and hence offer the potential to detect deletions and duplications at the single gene level, the limited number of SNPs within a particular locus may not be adequate to detect copy number changes or LOH at single exons.
Multiplex ligation dependent probe amplification (MLPA) is a PCR-based assay that allows for multiple specific nucleic acid sequences to be amplified simultaneously using a single PCR primer pair. Recently, high-density probe sets for chromosome 22q11.2 have been developed to better identify and localize deletions and duplications within the DiGeorge/Velo-cardio-facial Syndrome (DGS/VCFS) region in 22q11.2. We previously demonstrated that patients with constitutional and somatically acquired INI1 deletions could be detected with this high density probe set (24). A comprehensive MLPA probe set specific for INI1 (SMARCB1) has now been developed, which can be used to interrogate the copy number of each of the nine exons of the gene. We hypothesized that this MLPA kit would have the sensitivity to detect whole exon deletions and duplications in tumor tissues that would be missed by standard sequence analysis and yet could be below the sensitivity of the whole genome SNP based array.
Our previous studies suggested that the chromosome 22q11.2 breakpoints are often localized to low copy repeats (LCRs) in patients with germline deletions of INI1 (23). We expected, based on the increased density of the SNP arrays used herein, to more clearly refine the breakpoints in these sporadic tumors, and determine if they were also localized to LCR regions in 22q11.2.
In the present study, we employed a combination of FISH, PCR-based sequence and MLPA analysis, and whole genome based SNP array analysis with the Illumina 550K BeadChip to analyze a series of 51 primary rhabdoid tumors. The goals were to, first, achieve the most comprehensive analysis of INI1 that could be used in a clinical diagnostic setting for patient diagnosis and genetic counseling; second, detect additional recurrent alterations in the genome that could be used for identification of a second rhabdoid tumor locus; and third, determine the nature of the deletions, duplications or regions of LOH involving chromosome 22 in tumors from different anatomic locations.
Materials and Methods
Case selection
Tumor tissue was obtained from 51 patients for INI1 analysis according to procedures approved by the Institutional Review Board at The Children’s Hospital of Philadelphia (CHOP). Parental consent was obtained for genetic testing. The cases were specifically selected to include tumors for which we were previously unable to detect one or both of the inactivating deletions or mutations of INI1, and was therefore not designed to determine sensitivity or specificity of any individual assay. None of the patients had received prior chemotherapy or radiation before surgery. All of the cases were confirmed to be rhabdoid tumors by histology and/or immunohistochemistry with an antibody to INI1 (1). DNA was extracted from tumor tissue with a Puregene kit (Gentra Systems, Minneapolis, MN) according to the manufacturer’s protocol, and quantitated using a Nanodrop ND-1000 UV-Vis Spectrophotometer (Nanodrop Technologies, Wilmington, DE).
Fluorescence in Situ Hybridization (FISH)
Touch imprints from frozen tissue, formalin fixed tissue sections, or fixed cell pellets were analyzed by FISH. The INI1 probe was labeled by nick translation with ChromoTide® AlexaFluor 594-dUTP or ChromoTide®fluorescein-12-dUTP (Molecular Probes, Eugene OR). Probes for the Ewing’s sarcoma (EWS) region in 22q12 were used simultaneously with the probe for INI1 as an internal control. The probes were applied to slides of the tumor cells and co-denatured at 75°C on an Isotemp 125D heat block (Fisher Scientific, Pittsburgh, PA). Slides were incubated at 37°C overnight in a moist slide moat (Boekel Scientific, Feasterville, PA). They were then washed in a 0.4X SSC solution at 73°C for 2 minutes, followed by a 1-minute wash in 2X SSC/0.1% NP-40 and counterstained with DAPI (Sigma, St Louis, MO). Fluorescent signals from 100 to 200 cells were evaluated at 100X with a Nikon Eclipse E800 fluorescence microscope equipped with the proper filter sets. An Applied Imaging System (Santa Clara, CA) was used to record images of representative cells.
