Abstract
Helicobacter pylori γ-glutamyltranspeptidase (HpGT) is a general γ-glutamyl hydrolase and a demonstrated virulence factor. The enzyme confers a growth advantage to the bacterium, providing essential amino acid precursors by initiating the degradation of extracellular glutathione and glutamine. HpGT is a member of the N-terminal nucleophile (Ntn) hydrolase superfamily and undergoes autoprocessing to generate the active form of the enzyme. Acivicin is a widely used γ-glutamyltranspeptidase inhibitor that covalently modifies the enzyme, but its precise mechanism of action remains unclear. The time-dependent inactivation of HpGT exhibits a hyperbolic dependence on acivicin concentration with kmax = 0.033 ± 0.006 sec−1 and KI = 19.7 ± 7.2 μM. Structure determination of acivicin-modified HpGT (1.7 Å; Rfactor=17.9%; Rfree=20.8%) demonstrates that acivicin is accommodated within the γ-glutamyl binding pocket of the enzyme. The hydroxyl group of Thr 380, the catalytic nucleophile in the autoprocessing and enzymatic reactions, displaces chloride from the acivicin ring to form the covalently linked complex. Within the acivicin-modified HpGT structure, the C-terminus of the protein becomes ordered with Phe 567 positioned over the active site. Substitution or deletion of Phe 567 leads to a >10-fold reduction in enzymatic activity, underscoring its importance in catalysis. The mobile C-terminus is positioned by several electrostatic interactions within the C-terminal region, most notably a salt bridge between Arg 475 and Glu 566. Mutational analysis reveals that Arg 475 is critical for the proper placement of the C-terminal region, the Tyr 433 containing loop, and the proposed oxyanion hole.
Keywords: γ-glutamyltranspeptidase, Ntn-hydrolase, glutathione, acivicin, x-ray crystallography
H. pylori γ-glutamyltranspeptidase (HpGT) is a γ-glutamyl hydrolase with broad substrate specificity (1, 2), and is a member of the N-terminal nucleophile (Ntn) hydrolase superfamily (3, 4). The inactive precursor undergoes an intramolecular autoprocessing event, generating the mature and catalytically active heterotetramer. A conserved threonine residue, Thr 380, serves as the N-terminal nucleophile and is required for both maturation and enzymatic activity (1). HpGT has been shown to degrade extracellular glutathione and glutamine, providing a growth advantage to the bacterium within its microenvironment (2, 5, 6). Similarly, upregulation of human γ-glutamyltranspeptidase in cancer is thought to help supplement these rapidly dividing cells with essential amino acid precursors for glutathione and protein biosynthesis (7, 8). In mammalian systems, γ-glutamyltranspeptidase has been shown to be critical for the transport of cysteine for use in protein and glutathione biosynthesis (9, 10). The enzyme is required for normal glutathione metabolism, initiating extracellular glutathione degradation. Subsequent steps lead to the cellular uptake of the composite amino acids of glutathione: glutamate, cysteine, and glycine (11, 12).
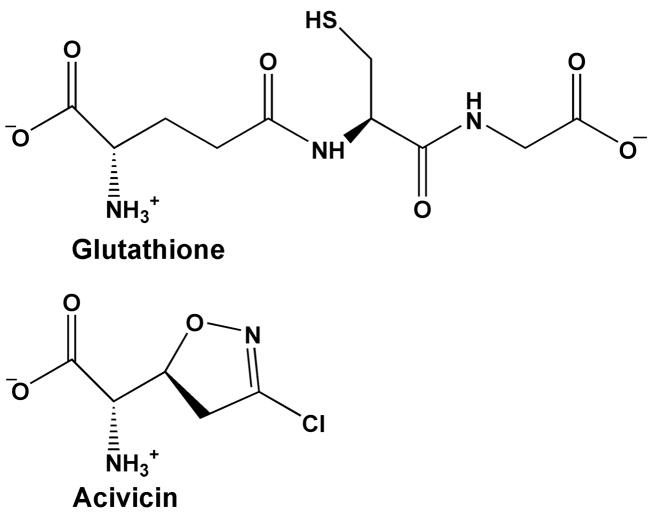
Acivicin is a commonly used mechanism-based inhibitor of γ-glutamyltranspeptidases (13, 14), although its precise mechanism of action has not been determined. Structurally, it resembles the γ-glutamyl group of known substrates of γ-glutamyltranspeptidases (Figure 1), and is predicted to react with the catalytic nucleophile of the enzyme. Previous studies by Meister and co-workers suggested that the site of reactivity is a hydroxyl group near the active site of the enzyme (15–17). Specifically, Thr 523 of the rat enzyme was covalently modified by the inhibitor (17), whereas Ser 406 of the human homologue was labeled (15). However, mutational analysis of the human enzyme, which shares nearly 80% sequence identity with rat γGT, indicated that neither Thr 524 (equivalent to Thr 523 of the rat enzyme) nor Ser 406 was required for enzymatic function (15). Furthermore, recent structural studies indicate that both residues are relatively far removed from the active site of the enzyme (18, 19).
Figure 1.
Comparison of glutathione and acivicin structures.
To reconcile these inconsistencies, a comprehensive examination of the inhibition of HpGT by acivicin was conducted. The kinetics of acivicin inhibition and the structure of the inhibited form of HpGT were determined. The data indicate that acivicin is accommodated within the γ-glutamyl binding pocket of the enzyme, with the catalytic nucleophile, Thr 380, as the site of covalent modification. The structure of the acivicin-modified HpGT also reveals residues within the C-terminal region of the protein that are critical for autoprocessing and/or catalysis.
MATERIALS AND METHODS
Expression and purification of wild-type and mutant HpGT
The preparation of HpGT has been described previously (1, 2). Briefly, recombinant HpGT was expressed in E. coli and purified from the soluble lysate by affinity chromatography using a nickel-chelating column (Novagen). Point mutations were introduced at residues R175, R475, R502, R513, E515, and F567 of the standard expression construct using the QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer’s protocol. For mutant proteins incapable of maturation, the equivalent substitutions were generated in a bicistronic HpGT expression construct (HpGT-Duet) designed to generate mature enzyme independently of self-processing. All constructs were verified by sequencing (Genomics Core at the University of Nebraska-Lincoln).
