Abstract
The formation of nitric oxide (NO) in biological systems has led to the discovery of a number of post-translational protein modifications that can affect biological conditions such as vasodilation. Studies both from our laboratory and others have shown that beside its effect on cGMP generation from soluble guanylate cylcase, NO can produce protein modifications through both S-nitrosylation of cysteine residues. Previously, we have identified the potential S-nitrosylation sites on endothelial NO synthase (eNOS). Thus, the goal of this study was to further increase our understanding of reactive nitrogen protein modifications of eNOS by identifing tyrosine residues within eNOS that are susceptible to nitration in vitro. To accomplish this, nitration was carried out using tetranitromethane followed by tryptic digest of the protein. The resulting tryptic peptides were analyzed by liquid chromatography/mass spectrometry (LC/MS) and the position of nitrated tyrosines in eNOS were identified. The eNOS sequence contains 30 tyrosine residues and our data indicate that multiple tyrosine residues are capable of being nitrated. We could identify 25 of the 30 residues in our tryptic digests and 19 of these were susceptible to nitration. Interstingly, our data identified four tyrosine residues that can be modified by nitration that are located in the region of eNOS responsible for the binding to heat shock protein 90 (Hsp90), which is responsible for ensuring efficient coupling of eNOS.
Introduction
The nitric oxide synthase (NOS) family of enzymes is responsible for NO production in vivo. NOS is a multigene family of at least three isoforms: inducible (iNOS), neuronal (nNOS), and endothelial (eNOS), of which eNOS is responsible for the production of endothelium-derived NO, an important mediator of pulmonary vascular tone and vascular reactivity.1,2 Endothelial NOS localizes in the Golgi complex of cultured bovine aortic endothelial cells, in human umbilical vein endothelial cells and in intact human blood vessels.3–6 Activation of eNOS occurs when endothelial cells are exposed to certain stimuli such as shear stress, resulting in increased NO production.2 The endothelial-targeted enhancement of eNOS activity appears particularly promising because it induces vasorelaxation, inhibits platelet aggregation and antagonizes microcirculatory disturbances which helps maintain vascular homeostasis.7 NO synthases have been characterized as cytochrome P450-like hemeproteins that require tetrahydrobiopterin (BH4), FMN and FAD as cofactors, and catalyze the NADPH-dependent oxidation of L-arginine to form NO and L-citrulline.8–10
The formation of reactive nitrogen species from •NO requires the presence of oxidants such as superoxide radicals (O2•−), hydrogen peroxide (H2O2) and transition metal centers, the concentration of which can be increased either by •NO itself or by the same mediators that up-regulate •NO production.11–14 These reactive nitrogen species (such as peroxynitrite ONOO−) are known to modify methionine, tryptophan, cysteine and tyrosine residues in proteins and peptides.12,14 One of the molecular footprints left by the reactions of reactive nitrogen species with biomolecules is the nitration itself (i.e., the substitution of a hydrogen atom for a nitro group, −NO2) of protein tyrosine residues to produce 3-nitrotyrosine.11 The formation of protein 3-nitrotyrosine was originally addressed in early protein chemistry studies with tetranitromethane (TNM) aimed at establishing the function of tyrosine residues in proteins.11,15 This now-established post-translational modification attracts considerable interest to biomedical research, because it can alter protein function, is associated with acute and chronic disease states and can be a predictor of disease risk.11
Our previous studies have shown that the presence of exogenous NO inhibits the activity of eNOS both in cultured cells,16 the purified eNOS protein17 and in lambs exposed to inhaled NO.16,18 We have also shown that the inhibitory effect of NO is mediated, at least in part, through the disruption of the eNOS dimer and that is associated with the release of zinc due to destruction of the zinc tetrathiolate cluster.19,20
Although there are a number of analytical techniques available for studying proteins, mass spectrometry (MS) is the premier tool in proteomics.21,22 Unique features of MS include good mass accuracy, excellent sensitivity and unparalleled specificity.23 Our group has successfully used mass spectrometry to investigate the S-nitrosylation both in short eNOS peptides24 and in the eNOS protein itself.25 Unlike the labile S-nitrosylation which requires several steps in MS-based analysis,25–27 tyrosine nitration is a stable, covalent modification. A single tyrosine nitration results in the mass increase of 45 Da in the tyrosine residue. In the presence of excess nitrating agent a double nitration (mass increase of 90 Da) is possible. Both singly and doubly nitrated tyrosine residues are stable under conventional MS/MS conditions (collision-induced dissociation and especially electron transfer dissociation). Thus, tyrosine-nitrated proteins can be studied by conventional proteomic techniques directly and there have been numerous mass spectrometry-based studies of this post-translational modification.28–32
The aim of this work was to identify the sites of tyrosine nitration in vitro using recombinant human eNOS protein.