High-Density SNP-based Oligonucleotide Array Analysis
The high-density oligonucleotide array analysis was performed using an Illumina Infinium whole-genome genotyping 550K Beadchip (Illumina, Inc, La Jolla, CA). 750ng of genomic DNA from the tumor sample was processed using reagents and protocols supplied by Illumina in the CHOP Center for Applied Genomics (25).
The data were analyzed using Beadstudio software (Illumina Inc.), which allows for the visualization of several different parameters relevant to the detection of copy number alterations (CNA), including the B allele frequency (BAF) and log R ratio (LRR). The B allele frequency represents the allelic copy ratio for the genomic SNPs. “Normal”, diploid DNA is expected to have three BAF clusters -- two for the homozygous SNPs (AA and BB) with BAFs close to 0 and 1, respectively, and one for the heterozygous SNPs (AB) with BAFs centered around 0.5. The LRR graph shows the log-normalized intensity ratio for each SNP in the test sample compared to a reference sample that includes DNA from 120 standard samples (from the HapMap set of 269). The presence of the expected two copies of any given SNP in a normal, diploid state would result in a LRR of ~0. Statistically significant deviations from 0 are interpreted as copy number changes. A positive LRR suggests gain in copy number and a negative LRR suggests a loss in copy number.
The BAF and LRR output data were also analyzed for copy number alterations using our Center for Biomedical Informatics Copy Number Analysis, Annotation and Visualization tool (CHOPPY). CHOPPY is a set of tools based on the Circular Binary Segmentation (CBS) algorithm which allows improved detection of copy number alterations (26). The visual output from the BeadStudio software was compared to the numerical output computed by CHOPPY. Heterozygous deletions and amplifications represented by fewer than 10 SNPs, with the exception of the INI1 locus, and copy neutral loss of heterozygosity (CN-LOH) events less than 5Mb in size were excluded from analysis. The breakpoints for the tumors with 22q11.2 heterozygous or homozygous deletions were localized with respect to the proximal and distal LCRs in 22q11.2 based on the Beadstudio and CHOPPY data. Results were compared to an in-house database of known, common copy number variations seen in 2,026 healthy controls detected with the same CHOPPY tools. All genomic positions were based upon NCBI build 36 of the human genome (hg18) from the UCSC Genome Browser (http://genome.ucsc.edu/).
The array results for chromosome 22 were then clustered using hierarchical clustering based on their copy number and LOH patterns, as previously described (27). For each sample, the copy number and LOH regions were first decomposed into individual SNPs within these regions. The distance between any two samples is measured as the total number of SNPs that do not belong to the same copy number and LOH categories between the two samples. Samples with similar copy number and LOH patterns on chromosome 22 are thus clustered close together.
MLPA
MLPA was performed with genomic DNA according to previously published methods based on the manufacturer’s protocol using the SALSA MLPA P258 (SMARCB1) kit (MRC-Holland; www.mrc-holland.com). This kit contains 2 probes for each of the 9 exons of INI1, probes for 9 other genes on chromosome 22, and 14 control probes from other chromosomes. The samples were processed and data analyzed as previously described (24).
PCR Sequencing Mutation Analysis
Oligonucleotide primers for exons 1–9 of the INI1 gene were designed from the intron/exon boundary sequences (GenBank accession nos. AP000349-350) for PCR. PCR products for individual exons were analyzed by direct sequencing as previously described (4).
Results
A total of 51 rhabdoid tumors were analyzed using the high-density Illumina 550K SNP oligonucleotide array. The samples included 36 CNS AT/RTs (34 brain, 2 spinal), 8 renal, and 7 extrarenal tumors (1 facial, 2 neck, 1 upper thigh, 2 liver, 1 lymph node). There were 326 CNAs detected in the 51 tumors (Supplemental Table 1). Of those CNAs, only 177 were thought to be potentially pathogenic based on comparison to in-house CNV databases as well as interrogation of the genomic regions using the UCSC genome browser. Ninety-seven of the 177 potentially pathogenic CNAs were localized to chromosome 22, and there were no other consistent abnormalities identified. Representative examples of the Beadstudio output for three tumors are shown in Figure 1. Deletions or LOH involving the INI1 region were identified in 49 of 51 cases by the SNP array, as shown in Figure 2. Among the 49 tumors with alterations detected by the array, 24 (12 brain, 1 spinal, 6 renal, and 5 extra-renal) had homozygous deletions, 11 (10 brain and 1 renal) had heterozygous deletions, and 14 (10 brain, 1 spinal, 1 renal, 2 soft tissue) had copy number neutral loss of heterozygosity involving chromosome 22q11.2. One brain tumor (05–188) did not have any detectable abnormality of the INI1 region by SNP array analysis, aside from a small duplication that was a known normal population variant. This tumor, however, did have a homozygous deletion of one exon that was only detectable by MLPA (Table 1).