Kinetic characterization of wild-type and mutant HpGT
Apparent kinetic constants for the autoprocessing of the HpGT precursor and the hydrolysis of the substrate analogue, L-glutamic acid γ-(4-nitroanilide) (GNA; Sigma) by mature HpGT were determined as described previously (1, 20). To examine the efficacy of acivicin ((αS, 5S)-α-amino-3-chloro-4,5-dihydro-5-isoxazoleacetic acid; Sigma) as a mechanism-based inhibitor, wild-type HpGT (0.1 mg/ml) was incubated in 20 mM Tris buffer, pH 7.4, at 4 °C with various concentrations of the inhibitor (5–30 μM). At the indicated time, an aliquot was removed and enzymatic activity was measured by monitoring the hydrolysis of GNA using a Cary 50 spectrophotometer (0.1 M Tris buffer, pH 8.0; 1 mM GNA; 25 °C). To investigate the effect of a protein ligand on the rate of inactivation, HpGT was preincubated with 1 mM glutamate prior to the addition of 10 μM acivicin. Data were plotted as a function of time and fit to a single-order decay with Prism (Graph Pad Software). Representative data from three or more determinations are presented with experimental errors determined from individual fits of the kinetic data.
Generation of the refined acivicin-modified HpGT structure
Wild-type HpGT crystals were grown as described previously (19). Prior to data collection, crystals were transferred to artificial mother liquor containing 30% polyethylene glycol 2000 monomethyl ether and 1 mM acivicin, a mechanism-based inhibitor of γ-glutamyltranspeptidase. The crystals were incubated in this solution for approximately ten minutes and then flash frozen in liquid nitrogen. Diffraction data (λ=0.9 Å; 100 K) were collected on Beamline 14-BM-C of BioCARS at Argonne National Laboratory’s Advanced Photon Source and analyzed using the HKL2000 software package (21). Wild-type HpGT apoenzyme (PDB ID 1NQO) was used as the initial model (19), and iterative rounds of model building and refinement were performed using the programs COOT (22) and REFMAC5 (23), respectively. An Rfree test set (10%) was maintained to monitor refinement, and TLSMD (24) was used to identify optimal multi-group TLS models for use in REFMAC (25). The acivicin-modified threonine residue was built in the Monomer Sketcher Library module of the CCP4 software package (26). As discussed below, both the sp3 and sp2-hybridization of C3 of acivicin were initially considered but ultimately the sp2-hybridization was chosen for subsequent refinement. Water molecules obeying proper hydrogen-bonding constraints with electron densities greater than 1.0 σ on a 2Fo–Fc map and 4.0 σ on an Fo–Fc map were also included in the final structure. Model geometry was monitored using MOLPROBITY (27) and figures were produced using Chimera (28).
RESULTS AND DISCUSSION
Inactivation of HpGT by acivicin
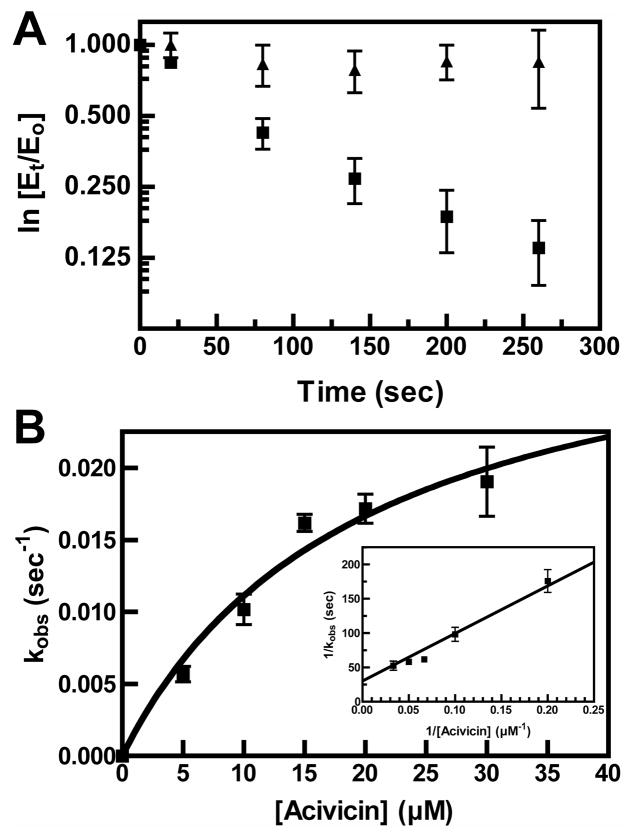
Incubation of HpGT with 10 μM acivicin at pH 7.4 and 4 °C resulted in a time-dependent inactivation of the enzyme (Figure 2A, squares), whereas the enzyme alone exhibited no loss of activity over the indicated time frame (data not shown). The inclusion of glutamate (1 mM; Figure 2A, triangles), a product of the hydrolysis reaction, resulted in an approximately 6-fold reduction in the inactivation rate (0.011 ± 0.001 sec−1 versus 0.002 ± 0.001 sec−1), suggesting that the acivicin binding site is coincident with the γ-glutamyl binding site. HpGT was incubated with various amounts of acivicin (5–30 μM) to determine the dependence of the rate of inactivation on reagent concentration (Figure 2B). The observed rate constants exhibited a hyperbolic dependence on the acivicin concentration, suggesting the formation of an enzyme-inhibitor complex prior to covalent modification. The observed rate constant (kobs) at a particular reagent concentration (R) can be described by
Figure 2. Inactivation of HpGT by acivicin.