Materials and methods
Materials
Trypsin was purchased from Promega (Madison, WI, USA). Ammonium bicarbonate, dithiothreitol, tetranitromethane, and iodoacetamide were all obtained from Sigma-Aldrich (St Louis, MO, USA). C18 Zip-Tips were obtained from Millipore (Bedford, MA, USA). BioSpin P-30 Tris columns were obtained from BioRad (Hercules, CA, USA). All solvents were of HPLC grade and purchased from Fisher Scientific (Fairborn, NJ, USA).
Expression and purification of human eNOS
The poly-His-pCWeNOS plasmid was transformed into the protease-deficient E. coli strain BL21 (DE3) pLysS (Novagen). Cells were grown in Luria broth with 1% glycerol containing 200 μg mL−1 ampicillin and 40 μg mL−1 chloramphenicol. Cultures were grown at 28°C until an OD600 of 0.8 was reached. Approximately one hour before that heme precursor δ-aminolevulinic acid (0.5 mM final concentration) was added. Cells were then induced by adding IPTG (0.8 mM final concentration), 0.5 mM ATP and 3 μM riboflavin were also added and the cells were then grown at 22°C for a further 48 h in the dark. Cells were then harvested by centrifugation (15 min at 4000 × g at 4°C). The cell pellet was resuspended in lysis buffer [40 mM N- (2-hydroxyethyl) piperazine-N- (3-Propane sulfonic acid) (EPPS), pH 7.6 containing 1 mg mL−1 lysozyme, 150 mM NaCl, 0.5 mM L-arginine, 4 μM H4B, 2 μM FAD, 10% glycerol and protease inhibitor cocktail (Sigma) were added according to the manufacturer’s recommendation. The bacterial suspension was incubated with mild shaking at 4°C for 30 min to ensure complete cell lysis. Cells were broken by sonication using three 25 s pulses followed by three cycles of freezing and thawing. Cell debris was removed by centrifugation at 30,000 × g for 30 min at 4°C. The supernatant was then applied to a Ni-NTA His-Bind Superflow (Novagen) column pre-equilibrated with Buffer A (40 mM EPPS, pH 7.6, containing 150 mM NaCl, 10% glycerol, and 0.5 mM L-Arginine. The column was washed with five bed volumes of buffer A followed by Buffer B (Buffer A with 25 mM imidazole). The bound protein was then eluted with Buffer C (Buffer A + 200 mM imidazole). The heme-containing fractions were pooled and concentrated using centriprep-100 YM-10 (Millipore). The concentrated protein was dialyzed against three changes of Buffer A containing 4 μM H4B and 1 mM DTT. The protein were further purified by using a 2′5′-ADP-sepharose column equilibrated with 40 mM tris-buffer pH 7.6, containing 1 mM L-arginine, 3 mM DTT, 4 μM H4B, 4 μM FAD, 10% glycerol and 150 mM NaCl (Buffer D) and washed with buffer D containing 400 mM NaCl to prevent non specific binding. eNOS was then eluted with Buffer E (Buffer D with 5 mM 2′AMP). The heme containing fractions were pooled, concentrated and dialyzed at 4°C against buffer D containing 1 mM DTT, 4 μM BH4, 4 μM FAD and 10% glycerol and stored at −80°C until used. The DTT, BH4 and FAD were removed by repeated buffer exchange using a centricon filter when required.
Nitration of eNOS
The eNOS samples were prepared in an ammonium bicarbonate buffer (pH 7.8) and nitrated using tetranitromethane (TNM). A 150 μL aliquot of eNOS protein (1 mg mL−1) was mixed with 15 μL of 50 mM TNM solution and incubated for 15 min at 37°C. Excess nitrating agent was removed by running the reaction mixture through a BioSpin column. Nitrated eNOS was then dried using Ar gas and reconstituted in the buffer of choice for the tryptic digest.
Tryptic digest
The sample was digested using a typical tryptic digest protocol described in the literature.22 The digested protein was dissolved in water (1 mg mL−1) and a 150 μL aliquot was used. First, 200 μL of 8 M urea in 0.4 M ammonium bicarbonate (pH 7.5–8.5) was added to the digestion buffer. Next, 50 μL of 45 mM dithiothreitol (DTT) was added and incubated at 50°C for 15 min to reduce the disulfide bonds. The reaction mixture was cooled to room temperature, then 50 μL of 100 mM iodoacetamide (IAA) was added and left in the dark for 15 min. The digestion buffer was then diluted to 2 M urea and 0.1 M ammonium bicarbonate by adding 350 μL of water, water:acetonitrile (80:20, v:v) or water:methanol (80:20, v:v). Finally, 20 μg of sequence-grade modified trypsin was added to 200 μL of 50 mM acetic acid. Then, 50 μL of that trypsin solution was added to the digestion buffer and incubated at 37°C overnight (times between 8 h to 24 h were used). The digestion was stopped by either freezing or acidifying the sample with TFA (100 μL of 3% TFA solution). Five eNOS digests were performed, varying the time and solvent composition.