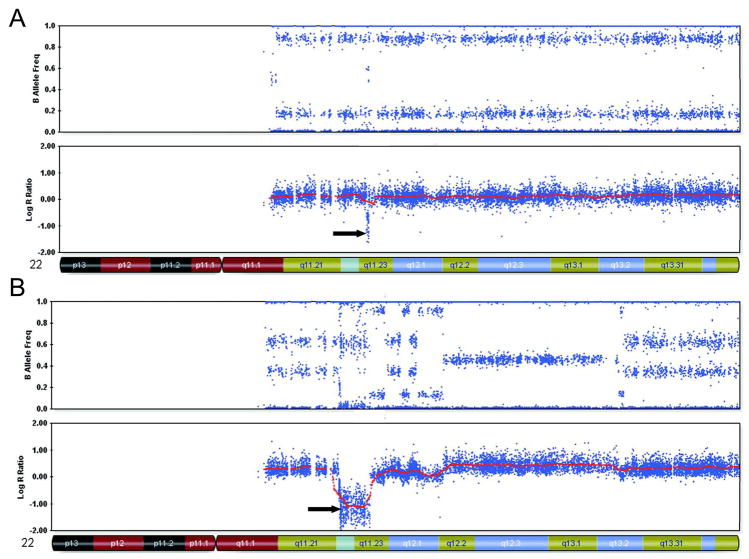
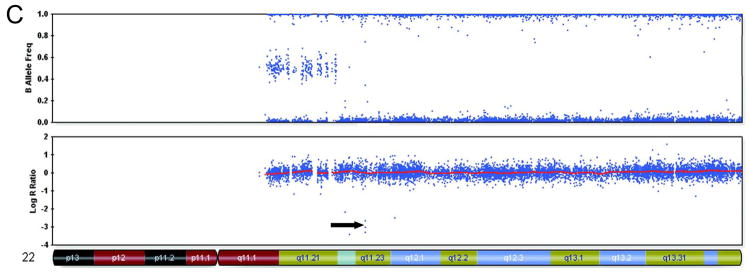
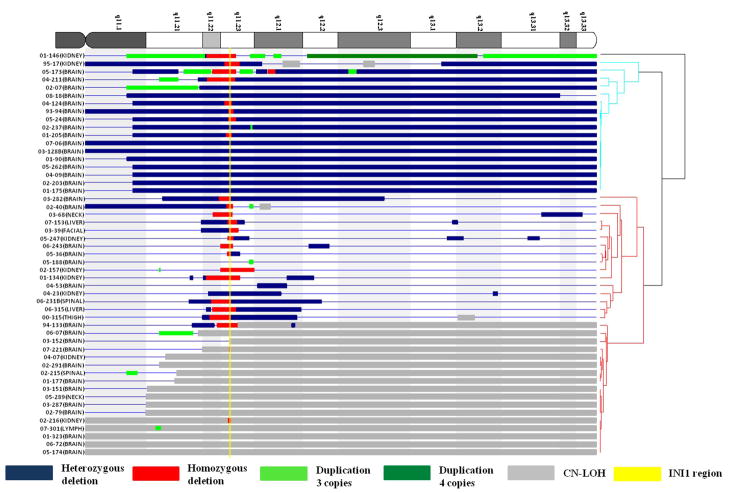
Figure 1.
