Summary
Xenopus tropicalis is rapidly being adopted as a model organism for developmental biology research and has enormous potential for increasing our understanding of how embryonic development is controlled. In recent years there has been a well-organized initiative within the Xenopus community, funded largely through the support of the National Institutes of Health in the US, to develop X. tropicalis as a new genetic model system with the potential to impact diverse fields of research. Concerted efforts have been made both to adapt established methodologies for use in X. tropicalis and to develop new techniques. A key resource to come out of these efforts is the genome sequence, produced by the US Department of Energy’s Joint Genome Institute and made freely available to the community in draft form for the past three years. In this review, we focus on how advances in X. tropicalis genetics coupled with the sequencing of its genome are likely to form a foundation from which we can build a better understanding of the genetic control of vertebrate development and why, when we already have other vertebrate genetic models, we should want to develop genetic analysis in the frog.
Keywords: Xenopus tropicalis, gene regulation, phylogenetic analysis, gene networks, genome, synteny, ChIP, trangenesis, meganuclease, TILLING
FORWARD GENETICS IN THE FROG
The pseudo-tetraploid frog Xenopus laevis has long been used for classical and molecular embryological studies because of the ease with which large numbers of easily manipulated embryos can be obtained and, as a result, we have learned more about early vertebrate development from the frog than from any other vertebrate model. While Xenopus tropicalis shares the positive features of its close relative, it has the added advantages of being a truly diploid species with a relatively short generation time, opening the door to developmental genetics. Forward genetic screens, aimed at identifying developmental mutants on the basis of their phenotype, are already being conducted and are beginning to yield interesting mutations (Goda et al., 2006). These mutations, induced by in vitro treatment of sperm with N-ethyl N-nitrosourea, are in addition to several naturally occurring mutations identified in the genetic backgrounds of both lab-bred and wild-caught frog populations (Grammer et al., 2005; Noramly et al., 2005). This type of mutant screen is likely to further shift the focus of Xenopus biologists from early developmental processes to later development and organogenesis. This shift had already begun as a result of the ability to block the translation of specific mRNAs by injection of synthetic morpholino oligonucleotides into the early embryo. As will be discussed later, initial attempts have been made to screen genes for developmental functions using morpholinos (Kenwrick et al., 2004; Rana et al., 2006), but classical forward genetics remains the most reliable and straightforward approach. The translucent nature of the Xenopus tadpole allows developing organs such as the heart to be easily observed, making the frog well-suited to phenotypic screens for mutations affecting the morphogenesis and function of organs, and other aspects of development such as left-right asymmetry. Phenotypic screens conducted so far have taken advantage of another useful feature of Xenopus biology, namely the ability to generate gynogenetic diploids (Noramly et al., 2005). Irradiated spermatozoa, which are not able to contribute to viable diploid embryos, can be used to fertilize eggs in which either the second meiotic or first mitotic division can be suppressed by exposure to high pressure or low temperature (“cold-shock”). The resulting embryos often survive through embryogenesis, sometimes reaching adulthood. Mutations carried in heterozygosity by the female from which the eggs are obtained are brought into homozygosity in as many as fifty percent of the gynogenetic diploid progeny, depending upon the technique used. The phenotypic effects of these mutations can be analyzed in these animals, allowing screening in half of the time required by standard three-generation screening strategies. Goda et al. have used this approach to isolate mutants with phenotypes affecting diverse aspects of embryogenesis including axial patterning, gut development, and organs such as the eye, ear, and heart (Goda et al., 2006). The genetic lesion underlying one such mutant, muzak, in which the heart has no beat, has been preliminarily mapped by bulk segregant analysis of amplified length polymorphisms and by simple sequence length polymorphism (SSLP) analysis.
The mapping of muzak and other mutations through the use of SSLP markers is being aided greatly by the availability of a simple sequence repeat (SSR) map for X. tropicalis (http://tropmap.biology.uh.edu/map.html). By looking for SSRs in the draft genome sequence of X. tropicalis and screening these for polymorphism between Ivory Coast and Nigerian strains, Amy Sater, Dan Wells and co-workers at the University of Houston and Baylor College of Medicine have produced a set of more than a thousand genetic markers. The map, generated by determining the co-segregation frequencies of these markers in mapping crosses, assembles the markers into ten linkage groups. Although a direct correspondence has not yet been demonstrated, these linkage groups likely correspond to the ten chromosomes of X. tropicalis. This valuable preliminary map is just one example of the type of resources made possible by the sequencing of the X. tropicalis genome.
SEQUENCING THE X. TROPICALIS GENOME
The effort to sequence the frog genome began at the Joint Genome Institute of the US Department of Energy in 2002 with the aim of sequencing its roughly 1.7 gigabases and assembling contiguous sequences into scaffolds large enough to contain not only the coding regions of genes but also their associated regulatory regions. At several points during the project, draft assemblies of the sequence data were released to the community and this has supported the development of a variety of sequence-based tools and research projects. Currently, assembly 4.1 of the genome sequence contains ~1.5 megabases and corresponds to around 95% of the known full-length X. tropicalis cDNAs. Half of this sequence is carried by just 272 large scaffolds of 1.56 Mb or more, making it likely that even distant regulatory sequences will be contiguous with the genes they regulate. Annotation of the genome sequence has been supported by large numbers of expressed sequence tagged (EST) clones for both X. tropicalis and X. laevis. These number more than a million clones in X. tropicalis alone. These are proving valuable for the annotation of the genome, providing positive evidence that gene models predicted on the basis of sequence features correspond to transcribed loci. However, of the roughly 28,000 gene models present, less than a third are currently supported by ESTs, perhaps due in large part to under-representation of low abundance transcripts or genes expressed in tissues or stages not represented in the sequenced cDNA libraries.