LC/MS/MS analysis
Peptide samples were desalted and concentrated by Zip-Tip, and then analyzed by on-line reverse-phase LC-MS/MS or nano-LC-MS/MS on a ThermoElectron (San Jose, CA, USA) LCQ Advantage quadrupole ion trap mass spectrometer. For capillary LC, about 100 pmoles of the eNOS digest were injected onto a 15 cm × 0.3 mm C18 column (Phenomenex). For nano-LC, approximately 4 pmoles of the modified eNOS digest was injected onto a 10 cm × 75 μm, C18-packed nanospray tip (particle size 3 μm). The flow rate was 4 μL min−1 (200 nL min−1 for nanospray) with a gradient of 3% to 80% B for 40 min. Solvent A was 0.1% formic acid in water (v/v) and solvent B was 100% Acetonitrile/0.1% (v/v) formic acid. Peptide ions were fragmented by collision-induced dissociation.
This raw data was then searched against a database of human nitric-oxide synthase with no enzyme assumptions but with possible modifications of single and double nitration on tyrosine (+45 Da for single nitration and +90 Da for double nitration) as well as alkylation of cysteines by IAA (+57 Da) and oxidation of cysteine and methionine (+1 Da) using ThermoElectron TurboSequest software according to published procedures.21–23
Analysis of 3D structure of eNOS
A three-dimensional structure of the oxygenase domain of eNOS was obtained from the Swissprot protein database (http://www.swissprot.ch) and inspected in the open-source software package, Pymol version 0.97 (DeLano Scientific LLC; http://pymol.sourceforge.net).
Results
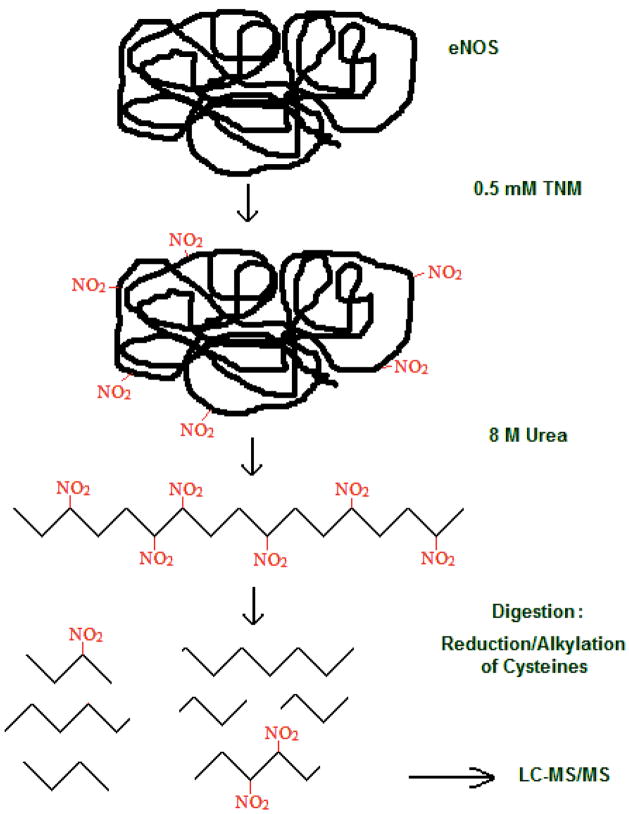
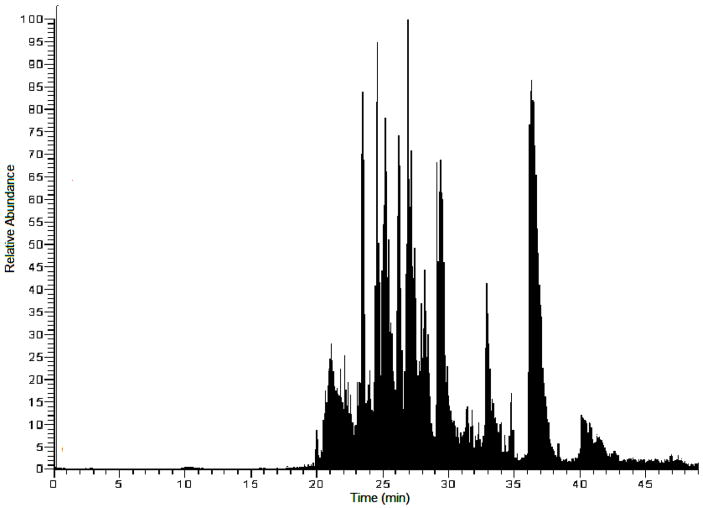
To determine the tyrosine nitration sites in eNOS, a standard protein nitration procedure followed by a tryptic digest and mass spectrometry analysis was employed, as shown in Figure 1. The eNOS peptides were desalted with C18 ZipTips and analyzed by LC-MS/MS or nanoLC-MS/MS. A typical analysis of the tryptic digest of nitrated eNOS gave a sequence coverage of 35–50%. A sample nano-LC total ion chromatogram is shown in Figure 2. A total of 33 eNOS peptides were identified from this chromatogram. By combining the results of five digests, a sequence coverage of almost 80% was achieved.