Representative samples of chromosome 22 from Beadstudio depicting various inactivating alterations of INI1 (indicated by arrows)
A. Case 02–216: CN-LOH with contamination involving most of the long arm, with a homozygous deletion that encompasses INI1
B. Case 01–146; complex duplications and deletions of chromosome 22 including a homozygous deletion in 22q11.2
C. Case 07–221; exon 7–9 deletion of INI1 identified by MLPA associated with a 3 SNP deletion.
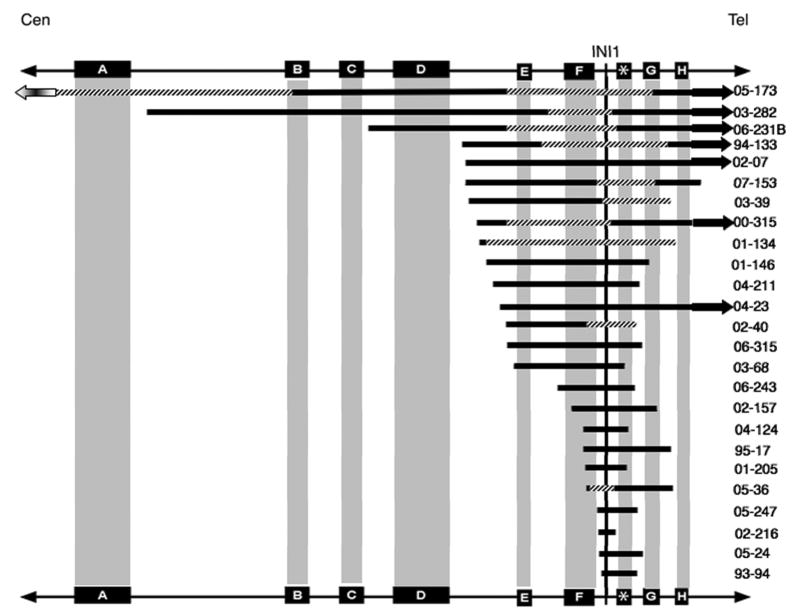
Figure 2.
Graphical representation of chromosome 22 copy number alterations demonstrated by SNP array analysis. Samples are arranged by hierarchical clustering, as described in the methods section. There are 3 major cluster groups: the top cluster, large deletions; the middle cluster, small deletions; and the bottom cluster, large regions of CN LOH. In general, tumors with large deletions or large regions of CN LOH were almost exclusively brain tumors. Soft tissue tumors were found primarily to have small deletions.
Table 1.
INI1 inactivating events in rhabdoid tumors without homozygous INI1 deletions
| ID | Array Result | Second hit modality | Second Event |
|---|---|---|---|
| 01-090 | heterozygous deletion | S* | exon 9 c.1143delG |
| 01-175 | heterozygous deletion | S | exon 5 c.601C>T |
| 02-07 | heterozygous deletion | S | exon 6 c.711delC |
| 02-203 | heterozygous deletion | S | exon 9 c.1143delG |
| 02-237 | heterozygous deletion | S | exon 9 c.1143delG |
| 03-128B | heterozygous deletion | S | exon 9 c.1143delG |
| 05-262 | heterozygous deletion | S | 3′ UTR c.1220T>G |
| 08-018 | heterozygous deletion | S | exon 9 c.1143delG |
| 01-323 | LOH | S | exon 2 c.118C>T |
| 02-079 | LOH | S | exon 5 c.601C>T |
| 02-215 | LOH | S | exon 5 c.539_546dup8 |
| 02-291 | LOH | S | exon 5 c.601C>T |
| 03-151 | LOH | S | exon 7 c.843G>A |
| 03-287 | LOH | S | exon 2 c.157C>T |
| 04-07 | LOH | S | exon 5 c.601C>T |
| 05-289 | LOH | S | exon 3 c.356_360delACCTC |
| 06-07 | LOH | S | exon 4 c.425T>G |
| 04-023 | heterozygous deletion | M | exon 7 homozygous deletion |
| 04-09 | heterozygous deletion | M | exon 6–9 deletion |
| 07-06 | heterozygous deletion | M | exon 6 duplication |
| 03-152 | LOH | M | exon 7 deletion |
| 05-174 | LOH | M | exon 6–7 duplication |
| 06-072 | LOH | M | exon 4–5 duplication |
| 07-301 | LOH | M | exon 1 deletion |
| 05-188 | Normal | M | exon 1 homozygous deletion |
| 04-053 | Normal | F, S | FISH 24% of cells with INI1 deletion, exon 9 c.1145delC |
| 07-221 | homozygous deletion | M | exon 7–9 homozygous deletion |
| 01-177 | LOH | NONE | NONE |
S- sequence analysis; M- MLPA; F- FISH
For rhabdoid tumor samples without homozygous INI1 deletions, FISH, MLPA and sequence analysis of the nine coding exons of the INI1 gene were used to identify the second inactivating event. As shown in Table 1, combined events inactivating both copies of INI1 (deletion, mutation, or copy number neutral LOH) were detected in 50 of 51 cases (Figure 2 and Table 1). One sample (01–177) had copy number neutral LOH identified by the array but no other coding sequence mutation or exon deletion/duplication. A second sample (05–262) had a heterozygous deletion by array and FISH, although the MLPA assay was non-informative. A single base