(A) HpGT (0.1 mg/ml) was incubated with 10 μM acivicin at pH 7.4 and 4 °C in the absence (squares) and presence (triangles) of 1 mM glutamate. The kobs for the reactions were determined from the slope of ln(Et/Eo) versus time, where Eo and Et are the enzymatic activities at time 0 and time t respectively. (B) Dependence of the rate of HpGT inactivation on acivicin concentration. HpGT was incubated with various concentrations of acivicin and the rate of inactivation determined as in Panel A. (Inset) Double reciprocal plot used to calculate kmax and KI.
where kmax is equal to the maximal inactivation rate at saturating concentrations of the reagent and KI= (k−1 + kmax)/k1 and represents the reagent concentration that results in half of the maximal inactivation rate (29). The double-reciprocal plot (Figure 2B, inset) was used to calculate kmax = 0.033 ± 0.006 sec−1 and KI = 19.7 ± 7.2 μM.
Identification of Threonine 380 as the site of modification by acivicin
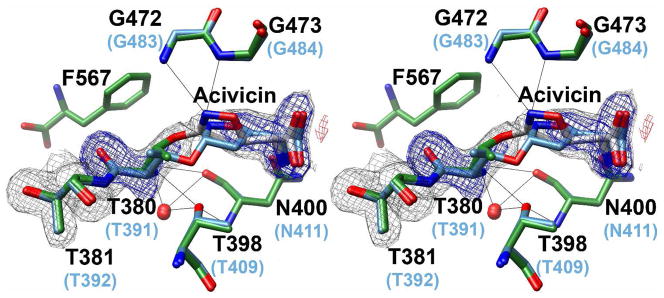
Structural studies were conducted to determine the mode of inhibition by acivicin. Wild-type HpGT crystals were transferred to artificial mother liquor containing 1mM acivicin, incubated for approximately 10 minutes, and flash frozen in liquid nitrogen. HpGT has been shown to be enzymatically active in the crystal lattice (20) and X-ray diffraction data were collected to a resolution of 1.7 Å (Table 1). A complete heterotetramer (2 large subunits, 2 small subunits) is contained within the asymmetric unit. Chains A and C correspond to the large subunits (residues 31 through 372 and 32 through 379, respectively), whereas chains B and D identify the small subunits (residues 380 through 567). The refined model has an overall Rfactor=17.9% (Rfree=20.8%) with >97% of all residues in the favored region of the Ramachandran plot (27). Prior to inclusion of Thr 380 and acivicin in the model, continuous electron density, corresponding to covalently attached inhibitor, was observed in the active site of the enzyme (Figure 3). A careful examination of the electron density map did not indicate the presence of a second site of modification. Electrospray mass spectrometry (University of Nebraska Mass Spectrometry Facilty) was used to confirm that only the small subunit of HpGT was modified by acivicin with an apparent 1:1 stoichiometry (data not shown).
Table 1.
Data collection and refinement statistics for the acivicin-modified H.pylori γ-glutamyltranspeptidase structure
| Data collection statistics | |
| Space group | P21 |
| Cell Dimensions (Å) | 54.7, 105.4, 91.9, β=91.7 |
| Resolution | 29.2–1.70 Å (1.76–1.70 Å) |
| Number of unique reflections | 110177 |
| Average Redundancy | 3.3 (3.3) |
| <I/σI> | 23.3 (1.6) |
| Completeness | 96.5% (95.1%) |
| Rmerge | 5.2% (64.1%) |
| Refinement statistics | |
| Number of atoms | 8687 |
| Protein atoms | 8128 |
| Solvent atoms | 737 |
| Ligand atoms | 20 |
| Rfactor | 17.9% (23.2%) |
| Rfree | 20.8% (27.9%) |
| Overall B factor | 22.6 Å2 |
| Protein B factor | 14.3 Å2 |
| Solvent B factor | 27.7 Å2 |
| Acivicin B factor | 29.8 Å2 |
| RMSD from ideal values | |
| Bond lengths | 0.013 Å |
| Bond angles | 1.45° |
| Estimated coordinate error | 0.073 Å |
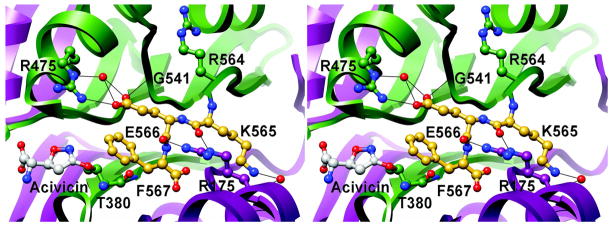
Figure 3. Electron density corresponding to the site of acivicin modification.
The refined model of acivicin-modified HpGT with pertinent active site residues is shown in stick representation and is superimposed with acivicin-modified E.coli γGT. Oxygen atoms are shown in red and nitrogen atoms in blue. Carbon atoms are colored green in HpGT, grey in acivicin, and blue in E.coli γGT. Residue numbers are noted in black (HpGT) and blue (E.coli γGT). Also shown are the calculated electron density maps after the initial round of refinement, prior to inclusion of Thr 380 and acivicin in the model. The relevant 2Fo-Fc electron density is contoured at 1.0 σ and illustrated in black. Positive and negative peaks in the difference map, contoured at 3.0 σ, are shown in blue and red respectively. Potential hydrogen bonds were identified with Chimera and are indicated as solid black lines.