Figure 1.
General procedure for identification of sites of nitration in eNOS protein.
Figure 2.
Representative total ion chromatogram of an eNOS tryptic digest. A total of 33 peptides were identified from this chromatogram.
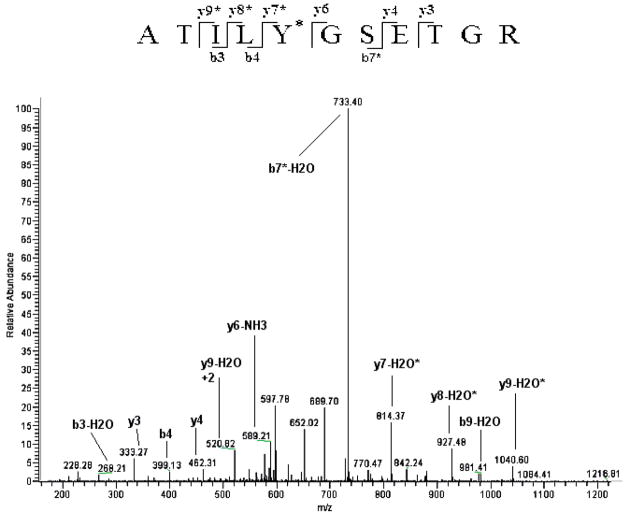
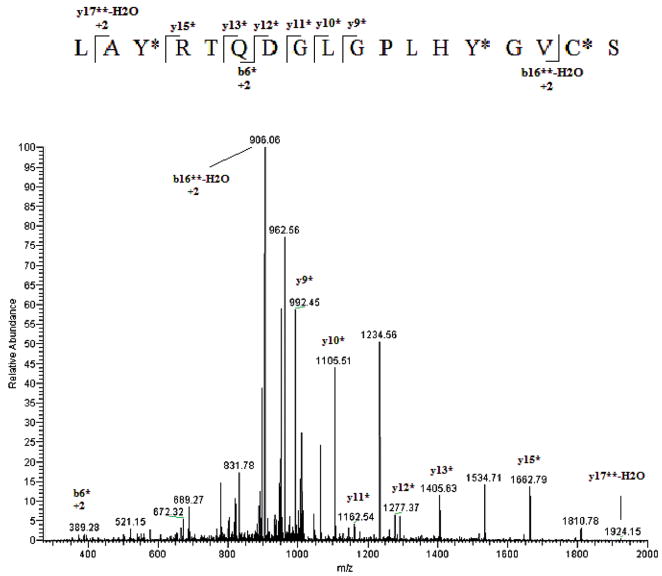
To determine which tyrosine residue had undergone nitration, MS/MS spectra of the eluting peptides were compared to the theoretical eNOS digest allowing for no nitration, single-or double nitration of each tyrosine residue. TNM has been shown to be selective in nitration of Tyr residues only.33–35 Figure 3 shows the fragmentation (MS/MS) of the ion with m/z 607.1 identified as the +2 ion of the eNOS peptide 520–530 (ATILY*GSETGR) showing nitration of Tyr 524. The tyrosine nitration was confirmed by the unmodified b4 and y6 ions and modified (addition of 45 Da) b5* and y7* ions. Another example of this approach is shown in Figure 4. There one can see the MS/MS spectrum of the ion with m/z 1028.7. This ion was matched to the eNOS peptide 960–977 (LAY*RTQDGLGPLHY*GVCS) in which both tyrosines, 962 and 973 were nitrated. The nitration of Tyr 962 is confirmed by the nitric oxide synthase (NOS) b6* ion while the nitration at Tyr 973 is indicated by the series of y9*–y15* ions.
Figure 3.
MS/MS spectrum of an eNOS peptide identified as ATILY*GSETGR by the database search. *Indicates the nitrated tyrosine residue identified as Tyr 524. Numbering includes the N-terminal methionine that is shown in parenthesis in Figure 5.
Figure 4.
MS/MS spectrum of an eNOS peptide identified as LAY*RTQDGLGPLHY*GVC§S by the database search. *Indicates the nitrated tyrosine residue. Notice that this peptide has two tyrosine residues that have been nitrated identified as Tyr 962 and 973. C§ denotes an oxidized cysteine.
A total of 30 tyrosine residues can be found in eNOS whose sequence is shown in Figure 5. Out of 30 tyrosine residues, 25 were located in the sequenced portion shown in grey in Figure 5. There are a total of six tyrosines that were sequenced, however, found not to be modified by nitration. They are Tyr 134, 135, 217, 597, 735 and 1057. (Numbering includes the N-terminal methionine that is shown in parenthesis in Figure 5.) The other 19 of the 25 sequenced tyrosines had the addition of one or two NO2 groups. Tyrosines 210, 357, 556, 609, 793, 973, 1087 and 1155 were found to be singly nitrated. The remaining 11 tyrosine residues, Tyr 81, 163, 331, 373, 410, 524, 534, 657, 939, 940, and 964, were found to be doubly nitrated in at least one experiment (some of them were found to be singly nitrated in one experiment and doubly nitrated in another).