alteration in the 3′ untranslated region (c.1220T>G) was identified in the tumor tissue from this patient. Matched normal tissue from the patient and parental blood samples were not available for comparison. Although it was presumed to be the second inactivating event, the biologic significance of this single base change is unknown. One case (04–53) required FISH to establish both inactivating events. Direct sequencing revealed a mutation in exon 9, but neither the array nor MLPA demonstrated a deletion at the INI1 locus (both demonstrated small chromosome 22 deletions distal to the INI1 locus). FISH, on the other hand, revealed a deletion of 22, with loss of both INI1 and EWS signals, in 24% of cells.
Further sublocalization of the chromosome 22 deletions within 22q11.2 was performed with respect to the low copy repeats (LCRs) in this region. The chromosome 22 breakpoints in cases with interstitial 22q11.2 deletions are depicted in Figure 3. In the present series of cases, only one patient was found to have a germline mutation (07–06) and no patient had evidence of a germline deletion. These numbers are a reflection of the case selection for this study, and should not be used to estimate the expected number of patients with germline alterations of INI1. As shown in the figure, the majority of breakpoints were located between LCR regions rather than within LCR regions or other recombination hotspots.
Figure 3.
Breakpoints for 22q11.2 deletions in relation to low copy repeat regions, designated by blocks A-H. The asterisk between F and G represents a common recombination hotspot. Changes from solid to hatched bars denote the start and end points for tumors with more than one alteration. For example, case 03–282 has a heterozygous deletion with a proximal breakpoint between LCR A and B and a distal breakpoint after LCR H. The second (homozygous) deletion starts between LCR E and F and ends between LCR F and the recombination hotspot.
Among the tumors with heterozygous deletions or copy number neutral LOH for 22q, mutations were identified in 18 cases by direct sequencing. Six of the CNS AT/RTs had a 1143delG or 1145delC in exon 9 that appears to be the most common hot spot for mutations in tumors of the CNS (4). Furthermore, 10 tumors had single or multiple exon deletions or duplications detected by MLPA. Case 07–221 had a homozygous deletion of exons 7 to 9 that was detected by both SNP array and MLPA, and 05–174 had a duplication of exon 6–7 that was detected by MLPA. The exon 4–9 deletion identified by MLPA in 93–94 was not detected on the SNP array, possibly due to contamination with normal tissue. The smaller exon deletions or duplications revealed by MLPA in the remaining cases were not observed by the array analysis.
Discussion
In the present study, 51 rhabdoid tumors from a variety of anatomic locations were studied using four different testing modalities to identify potential underlying genetic changes leading to the development of malignant rhabdoid tumors. Notably, despite the differences in anatomic location, half of the tumors (24/51) were characterized by homozygous deletions of INI1. This included over one third (13 of 36) of the CNS AT/RTs as well as the majority of both renal (6/8) and extra renal (5/7) tumors.