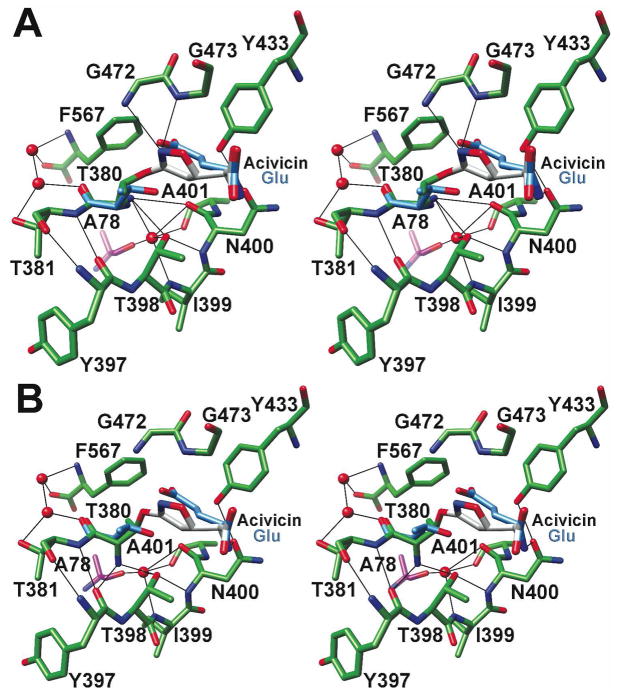
An examination of the final model indicates that HpGT binds acivicin and glutamate in similar orientations (Figure 4). The structurally equivalent carboxylate and amino groups of bound glutamate and acivicin adopt nearly identical conformations and form comparable hydrogen bonds with the enzyme. The active site of HpGT readily accommodates the dihydroisoxazole ring of acivicin, which superimposes reasonably well with the glutamate side chain. In the covalent complex, the chlorine of acivicin has been displaced by the nucleophilic attack of the Thr 380 hydroxyl group, the catalytic nucleophile in both the autoprocessing and hydrolysis reactions. Interestingly, two alternate conformations of the covalently modified Thr 380 are observed. In one heterodimer (chain ids A and B), Thr 380 adopts a conformation similar to that observed for the HpGT-glutamate product complex (Figure 4A). Its amino terminal group is within hydrogen bond distance of a structurally conserved water molecule, the backbone carbonyl oxygen of Asn 400, and the side chain oxygen of Thr 398. In the second heterodimer (chain ids C and D), the Thr 380 amino group has rotated approximately 90 degrees, disrupting its interaction with Asn 400 and forming a new hydrogen bond with the backbone carbonyl oxygen of Tyr 397 (Figure 4B). The functional significance of this alternate Thr 380 conformation is not evident from the current studies. However, this observation suggests that Thr 380 may adopt alternate conformations during maturation and catalysis. These structural changes may impact the pKa of the α-amino group of Thr 380 and allow it to serve as a general base during catalysis.
Figure 4. Alternate conformations of Thr 380 following acivicin modification.
Crystals of acivicin-modified HpGT contain a heterotetramer within the asymmetric unit, and provide two separate active site models. The active site regions of the refined structure are colored as in Figure 3. For reference, Thr 380 and bound glutamate from the previously determined HpGT-glutamate complex (blue) have been superimposed (20). The dihydroisoxazole ring of acivicin superimposes reasonably well with the glutamate side chain, and nearly identical placements of active site residues are observed with the exception of the slight displacement of Thr 380. (A) Within one active site, formed by the A and B subunits of the heterotetramer, Thr 380 adopts a conformation comparable to previous HpGT structures, with its α-amino group within hydrogen bond distance to the backbone carbonyl group of Asn 400, the hydroxyl group of Thr 398, and a tightly bound water molecule. (B) In the second active site comprised of subunits C and D, Thr 380 has rotated such that its α-amino group is within hydrogen bond distance to the backbone carbonyl group of Tyr 397 and the hydroxyl group of Thr 398. A nearly identical placement of acivicin is observed in both active sites.
Previous studies with mammalian γ-glutamyltranspeptidases suggested that acivicin reacts with a hydroxyl group within the 20 kDa subunit. For human γGT, the site of modification was identified as Ser 406 (15) whereas Thr 523 was modified in rat γGT (17). The equivalent residues in HpGT, Ser 405 and Lys 524, are more than 14 Å and 26 Å, respectively, from the reactive center of bound acivicin, and there is no evidence for their modification within the electron density maps. This observation suggests that the previously identified targets of acivicin modification likely represent secondary sites of reactivity. Although it is possible that there are structural differences between HpGT and mammalian γGT that may account for the observed differences, recent studies using the related mechanism-based inhibitor, 2-amino-4-[mono(4-cyanophenyl)phosphono]butanoic acid (30, 31), demonstrated the N-terminal threonine residue of human γGT, Thr 381, is clearly modified (32). Furthermore, replacement of either Thr 524 or Ser 406 with an alanine residue in human γGT had no impact on enzymatic activity, indicating that neither residue is directly involved in substrate binding and catalysis.
Recently, the structure of acivicin-modified E. coli γGT was reported and Thr 391, equivalent to Thr 380 in HpGT, was identified as the site of modification (33). The E. coli and the H. pylori structures were determined to similar resolutions (1.65 Å and 1.7 Å respectively) and are comparable with respect to the overall acivicin binding site (Figure 3). However, dissimilarities with respect to the dihydroisoxazole ring are evident. In the HpGT structure, the dihydroisoxazole ring is nearly perpendicular to that observed in the E. coli structure. This orientation allows the N2 nitrogen of acivicin to interact directly with the backbone amides of Gly 472 and Gly 473 in HpGT. In E. coli γGT, a bridging water molecule is required to interact with the backbone amide of Gly 483 (33). In addition, the hybridization at C3, the site of attachment to the Oγ of the catalytic threonine, is markedly different. In the E. coli structure, the Oγ of Thr 391 is significantly out of plane with respect to the dihydroisoxazole ring, suggesting that C3 is sp3-hybridized. In acivicin-modified HpGT, the Oγ of Thr 380 is co-planar with respect to the dihydroisoxazole ring, indicating that C3 is likely sp2-hybridized. These observed differences are significant with respect to the precise mechanism of modification.