Figure 5.
Total sequence of eNOS protein from SwissProt. The portions of sequence identified by LC/MS analysis are highlighted in grey. Tyrosine residues found nitrated are in bold and underlined (single nitration) or shaded in black (double nitration).
Discussion
Nitrating reagents such as tetranitromethane (TNM) or peroxynitrite (ONOO−) have the potential to induce tyrosine nitration in proteins (along with other modifications).12 Peroxynitrite is known to be an excellent nitrating reagent in vivo and also has been used for in vitro nitrations.36,37 The effect of ONOO− on proteins does, however, involve very complex chemistry that is very sensitive to the experimental conditions used (temperature, light, etc.). Because of these complications and the multiple reaction pathways possible,38 ONOO− itself was not chosen as nitrating reagent in our experiments. Rather, we utilized tetranitromethane (TNM). TNM shows very good nitration efficiency and is also chemically more stable than ONOO−. TNM is also a specific reagent for nitration of tyrosine residues,34,35 which eliminates unwanted side processes. Thus, for these reasons, TNM was used as the nitrating reagent in our experiments.
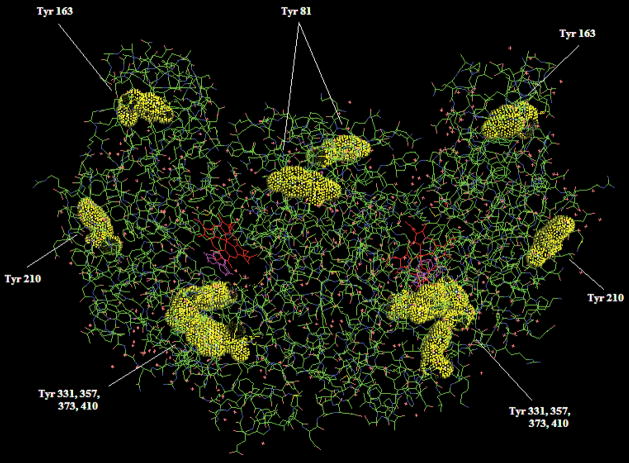
A typical sequence coverage for the analysis of nitrated eNOS digest was in the range of 30–50%. This is common for a protein of eNOS size (~260 kDa as a dimer). The variation between digests is not uncommon39 especially since organic modifiers were used to aid in digestion as proposed by Russell and co-workers40 and the time of digestion was varied. Five different digests produced a total sequence coverage of almost 80%. Some of the tyrosine residues were found to be doubly nitrated. These double nitration events are an indication of a higher time of exposure to the nitrating reagent which suggests that these tyrosines are either located on the protein surface or are otherwise more easily accessible to TNM. While the levels of tyrosine nitration with TNM in vitro are much higher than those expected in vivo, numerous studies suggest that the nitration of tyrosine residues does not affect the folding of the protein significantly,13,30,41,42 including one example of an in vitro nitration by TNM.43 To investigate the relationship of these tyrosine residues with the 3D structure of eNOS, we plotted the protein X-ray structure available in the Swissprot protein database using Pymol44 software. Figure 6 shows the X-ray structure of the eNOS dimer, oxygenase domain only (the only domain of the human eNOS available in the database). Many of the surface tyrosines (Tyr 81, Tyr 163, Tyr 210, Tyr 331, Tyr 357, Tyr 373 and Tyr 410) were identified as sites of nitration in our studies except for Tyr 217 which was detected in the unmodified form. In the reductase domain we identified nitration of tyrosine residues within regions which are known to be responsible for binding FMN, FAD and NADPH (Tyr 657, 793, 1087 and 1155).45
Figure 6.
3-D crystal structure of the oxygenase domain of the eNOS dimer. All tyrosine residues that have been identified as nitrated are shown in yellow with space filling and their positions labeled. Numbering includes the N-terminal methionine that is shown in parenthesis in Figure 5.