As shown in Figure 2, there were some differences in the patterns of structural changes of chromosome 22 in tumors from different sites made apparent by the hierarchical clustering. This series included a predominant number of CNS tumors (36/51) and the frequencies of CNAs versus copy number neutral LOH were fairly evenly distributed (13 tumors with homozygous deletions, 10 with heterozygous deletions, and 11 with copy number neutral LOH). Tumors with whole or large chromosome 22 deletions were exclusively from the brain. Tumors with whole arm or large regions of chromosome 22 copy number neutral LOH were mostly from brain as well. In contrast, the soft tissue rhabdoid tumors were more likely to have smaller deletions in 22q11.22 to 22q11.23. The renal rhabdoid tumors had either copy number neutral LOH or complex CNAs. Despite these trends, due to the high prevalence of CNS tumors in these samples, and the low numbers of other tumor types, it is difficult to draw conclusions about the type of inactivating events as related to anatomic location. In addition, although some CNAs involving chromosomes other than 22 were observed more than once, there were no other non-random patterns of alteration among or between the tumors from different anatomic sites.
Previous studies of rhabdoid tumors have shown that these tumors are mostly diploid, with few recurrent regions of LOH or copy number changes other than chromosome 22 (12). Our data supports the fact that most of the tumors are diploid, as there was only one hyperdiploid sample (01–323) among the 51 tumors tested. However, the complex nature of some of the rearrangements of chromosome 22 was somewhat unexpected, as illustrated by case 01–146 (Figure 1B). Although most of the genome appeared balanced, chromosome 22 was disrupted by a number of events leading to 3 or 4 copies of the chromosome for some regions, as well as heterozygous and homozygous deletions that ultimately resulted in loss of the INI1 region. In this sample, numerous genes would be subjected to dosage changes, the net effect of which is unclear.
The high resolution SNP array data may define the approximate locations of the breakpoints leading to deletions and duplications that can yield insight into the mechanism of the inactivating events. Our previous study of patients with rhabdoid tumors and germline 22q11.2 deletions suggested that some of the underlying genetic changes could be related to recombination events mediated by LCRs located in chromosome 22 (23). The present cohort, on the other hand, was comprised predominantly of patients with sporadic tumors, as only one patient had a germline mutation (07–06) and no patient had a germline deletion. As shown in Figure 3, the majority of the breakpoints in the cases described here were between and not within the LCR regions. This finding is not unexpected as LCR-mediated rearrangements in the germline are believed to occur via non-allelic homologous recombination during meiosis (28, 29). The deletion events in the sporadic tumors are somatic events and may be the result of alternate mechanisms such as non-homologous end joining. Thus, our analysis suggests that there may be distinct mechanisms of deletion in the setting of a germline deletion versus a sporadic event.
The degree of complexity for the alterations of chromosome 22 in a subset of the tumors suggests that these deletions are, indeed, taking place by a variety of mechanisms, which are likely to be unbalanced translocations rather than simple LCR mediated recombinations. Cytogenetically balanced translocations involving 22q11.2 have been shown by FISH to be associated with submicroscopic deletions of INI1, as previously described (4). The partner chromosomes are variable and only limited numbers of tumors have been studied to determine whether the derivative chromosome is also deleted or if specific genes have been interrupted by the translocations. One case (00–315) had an apparently balanced t(7;22)(p15;q11.2), yet a homozygous deletion of INI1 was identified by FISH and subsequently confirmed by high density SNP array analysis. This array also revealed a 2.5Mb deletion in 7p15, indicating that the translocation partner chromosome was also deleted as a result of the translocation. The HOXA gene cluster and the CREB5 genes were included in the region of loss. Whether such additional changes are ultimately related to the clinical phenotype of the patient, their response to therapy, or their clinical outcome remains to be seen.
The current study shows that by employing a combination of SNP array analysis, MLPA, FISH, and direct sequencing, the inactivating deletions and mutations of INI1 can be identified in the vast majority of pediatric rhabdoid tumors. All 51 tumors described here had at least one detectable inactivating event, and both inactivating events were identified in 50 (98%) tumors. In the remaining case (01–177), there was loss of INI1 expression by immunohistochemistry, suggesting that the second unidentified inactivating event may have been associated with a mutation or epigenetic modification of a non-coding region of the gene.