Largely based on the unexpected sp3 geometry for C3 in E. coli γGT, the authors proposed an unusual inactivation mechanism that would require the oxygen-nitrogen bond of the dihydroisoxazole ring to break and reform, resulting in a net migration of a double bond from N2-C3 to C4-C5 (33). Such a mechanism would likely be facilitated by the presence of a general base near C4 and C5, however the identity of such a functional group is not readily apparent in the E. coli γGT structure. In contrast, the observed sp2 geometry for C3 in the HpGT structure is consistent with simple chloride displacement from acivicin by Thr 380 and maintenance of the dihydroisoxazole ring, analogous to the mechanism proposed for the acivicin mediated inactivation of carbamoyl phosphate synthetase (34). The nucleophilic attack of Thr 380 Oγ on C3 of acivicin would be facilitated by the backbone amides of Gly 472 and Gly 473, which would stabilize the developing negative charge on N2. Collapse of the tetrahedral intermediate would then lead to displacement of the chloride ion. Confidence in the assigned sp2 geometry for the HpGT structure is considerable, as attempts to model C3 of acivicin with an sp3 geometry resulted in several significant difference peaks around the Thr 380 Oγ - C3 bond (data not shown). Given that mature HpGT and E. coli γGT share >50% sequence identity and maintain nearly identical active site architectures, it is unlikely that acivicin modification would proceed via different mechanisms.
The C-terminus of HpGT contributes to formation of the active site
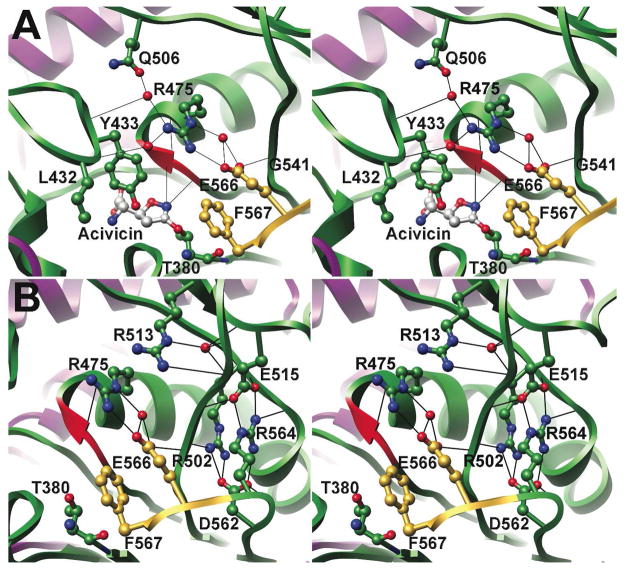
In previously described HpGT structures, the final three residues of the 20 kDa subunit were not visible within the electron density maps and were not included in the final models. Modification of HpGT with acivicin leads to ordering of Lys 565, Glu 566, and Phe 567 (Figure 5A), with Phe 567 positioned proximal to the substrate binding site and adjacent to Thr 380, the site of acivicin modification (Figure 5B). Removal of the benzyl ring (F567A) or the entire residue (F567Stop) results in an approximately 12-fold reduction in enzymatic activity (Table 2) without significantly impacting either Km or the rate of autoprocessing. These observations suggest that the bulky ring of Phe 567 may occlude solvent access to the active site and facilitate catalysis in a fashion similar to Tyr 433. Previously, we have shown that Tyr 433 is located on a mobile loop that clamps down over the substrate, forming a wall of the active site (20). Similarly, the Y433A mutant had approximately 25-fold less activity with no significant impact on Km or autoprocessing.
Figure 5. The C-terminus is ordered in the acivicin-modified HpGT structure.
A ribbon stereodiagram of HpGT is shown with the 40 kDa subunit colored in purple and the 20 kDa subunit in green. Relevant active site residues are shown in ball and stick representation. Carbon atoms are colored in green (20 kDa subunit), purple (40 kDa subunit), grey (acivicin) and yellow (the ordered C-terminus). Oxygen atoms are shown in red and nitrogen atoms in blue. In each of the previously determined HpGT structures, residues Lys 565, Glu 566, and Phe 567 were disordered. In the acivicin-modified HpGT structure, unambiguous electron density corresponding to the C-terminus of the protein was observed (not shown). Residues Arg 175 and Arg 475 appear to stabilize this region, positioning Phe 567 over the catalytic nucleophile, Thr 380.
Table 2.
Apparent kinetic constants for wild-type and mutant HpGT.
| Hydrolysis | Processing | ||
|---|---|---|---|
| Enzyme | Km GNA(μM) | Vmax (μmol/min/mg) | t1/2 (minutes) |
| HpGT | 22.1 ± 1.4 | 5.18 ± 0.08 | 49.3 ± 5.3 |
| R175L | 9.5 ± 0.9 | 0.77 ± 0.02 | 47.7 ± 4.0 |
| R475L | 2.9 ± 0.5 | 0.062 ± 0.003 | 45.6 ± 3.1 |
| R502L | N/A | N/A | No processing |
| R502L-Duet | 5.0 ± 0.3 | 0.068 ± 0.001 | N/A |
| R513L | 3.8 ± 0.3 | 0.28 ± 0.01 | 227 ± 16 |
| E515Q | N/A | N/A | No processing |
| E515Q-Duet | 2.7 ± 0.2 | 0.025 ± 0.001 | N/A |
| F567A | 15.9 ± 1.8 | 0.44 ± 0.01 | 27.1 ± 4.8 |
| F567Stop | 17.3 ± 2.4 | 0.43 ± 0.02 | 34.5 ± 3.3 |
As with the mobile loop containing Tyr 433, the C-terminus of the 20 kDa subunit is positioned by several hydrogen bonds (Figure 5). The side chain of Arg 175 forms hydrogen bonds with the backbone carbonyls of Lys 565 and Glu 566. The backbone amide nitrogen of Arg 175 also interacts via a water molecule with the ε amino group of Lys 565. Substitution with a leucine (R175L) results in a comparable ~7-fold reduction in catalytic activity with no adverse effects on substrate binding or enzyme maturation (Table 2). In addition, Glu 566 can form hydrogen bonds directly and through a bound water molecule with Arg 475 as well as the backbone amide of Gly 541. As will be discussed below, disruption of these interactions dramatically reduces enzymatic activity.