Of particular interest was the identification of the nitration of Tyr 331, 357 and 373 which are all located in the Hsp90-binding domain of eNOS. Hsp90, a 90-kDa, mostly cytosolic, heat shock protein is expressed at high levels (accounting for up to 1% to 2% of total cellular protein content), even in unstressed conditions and is a chaperone protein involved in the folding of a number of specific protein substrates including signal transducing molecules such as the Src-kinase family of non-receptor tyrosine kinases, Raf, other serine/threonine kinases, transcription factors such as steroid hormone receptors, p53 and eNOS, among others.44,46 The Hsp90-binding domain of eNOS encompasses residues 300–400.44 Within this region there are three tyrosine residues (331, 357 and 373). All three of these residues have been detected in our experiments and identified as being nitrated. This may potentially indicate that in vivo, eNOS nitration by RNS may affect the proper function of this domain with respect to its interaction with HSP90. This could potentially have a negative influence on the activity of the eNOS protein itself as we, and others, have demonstrated that the interaction of eNOS with Hsp90 is critical for efficient NO generation.7,44,46
In conclusion, we have identified multiple tyrosine residues of recombinant human eNOS protein that are capable of undergoing both single- and/or double nitration events in the presence of an excess of the nitrating agent, TNM. Out of 30 Tyr residues present in eNOS, we were able to detect 25 within the sequenced peptides. Nineteen of these were found to be capable of being nitrated. According to the crystal structure of the oxygenase domain of eNOS, most of these tyrosine residues are located on the surface of the protein. Among the residues found to undergo in vitro nitration are Tyr 331, 357 and 373, which are located in the Hsp90-binding domain of eNOS.44
In conclusion, this study complements our previous work on the identification of the S-nitrosylation sites in eNOS44,46 and lays a foundation for potential mutation experiments which may shed light on the role of specific tyrosine residue nitration events on eNOS protein function.
Acknowledgments
This research was supported in part with funding from the Medical College of Georgia under the National Institute of Health, National Heart, Lung, and Blood Institute Grant No. R01 HL070061 (to SMB) as well as a Network Grant from Fondation LeDucq (to SMB). The authors also thank Dr. Kristy Brown for the help with nano-LC/MS/MS experiments. Generous support of the National Science Foundation (Grant CHE-0130635) for the purchase of ThermoElectron LCQ Advantage mass spectrometer at NIU is greatly acknowledged.
References
- 1.Forstermann U, Kleinert H. Nitric oxide synthase: expression and expressional control of the three isoforms. Naunyn Schmiedebergs Arch Pharmacol. 1995;352:351–364. doi: 10.1007/BF00172772. [DOI] [PubMed] [Google Scholar]
- 2.Wedgwood S, McMullan DM, Bekker JM, Fineman JR, Black SM. Role for endothelin-1-induced superoxide and peroxynitrite production in rebound pulmonary hypertension associated with inhaled nitric oxide therapy. Circ Res. 2001;89(4):357–364. doi: 10.1161/hh1601.094983. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Garcia-Cardena G, Sessa WC. Palmitoylation of endothelial nitric oxide synthase is necessary for optimal stimulated release of nitric oxide: implications for caveolae localization. Biochem J. 1996;35(41):13277–13281. doi: 10.1021/bi961720e. [DOI] [PubMed] [Google Scholar]
- 4.Morin AM, Stanboli A. Nitric oxide synthase in cultured endothelial cells of cerebrovascular origin: cytochemistry. J Neurosci Res. 1993;36:272–279. doi: 10.1002/jnr.490360305. [DOI] [PubMed] [Google Scholar]
- 5.O’Brien HA. Nitric oxide synthase is localized predominantly in the Golgi apparatus and cytoplasmic vesicles of vascular endothelial cells. Histochem Cell Biol. 1995;103:221–225. doi: 10.1007/BF01454027. [DOI] [PubMed] [Google Scholar]
- 6.Sessa WC, Cardea-Garca G, Liu J, Keh A, Pollock JS, Bradley J, Thiru S, Braverman IM, Desai KM. The Golgi association of endothelial nitric oxide synthase is necassary for the efficient synthesis of nitric oxide. J Biol Chem. 1995;270:17641–17644. doi: 10.1074/jbc.270.30.17641. [DOI] [PubMed] [Google Scholar]
- 7.Kupatt C, Dessy C, Hinkel R, Raake P, Daneau G, Bouzin C, Boekstegers P, Feron O. Heat shock protein 90 transfection reduces ischemia-reperfusion-induced myocardial dysfunction via reciprocal endothelial no synthase serine 1177 phosphorylation and threonine 495 dephosphorylation. Arterioscl Throm Vasc Biol. 2004;24(8):1435–1441. doi: 10.1161/01. ATV.0000134300.87476.d1. [DOI] [PubMed] [Google Scholar]