These data strongly support the fact that INI1 is the primary gene responsible for the development of rhabdoid tumors. Although the high-density SNP array analysis did not reveal any other consistently altered regions, clinical correlative studies including outcome data are in progress to determine if some of the less prevalent changes predict differences in prognosis and response to treatment. At the present time, the SNP-based oligonucleotide array can be used to refine disease associated CNAs, and distinguish for example, a medulloblastoma with an isochromosome 17q from a primary rhabdoid tumor with loss of 22q11.2. When indicated, molecular analysis of INI1 using MLPA and direct sequencing may then be employed. Once the tumor associated changes are found, an analysis of germline DNA from the patient and parents can be analyzed to rule out an inherited or de novo germline mutation or deletion of INI1, so that appropriate recurrence risk assessments can be made.
Supplementary Material
Supplemental Table 1: Copy number alterations demonstrated by SNP array for all chromosomes
Acknowledgments
This work was supported by a grant from the NIH (CA46274) to J.A.B and (GM081519) to T.H.S. E.M.J. received salary support from the Neurosurgery Research and Education Foundation. We gratefully acknowledge support from Emma’s Hope, Shamrocks for Shannon, and the Kevin J. Snyder Memorial Foundation. The authors wish to thank Luanne M. Wainwright and Fan Zhang for technical assistance; and the following individuals for contributing patient samples for this study:
Jeffrey Allen, Dan Bowers, Philip Boyer, Jeffrey S. Dome, Harriet Druker, Ann-Christine Duhaime, Morris Edelman, Mark Edgar, Jonathan Eisenstat, Linda Ernst, Ralph Franciosi, Renuka Gera, Jeffrey Golden, Michael Goldfischer, Robert Goldsby, Jeffrey Goldstein, Ignacio Gonzalez-Gomez, Peter Helseth, Michael Isakoff, Mark Kieran, Patricia Kirby, Lisa Kohorn, Michael LaQuaglia, Jenny Libien, Jeffrey Lobel, Amy Lowichik, David Malkin, Mark Matthews, Claire Mazewski, Elizabeth Mroczek-Musulman, Bruce Pawel, Heather Prashner, Ted Pysher, R. H. Rhodes, Vilmarie Rodriguez, Lucy Rorke-Adams, Pierre Russo, Ana Sotrel, Phillip Jay Storm, Leslie Sutton, Karen Tsuchiya, Elsa Valderrama, Cynthia Wetmore, Anthony Yachnis, David Zagzag, Holly Zhou, and Craig Zuppan.
References
- 1.Judkins AR, Mauger J, Ht A, Rorke LB, Biegel JA. Immunohistochemical analysis of hSNF5/INI1 in pediatric CNS neoplasms. Am J Surg Path. 2004;28:644–50. doi: 10.1097/00000478-200405000-00013. [DOI] [PubMed] [Google Scholar]
- 2.Biegel JA, Rorke LB, Packer RJ, Emanuel BS. Monosomy 22 in rhabdoid or atypical tumors of the brain. J Neurosurg. 1990;73:710–4. doi: 10.3171/jns.1990.73.5.0710. [DOI] [PubMed] [Google Scholar]
- 3.Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–9. [PubMed] [Google Scholar]
- 4.Biegel JA, Tan L, Zhang F, Wainwright L, Russo P, Rorke LB. Alterations of the hSNF5/INI1 gene in central nervous system atypical teratoid/rhabdoid tumors and renal and extrarenal rhabdoid tumors. Clin Cancer Res. 2002;8:3461–7. [PubMed] [Google Scholar]
- 5.Versteege I, Sevenet N, Lange J, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 6.Modena P, Lualdi E, Facchinetti F, et al. SMARCB1/INI1 tumor suppressor gene is frequently inactivated in epithelioid sarcomas. Cancer Res. 2005;65:4012–9. doi: 10.1158/0008-5472.CAN-04-3050. [DOI] [PubMed] [Google Scholar]
- 7.Cheng JX, Tretiakova M, Gong C, Mandal S, Krausz T, Taxy JB. Renal medullary carcinoma: rhabdoid features and the absence of INI1 expression as markers of aggressive behavior. Mod Pathol. 2008 doi: 10.1038/modpathol.2008.44. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Hulsebos TJ, Plomp AS, Wolterman RA, Robanus-Maandag EC, Baas F, Wesseling P. Germline mutation of INI1/SMARCB1 in familial schwannomatosis. Am J Hum Genet. 2007;80:805–10. doi: 10.1086/513207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO reports. 2000;1:500–6. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biggar SR, Crabtree GR. Continuous and widespread roles for the Swi-Snf complex in transcription. Embo J. 1999;18:2254–64. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muchardt C, Yaniv M. The mammalian SWI/SNF complex and the control of cell growth. Sem Cell Dev Biol. 1999;10:189–95. doi: 10.1006/scdb.1999.0300. [DOI] [PubMed] [Google Scholar]