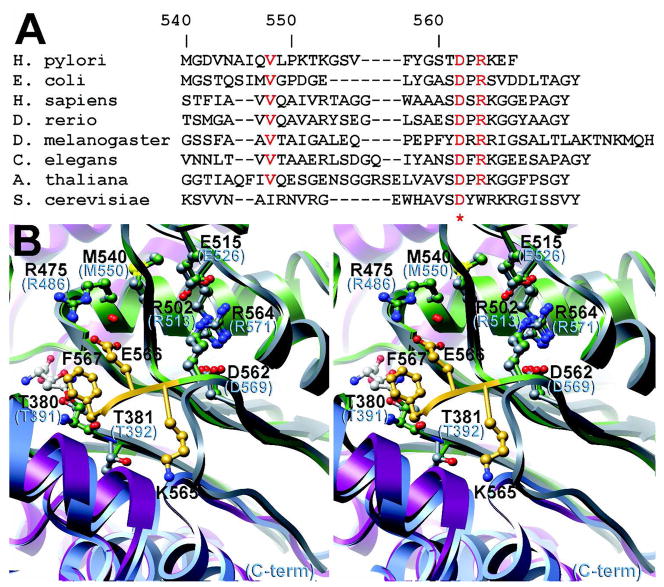
Although the C-terminus of HpGT is clearly contributing to the overall enzymatic efficiency of the enzyme, this region of the protein exhibits low sequence conservation. Representative γGT sequences corresponding to the C-terminal region are aligned in Figure 6. Asp 562 and Arg 564 are highly conserved across numerous species but beyond these two residues, considerable sequence variability exists. Although a general trend towards an aromatic residue at the ultimate C-terminus is observed, structural alignment of E. coli γGT and HpGT indicates that it is unlikely this residue is positioned similarly in all γGT (Figure 6A). Overall, the two structures are highly homologous and maintain similar positions for Asp 562 and Arg 564 (Asp569 and Arg 571 in E. coli γGT). However, the C-terminus of HpGT extends into the active site with Phe 567 in close proximity to the catalytic nucleophile, whereas the C-terminus of E. coli γGT loops out of the active site towards the surface of the protein (Figure 6A). Thus the C-terminal capping of the HpGT active site may be a unique structural feature of this enzyme that could be exploited in the design of HpGT selective inhibitors.
Figure 6. The truncated C-terminus of HpGT is unique among.
γ-glutamyltranspeptidases. (A) An alignment of representative γ-glutamyltranspeptidase sequences indicates that an aromatic residue is typically found at the C-terminal position. HpGT is considerably shorter than other γ-glutamyltranspeptidases, suggesting that the active site capping by Phe 567 observed in the acivicin-modified HpGT structure may be unique among γ-glutamyltranspeptidases. Sequences used in the alignment are H. pylori (O25743), E. coli (P18956), H. sapiens (P19440), D. rerio (Q7T2A1), D. melanogaster (Q9VWT3), C. elegans (Q9N5V4), A. thaliana (Q8VYW6), and S. cerevisiae (Q05902). (B) The superimposed structures of HpGT and E. coli γGT are shown in ribbon representation. HpGT is colored as in Figure 4 and the 40 kDa and 20 kDa subunits of E. coli γGT are colored in blue and grey respectively. Whereas the C-terminus of HpGT (yellow) extends over the active site, the C-terminus of E. coli γGT reverses and extends back to the surface of the protein, creating a more open active site pocket.
Arginine 475 is required for proper placement of the Tyr 433 loop and the C-terminal tail of HpGT
As indicated above, Glu 566 is positioned in part through interactions with Arg 475 and likely stabilizes the C-terminus of the 20 kDa subunit. Both residue positions are highly variable, although Arg 475 is located in a GXXGGXXI motif that is remarkably conserved within the γGT family (residues 469–476). Gly 472 and Gly 473 of this motif form the oxyanion hole that stabilizes the transition state of the reaction (Figures 3 and 4A). Arg 475, via bridging water molecules, also interacts with the backbone carbonyls of Leu 432 and Tyr 433 of the active site loop (Figure 7A). To examine the importance of this residue, a leucine mutant was generated (R475L; Table 2). This substitution resulted in a >80-fold reduction in enzymatic activity without impacting autoprocessing and illustrates the central role of this residue in catalysis. In addition, the R475L mutant exhibits an ~8-fold reduction in Km suggesting that the local substrate binding environment is also affected by disruption of the Arg 475 interactions. These observed kinetic differences indicate that Arg 475 is important for the proper positioning of Phe 567 and Tyr 433, which work in concert to exclude bulk solvent from the HpGT active site. In addition, these interactions likely lead to optimal formation of the oxyanion hole. Additional mutational analyses and pre-steady state kinetic studies to delineate the individual contributions of the Tyr 433 loop, the C-terminal cap, and Arg 475 to catalysis are ongoing.
Figure 7. An extensive hydrogen bond network in the C-terminal region of the 20 kDa subunit of HpGT is required for autoprocessing and catalytic activity.
In the stereodiagrams, a ribbon illustration of HpGT is presented with relevant residues shown in ball and stick representation. Atoms are colored as in Figure 4 and potential hydrogen bonds were identified with Chimera (indicated as solid black lines). Gly 472 and Gly 473, whose backbone amides form the proposed oxyanion hole, are highlighted as a red ribbon. (A) Arg 475 is critical for positioning of both the Tyr 433 loop and the Phe 567 C-terminal tail, as well as the backbone amides of Gly 472 and Gly 473. (B) The Glu 515/Arg 564 and the Asp 562/Arg 502 salt bridges stabilize the active site region and are critical for autoprocessing and enzymatic activity. Arg 513 also contributes to the overall integrity of the active site.
Disruption of conserved salt-bridges in the C-terminal region of the 20 kDa subunit significantly impairs autoprocessing and hydrolase activities
As noted above, Asp 562 and Arg 564 are conserved among γGT family members (Figure 6A). An examination of the HpGT structure indicates that the carboxylate of Asp 562 interacts with Arg 564 as well as another conserved arginine residue, Arg 502 (Figure 7B). Similarly, Arg 564 forms a salt bridge with a conserved glutamate residue, Glu 515. Given the extensive hydrogen bond network involving these residues and their remote location relative to the catalytic nucleophile (>12 Å), it is likely that these four highly conserved residues have a structural role. To test this possibility, the Glu 515/Arg 564 and the Arg 502/Asp 562 salt bridges were disrupted by site directed mutagenesis. The isosteric E515Q substitution abolished processing, indicating that strict conservation of the electrostatic character as well as the ability to form hydrogen bonds is required for autoprocessing (Table 2). Substitution of Arg 502 with a leucine (R502L) similarly abolished maturation of the protein (Table 2). Importantly, both mutants were expressed in the soluble fraction and were stable at 37 °C for > 14 days (data not shown), suggesting that gross disruption of protein structure does not occur as a result of these substitutions.