- 8.Marsden PA, Heng HH, Scherer SW, Stewart RJ, Hall AV, Shi XM, Tsui LC, Schappert KT. Structure and chromosomal localization of the human constitutive endothelial nitric oxide synthase gene. J Biol Chem. 1993;268(23):17478–17488. [PubMed] [Google Scholar]
- 9.Stuehr DJ, Ikeda-Saito M. Spectral characterization of brain and macrophage nitric oxide synthases. J Biol Chem. 1992;267(29):20547–20550. [PubMed] [Google Scholar]
- 10.White KA, Marletta MA. Nitric oxide synthase is a cytochrome p-450 type hemoprotein. Biochem J. 1992;31(29):6627–6631. doi: 10.1021/bi00144a001. [DOI] [PubMed] [Google Scholar]
- 11.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci USA. 2004;101(12):4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn DM, Sadidi M, Liu X, Kreipke C, Geddes T, Borges C, Throck Watson J. Peroxynitrite-induced nitration of tyrosine hydroxylase. J Biol Chem. 2002;277(16):14336–14342. doi: 10.1074/jbc. M200290200. [DOI] [PubMed] [Google Scholar]
- 13.Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem Biophys Res Commun. 2003;305:776–783. doi: 10.1016/S0006-291X(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 14.Berlett BS, Friguet B, Yim MB, Chock PB, Stadtman ER. Peroxynitrite-mediated nitration of tyrosine residues in Escherichia coli glutamine synthetase mimics adenylylation: Relevance to signal transduction. Proc Natl Acad Sci USA. 1996;93:1776–1780. doi: 10.1073/pnas.93.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokolovsky M, Riordan JF, Vallee BL. Tetranitromethane. A reagent for the nitration of tyrosyl residues in proteins. Biochemistry. 1966;5(11):3582–3589. doi: 10.1021/bi00875a029. [DOI] [PubMed] [Google Scholar]
- 16.Wedgewood S, Steinhorn RH, Bunderson M, Wilham J, Lakshminrusimha S, Brennan LA, Black SM. Increased hydrogen peroxide downregulates soluble guanylate cyclase in lungs of lambs with peprsistent hypertension of the newborn. Am J Physiol Lung Cell Mol Physiol. 2005;289:L660–L666. doi: 10.1152/ajplung.00369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravi K, Brennan LA, Levic S, Ross PA, Black SM. S-nitrosylation of endothelial nitric oxide synthase is associated with monomerization and decreased enzyme activity. Proc Natl Acad Sci USA. 2004;101(8):2619–2624. doi: 10.1073/pnas.0300464101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mata-Greenwood E, Jenkins C, Farrow KN, Konduri G, Russell J, Lakshminrusimha S, Black SM, Steinhorn RH. eNOS function is developmentally regulated: uncoupling of eNOS occurs postnatally. Am J Physiol Lung Cell Mol Physiol. 2006;290:L232–L241. doi: 10.1152/ajplung.00393.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govers R, Rabelink TJ. Cellular regulation of endothelial nitric oxide synthase. Am J Physiol Renal Physiol. 2001;280(2):F193–F206. doi: 10.1152/ajprenal.2001.280.2.F193. [DOI] [PubMed] [Google Scholar]
- 20.Hemmens B, Goessler W, Schmidt K, Mayer B. Role of bound zinc in dimer stabilization but not enzyme activity of neuronal nitric-oxide synthase. J Biol Chem. 2000;275(46):35786–91. doi: 10.1074/jbc. M005976200. [DOI] [PubMed] [Google Scholar]
- 21.Budde WL. Analytical Mass Spectrometry: Strategies for Environmental and Related Applications. Oxford University Press; Washington, DC, USA: 2001. [Google Scholar]
- 22.Dass C. Principles and Practice of Biological Mass Spectrometry. John Wiley & Sons Inc; New York, USA: 2001. [Google Scholar]
- 23.Hoffmann ED, Stroobant V, editors. Mass Spectrometry: Principles and Applications. 2. John Wiley & Sons Ltd.; Chichester, UK: 2002. [Google Scholar]
- 24.Taldone FS, Tummala M, Goldstein EJ, Ryzhov V, Ravi K, Black SM. Studying the S-nitrosylation of model peptides and eNOS protein by mass spectrometry. Nitric Oxide. 2005;13(3):176–187. doi: 10.1016/j.niox.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Tummala M, Ryzhov V, Ravi K, Black SM. DNA Cell Biol. doi: 10.1089/dna.2007.0655. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3(2):193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 27.Jaffrey SR, Fang M, Snyder SH. Nitrosopeptide mapping: a novel methodology reveals s-nitrosylation of dexras1 on a single cysteine residue. Chem Biol. 2002;9(12):1329–35. doi: 10.1016/S1074-5521(02)00293-4. [DOI] [PubMed] [Google Scholar]