- 12.Biegel JA. Molecular genetics of atypical teratoid/rhabdoid tumor. Neurosurg Focus. 2006;20:E11. doi: 10.3171/foc.2006.20.1.12. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, Tan L, Wainwright LM, Bartolomei MS, Biegel JA. No evidence for hypermethylation of the hSNF5/INI1 promoter in pediatric rhabdoid tumors. Genes Chromosomes Cancer. 2002;34:398–405. doi: 10.1002/gcc.10078. [DOI] [PubMed] [Google Scholar]
- 14.Bourdeaut F, Freneaux P, Thuille B, et al. hSNF5/INI1-deficient tumours and rhabdoid tumours are convergent but not fully overlapping entities. J Pathol. 2007;211:323–30. doi: 10.1002/path.2103. [DOI] [PubMed] [Google Scholar]
- 15.Fruhwald MC, Hasselblatt M, Wirth S, et al. Non-linkage of familial rhabdoid tumors to SMARCB1 implies a second locus for the rhabdoid tumor predisposition syndrome. Pediatr Blood Cancer. 2005 doi: 10.1002/pbc.20526. [DOI] [PubMed] [Google Scholar]
- 16.Bignell GR, Huang J, Greshock J, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004;14:287–95. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman JM, Baross A, Delaney AD, et al. Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet. 2006;79:500–13. doi: 10.1086/507471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janne PA, Li C, Zhao X, et al. High-resolution single-nucleotide polymorphism array and clustering analysis of loss of heterozygosity in human lung cancer cell lines. Oncogene. 2004;23:2716–26. doi: 10.1038/sj.onc.1207329. [DOI] [PubMed] [Google Scholar]
- 19.Ming JE, Geiger E, James AC, et al. Rapid detection of submicroscopic chromosomal rearrangements in children with multiple congenital anomalies using high density oligonucleotide arrays. Hum Mutat. 2006;27:467–73. [Google Scholar]
- 20.Zhao X, Li C, Paez JG, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–71. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- 21.Langdon JA, Lamont JM, Scott DK, et al. Combined genome-wide allelotyping and copy number analysis identify frequent genetic losses without copy number reduction in medulloblastoma. Genes Chromosomes Cancer. 2006;45:47–60. doi: 10.1002/gcc.20262. [DOI] [PubMed] [Google Scholar]
- 22.Maris JM, Hii G, Gelfand CA, et al. Region-specific detection of neuroblastoma loss of heterozygosity at multiple loci simultaneously using a SNP-based tag-array platform. Genome Res. 2005;15:1168–76. doi: 10.1101/gr.3865305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson EM, Shaikh TH, Gururangan S, et al. High-density single nucleotide polymorphism array analysis in patients with germline deletions of 22q11.2 and malignant rhabdoid tumor. Hum Genet. 2007;122:117–27. doi: 10.1007/s00439-007-0386-3. [DOI] [PubMed] [Google Scholar]
- 24.Jalali GR, Vorstman JA, Errami A, et al. Detailed analysis of 22q11.2 with a high density MLPA probe set. Hum Mutat. 2007;29:433–40. doi: 10.1002/humu.20640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peiffer DA, Le JM, Steemers FJ, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–48. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 27.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emanuel BS, Shaikh TH. Segmental duplications: an ‘expanding’ role in genomic instability and disease. Nat Rev Genet. 2001;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- 29.Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet. 2004;13 Spec No 1:R57–64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: Copy number alterations demonstrated by SNP array for all chromosomes