To test the importance of the Glu 515/Arg 564 and the Arg 502/Asp 562 salt bridges in catalysis, mature E515Q and R502L mutants were generated using a bicistronic HpGT expression system. Using this system, an N-terminal histidine tag is incorporated into the 40 kDa subunit and coexpressed with the untagged 20 kDa subunit. Both the E515Q-Duet and the R502L-Duet mutants were expressed in the soluble fraction as an intact heterotetramer, suggesting that the amino acid substitutions do not dramatically impact protein structure. Strikingly, both E515Q-Duet and the R502L-Duet mutants exhibited kinetic constants comparable to the mature R475L mutant. Vmax values for E515Q-Duet and R502L-Duet were reduced >200-fold and >80-fold relative to wild-type HpGT, respectively. Similar 6 to 8-fold reductions in Km were also observed. To further probe the structural significance of this region, a third mutant, R513L was constructed. Arg 513 is not a conserved residue but is located within the loop connecting Arg 502 and Glu 515, next to Arg 475 (Figure 7B). The R513L mutant exhibited an intermediate phenotype, with a ~5-fold reduction in processing rate and a ~20-fold reduction in Vmax. As both Glu 515 and Arg 502 are considerably removed from the active site, these results are consistent with a structural role for these two residues. Disruption of Glu 515/Arg 564 and the Arg 502/Asp 562 salt bridges likely impacts local protein structure and/or dynamics, negatively impacting autoprocessing and enzymatic activity.
In summary, the structure of acivicin-modified HpGT is consistent with the nucleophilic attack of Thr 380 Oγ at C3 of acivicin and the displacement of chloride. In contrast to the inactivation of E. coli γGT (33), the integrity of the dihydroisoxazole ring is likely maintained during the reaction, as C3 retains its sp2 hybridization. The unique C-terminal capping of the active site we observed within the acivicin-modified HpGT structure, coupled with our mutagenesis studies, demonstrates that Phe 567 contributes to the overall catalytic efficiency of the enzyme. Further examination of the contributions of residues within the C-terminal domain of the 20 kDa subunit indicate that local structural motifs are critical for optimal enzymatic activity and autoprocessing. These new insights into HpGT structure and function relationships will facilitate the design of selective γGT inhibitors.
Acknowledgments
This publication was made possible by NIH Grant Numbers P20 RR-17675 from the National Center for Research Resources and 1R01 GM077289 (JJB). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. Use of BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources, under grant number RR07707.
The authors would like to thank the BioCARS staff for assistance in x-ray data collection, Dr. Mark Wilson (University of Nebraska) for helpful discussions, and Dr. Melanie Simpson (University of Nebraska) for her thoughtful insights and review of the manuscript. The authors also acknowledge Gina Boanca for preparation of the HpGT crystals.
The abbreviations used are
- HpGT
Helicobacter pylori γ-glutamyltranspeptidase
- γGT
γ-glutamyltranspeptidase
- GNA
L-glutamic acid γ-(4-nitroanilide)
Footnotes
Coordinates and structure factors have been deposited in the PDB under accession code 3FNM.
References
- 1.Boanca G, Sand A, Barycki JJ. Uncoupling the enzymatic and autoprocessing activities of Helicobacter pylori gamma-glutamyltranspeptidase. J Biol Chem. 2006;281:19029–19037. doi: 10.1074/jbc.M603381200. [DOI] [PubMed] [Google Scholar]
- 2.Chevalier C, Thiberge JM, Ferrero RL, Labigne A. Essential role of Helicobacter pylori gamma-glutamyltranspeptidase for the colonization of the gastric mucosa of mice. Mol Microbiol. 1999;31:1359–1372. doi: 10.1046/j.1365-2958.1999.01271.x. [DOI] [PubMed] [Google Scholar]
- 3.Brannigan JA, Dodson G, Duggleby HJ, Moody PC, Smith JL, Tomchick DR, Murzin AG. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki H, Kumagai H. Autocatalytic processing of gamma-glutamyltranspeptidase. J Biol Chem. 2002;277:43536–43543. doi: 10.1074/jbc.M207680200. [DOI] [PubMed] [Google Scholar]
- 5.McGovern KJ, Blanchard TG, Gutierrez JA, Czinn SJ, Krakowka S, Youngman P. gamma-Glutamyltransferase is a Helicobacter pylori virulence factor but is not essential for colonization. Infect Immun. 2001;69:4168–4173. doi: 10.1128/IAI.69.6.4168-4173.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shibayama K, Wachino J, Arakawa Y, Saidijam M, Rutherford NG, Henderson PJ. Metabolism of glutamine and glutathione via gamma-glutamyltranspeptidase and glutamate transport in Helicobacter pylori: possible significance in the pathophysiology of the organism. Mol Microbiol. 2007;64:396–406. doi: 10.1111/j.1365-2958.2007.05661.x. [DOI] [PubMed] [Google Scholar]
- 7.Godwin AK, Meister A, O’Dwyer PJ, Huang CS, Hamilton TC, Anderson ME. High resistance to cisplatin in human ovarian cancer cell lines is associated with marked increase of glutathione synthesis. Proc Natl Acad Sci U S A. 1992;89:3070–3074. doi: 10.1073/pnas.89.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanigan MH, Gallagher BC, Townsend DM, Gabarra V. Gamma-glutamyl transpeptidase accelerates tumor growth and increases the resistance of tumors to cisplatin in vivo. Carcinogenesis. 1999;20:553–559. doi: 10.1093/carcin/20.4.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Will Y, Fischer KA, Horton RA, Kaetzel RS, Brown MK, Hedstrom O, Lieberman MW, Reed DJ. gamma-glutamyltranspeptidase-deficient knockout mice as a model to study the relationship between glutathione status, mitochondrial function, and cellular function. Hepatology. 2000;32:740–749. doi: 10.1053/jhep.2000.17913. [DOI] [PubMed] [Google Scholar]