- 28.Amoresano A, Chiappetta G, Pucci P, D’Ischia M, Marino G. Bidimensional tandem mass spectrometry for selective identification of nitration sites in proteins. Anal Chem. 2007;79:2109–2117. doi: 10.1021/ac0620361. [DOI] [PubMed] [Google Scholar]
- 29.Borges C, Kuhn DM, Watson JT. Mass mapping sites of nitration in tyrosine hydroxylase: random vs. selective nitration of three tyrosine residues. Chem Res Toxicol. 2003;16:536–540. doi: 10.1021/tx0256681. [DOI] [PubMed] [Google Scholar]
- 30.Ischiropoulos H. Biological tyrosine nitration: A patho-physiological function of nitric oxide and reactive oxygen species. Arch Biochem Biophys. 1998;356:1–11. doi: 10.1006/abbi.1998.0755. [DOI] [PubMed] [Google Scholar]
- 31.Walcher W, Franze T, Weller MG, Pöschl U, Huber CG. Liquid- and gas-phase nitration of bovine serum albumin studied by LC-MS and LC-MS/MS using monolithic columns. J Proteom Res. 2003;2:534–542. doi: 10.1021/pr034034s. [DOI] [PubMed] [Google Scholar]
- 32.Willard BB, Ruse CI, Keightley JA, Bond M, Kinter M. Site-specific quantitation of protein nitration using liquid chromatography/tandem mass spectrometry. Anal Chem. 2003;75:2370–2376. doi: 10.1021/ac034033j. [DOI] [PubMed] [Google Scholar]
- 33.Sokolovsky M, Riordan JF, Vallee BL. Tetranitromethane. A reagent for the nitration of tyrosyl residues in proteins. Biochemistry. 1966;5:3582. doi: 10.1021/bi00875a029. [DOI] [PubMed] [Google Scholar]
- 34.Riordan JF, Sokolovsky M, Vallee BL. Tetranitromethane. A reagent for the nitration of tyrosine and tyrosyl residues of proteins. J Am Chem Soc. 1966;88(17):4104–4105. doi: 10.1021/ja00969a046. [DOI] [PubMed] [Google Scholar]
- 35.Rebrin I, Bregere C, Kamzalov S, Gallaher TK, Sohal RS. Nitration of tryptophan 372 in succinyl-coa: 3-detoacid coa transferase during aging in rat heart mitochondria. Biochemistry. 2007;46:10130–10144. doi: 10.1021/bi7001482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Batthyány C, Souza JM, Durán R, Cassina A, Cerveñansky C, Radi R. Time course and site(s) of cytochrome c tyrosine nitration by peroxynitrite. Biochemistry. 2005;44:8038–8046. doi: 10.1021/bi0474620. [DOI] [PubMed] [Google Scholar]
- 37.Dan Yang H-LW, Sun Z-N, Chung N-W, Shen J-G. A highly selective fluorescent probe for the detection and imaging of peroxynitrite in living cells. J Am Chem Soc. 2006;128:6004–6005. doi: 10.1021/ja0603756. [DOI] [PubMed] [Google Scholar]
- 38.Koppenol WH. Peroxynitrite uncloaked? Chem Res Toxicol. 1998;11:716–717. doi: 10.1021/tx9800607. [DOI] [PubMed] [Google Scholar]
- 39.Hagman C, Ramstrom M, Jansson M, James P, Hakansson P, Bergquist J. Reproducibility of tryptic digestion investigated by quantitative Fourier transform ion cyclotron resonance mass spectrometry. J Proteom Res. 2005;4:394–399. doi: 10.1021/pr049809r. [DOI] [PubMed] [Google Scholar]
- 40.Russell WK, Park ZY, Russell DH. Proteolysis in mixed organic-aqueous solvent systems: applications for peptide mass mapping using mass spectrometry. Anal Chem. 2001;73:2682–2685. doi: 10.1021/ac001332p. [DOI] [PubMed] [Google Scholar]
- 41.Turko IV, Murad F. Protein nitration in cardiovascular diseases. Pharmacol Rev. 2002;54:619–634. doi: 10.1124/pr.54.4.619. [DOI] [PubMed] [Google Scholar]
- 42.Kanski J, Hong SJ, Schoneich C. Proteomic analysis of protein nitration in aging skeletal muscle and identification of nitrotyrosine-containing sequences in vivo by nanoelectrospray ionization tandem mass spectrometry. J Biol Chem. 2005;280(25):24621–24626. doi: 10.1074/jbc.M501773200. [DOI] [PubMed] [Google Scholar]
- 43.Uversky VN, Yamin G, Munishkina LA, Karymov MA, Millett IS, Doniach S, Lyubchenko YL, Fink AL. Effects of nitration on the structure and aggregation of alpha-synuclein. Mol Brain Res. 2005;134:84–102. doi: 10.1016/j.molbrainres.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 44.Balligand JL. Heat shock protein 90 in endothelial nitric oxide synthase signaling: following the lead(er)? Circ Res. 2002;90(8):838–841. doi: 10.1161/01. RES.0000018173.10175.FF. [DOI] [PubMed] [Google Scholar]
- 45.Mardsen PA, Schappert KT, Chen HS, Flowers M, Sundell CL, Wilcox JN, Lamas S, Michel T. Molecular cloning and characterization of human endothelial nitric oxide synthase. FEBS Lett. 1992;307:287–293. doi: 10.1016/0014-5793(92)80697-F. [DOI] [PubMed] [Google Scholar]
- 46.Ou J, Ou Z, McCarver DG, Hines RN, Oldham KT, Ackerman AW, Pritchard KA., Jr Trichloroethylene decreases heat shock protein 90 interactions with endothelial nitric oxide synthase: implications for endothelial cell proliferation. Toxicol Sci. 2003;73:90–97. doi: 10.1093/toxsci/kfg062. [DOI] [PubMed] [Google Scholar]