- 10.Lieberman MW, Wiseman AL, Shi ZZ, Carter BZ, Barrios R, Ou CN, Chevez-Barrios P, Wang Y, Habib GM, Goodman JC, Huang SL, Lebovitz RM, Matzuk MM. Growth retardation and cysteine deficiency in gamma-glutamyl transpeptidase-deficient mice. Proc Natl Acad Sci U S A. 1996;93:7923–7926. doi: 10.1073/pnas.93.15.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orlowski M, Meister A. The gamma-glutamyl cycle: a possible transport system for amino acids. Proc Natl Acad Sci U S A. 1970;67:1248–1255. doi: 10.1073/pnas.67.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffith OW, Bridges RJ, Meister A. Evidence that the gamma-glutamyl cycle functions in vivo using intracellular glutathione: effects of amino acids and selective inhibition of enzymes. Proc Natl Acad Sci U S A. 1978;75:5405–5408. doi: 10.1073/pnas.75.11.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capraro MA, Hughey RP. Use of acivicin in the determination of rate constants for turnover of rat renal gamma-glutamyltranspeptidase. J Biol Chem. 1985;260:3408–3412. [PubMed] [Google Scholar]
- 14.Hill KE, Von Hoff DD, Burk RF. Effect of inhibition of gamma-glutamyltranspeptidase by AT-125 acivicin on glutathione and cysteine levels in rat brain and plasma. Invest New Drugs. 1985;3:31–34. doi: 10.1007/BF00176821. [DOI] [PubMed] [Google Scholar]
- 15.Smith TK, Ikeda Y, Fujii J, Taniguchi N, Meister A. Different sites of acivicin binding and inactivation of gamma-glutamyl transpeptidases. Proc Natl Acad Sci U S A. 1995;92:2360–2364. doi: 10.1073/pnas.92.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stole E, Smith TK, Manning JM, Meister A. Interaction of gamma-glutamyl transpeptidase with acivicin. J Biol Chem. 1994;269:21435–21439. [PubMed] [Google Scholar]
- 17.Stole E, Seddon AP, Wellner D, Meister A. Identification of a highly reactive threonine residue at the active site of gamma-glutamyl transpeptidase. Proc Natl Acad Sci U S A. 1990;87:1706–1709. doi: 10.1073/pnas.87.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada T, Suzuki H, Wada K, Kumagai H, Fukuyama K. Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate. Proc Natl Acad Sci U S A. 2006;103:6471–6476. doi: 10.1073/pnas.0511020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boanca G, Sand A, Okada T, Suzuki H, Kumagai H, Fukuyama K, Barycki JJ. Autoprocessing of Helicobacter pylori gamma-glutamyltranspeptidase leads to the formation of a threonine-threonine catalytic dyad. J Biol Chem. 2007;282:534–541. doi: 10.1074/jbc.M607694200. [DOI] [PubMed] [Google Scholar]
- 20.Morrow AL, Williams K, Sand A, Boanca G, Barycki JJ. Characterization of Helicobacter pylori gamma-glutamyltranspeptidase reveals the molecular basis for substrate specificity and a critical role for the tyrosine 433-containing loop in catalysis. Biochemistry. 2007;46:13407–13414. doi: 10.1021/bi701599e. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. Journal of Applied Crystallography. 2006;39:109–111. [Google Scholar]
- 25.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 26.Collaborative. The CCP4 suite: programs for protein crystallography. Acta Crystallographica Section D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 27.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 28.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Huang YC, Colman RF. Affinity labeling of the allosteric ADP activation site of NAD-dependent isocitrate dehydrogenase by 6-(4-bromo-2,3-dioxobutyl)thioadenosine 5′-diphosphate. J Biol Chem. 1984;259:12481–12488. [PubMed] [Google Scholar]
- 30.Han L, Hiratake J, Kamiyama A, Sakata K. Design, synthesis, and evaluation of gamma-phosphono diester analogues of glutamate as highly potent inhibitors and active site probes of gamma-glutamyl transpeptidase. Biochemistry. 2007;46:1432–1447. doi: 10.1021/bi061890j. [DOI] [PubMed] [Google Scholar]
- 31.Han L, Hiratake J, Tachi N, Suzuki H, Kumagai H, Sakata K. gamma-(Monophenyl)phosphono glutamate analogues as mechanism-based inhibitors of gamma-glutamyl transpeptidase. Bioorg Med Chem. 2006;14:6043–6054. doi: 10.1016/j.bmc.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Castonguay R, Halim D, Morin M, Furtos A, Lherbet C, Bonneil E, Thibault P, Keillor JW. Kinetic characterization and identification of the acylation and glycosylation sites of recombinant human gamma-glutamyltranspeptidase. Biochemistry. 2007;46:12253–12262. doi: 10.1021/bi700956c. [DOI] [PubMed] [Google Scholar]
- 33.Wada K, Hiratake J, Irie M, Okada T, Yamada C, Kumagai H, Suzuki H, Fukuyama K. Crystal structures of Escherichia coli gamma-glutamyltranspeptidase in complex with azaserine and acivicin: novel mechanistic implication for inhibition by glutamine antagonists. J Mol Biol. 2008;380:361–372. doi: 10.1016/j.jmb.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 34.Miles BW, Thoden JB, Holden HM, Raushel FM. Inactivation of the amidotransferase activity of carbamoyl phosphate synthetase by the antibiotic acivicin. J Biol Chem. 2002;277:4368–4373. doi: 10.1074/jbc.M108582200. [DOI] [PubMed] [Google Scholar]