IDENTIFYING VERTEBRATE GENE NETWORKS
One of the most important challenges facing developmental biologists is to understand how developmental genes are regulated. Turning genes on or off in particular tissues or at particular times in development is a complex process and is vital for differentiation of the numerous cell types that make up the animal. Studies of the downstream targets of particular transcription factors, and of the upstream regulation of genes are slowly building a picture of the networks of regulatory interactions that constitute the transcriptional programs underlying cellular differentiation. A number of studies have identified downstream target genes for a handful of transcription factors, but in the past this has often been difficult. Xenopus is an invaluable system for these studies, because of the ability to look at the immediate-early transcriptional responses of explanted tissue when the function of a recombinant, hormone-activated form of a transcription factor is triggered in the presence of cycloheximide, an inhibitor of translation (Kolm and Sive, 1995; Tada et al., 1997). Increasingly, this approach is being coupled with expression microarray analysis to look at transcriptional changes on a genome-wide scale, thereby avoiding the guesswork associated with assaying changes in the expression of candidate targets (Taverner et al., 2005). Chromatin immunoprecipitation (ChIP), in which a protein is precipitated together with its cross-linked native DNA targets, is an alternative means of identifying the targets of a transcription factor, and benefits in Xenopus from the ease with which large numbers of embryos can be generated (Stewart et al., 2006). ChIP is also starting to be used in conjunction with promoter microarrays (‘ChIP-on-chip’), an approach which has been successfully used for genome-wide analysis in other systems (Denissov et al., 2007; Wardle et al., 2006). Genomic DNA sequence data forms the basis of this type of array, in which sets of oligonucleotides are designed to correspond to short regions of sequence distributed periodically across the upstream and downstream regions of genes. Both expression array studies and ChIP-on-chip will inevitably accelerate the identification of the downstream targets of transcription factors and also have the potential to identify unanticipated regulatory pathways.
Sequencing the X. tropicalis genome presents the possibility of searching for regulatory sequences directly, using bioinformatics approaches. The DNA binding specificities of numerous transcription factors have been experimentally determined by binding site selection studies etc. and provide a potential starting point from which to search for downstream target genes of these trans-acting regulators. Databases such as TRANSFAC and JASPAR have been developed to collect transcription factor binding sequence (TFBS) data, usually in the form of position weight matrices representing the statistical likelihood of each nucleotide being present at each position in the binding sequence (Matys et al., 2006; Vlieghe et al., 2006). The information contained in these databases can be used by other programs designed to predict potential binding sites on a genome-wide scale (e.g. MAPPER, ABS, TRED; see web resource list given in the appendix). However, the short length of binding sequences (usually only 5–12bp), coupled with the tolerance of transcription factors for sequence variation, means that such programs often predict vast numbers of binding sites, of the order of 106 or more per genome, that significantly match the experimentally-determined TFBS. These predictions may be a thousand-fold higher than the actual number of sites with biological functions (Wasserman and Sandelin, 2004). Furthermore, the accuracy of the predictions is reliant upon the quality of the underlying experimental data for each transcription factor. Recent work looking for regulatory sequences in the human and Drosophila melanogaster genomes has focused on the clustering of predicted TFBSs as a means of finding cis-regulatory modules (CRMs) with biological functions (Berman et al., 2002; Blanchette et al., 2006). This is based upon the experimental observation that factors affecting the rate of transcription at a particular locus do not operate in isolation. Often, interactions between different DNA-binding transcription factors bound to neighboring sites nucleate the formation of transcriptional complexes on promoters and, at least in some cases, the rate of transcription appears to be determined by a balance of activating and repressing (or “quenching”) factors (Janssens et al., 2006). Clustered sites can be identified within genomic sequence using the recently developed Enhancer Element Locator (EEL) and Ahab/Stubb/Argos programs (Hallikas et al., 2006; Palin and Ukkonen, 2006; Rajewsky et al., 2002; Schroeder et al., 2004; Sinha et al., 2006). Although attempts at identifying functional regulatory regions by looking for clustered transcription factor binding sites can be successful for some genes, it is unclear how prevalent this type of CRM clustering is (Halfon, 2006). Genome-wide analysis of such clusters suggests that they are more commonly associated with genes encoding developmental regulators than with other gene classes (Blanchette et al., 2006). Furthermore, studies in several model systems have demonstrated that transcription can be influenced by cis-acting enhancer elements situated long distances from the promoter of the gene they regulate. For example, a key mammalian enhancer regulating the expression of sonic hedgehog has been identified ~1 megabase from the promoter of the gene (Lettice et al., 2002). Similarly, transcription may be regulated by multiple sites scattered over relatively large regions, as found for the endoderm-specific Endo16 gene of the sea urchin S. purpuratus, in which expression is controlled by a 2.3 kilobase cis-regulatory element with more than thirty functional binding sites (Yuh et al., 1994, 1998). Such observations suggest that clustering of transcription factor binding sites within short regions of the genome is not always a necessary requirement for function and highlight the potential difficulty of defining distinct functional clusters.
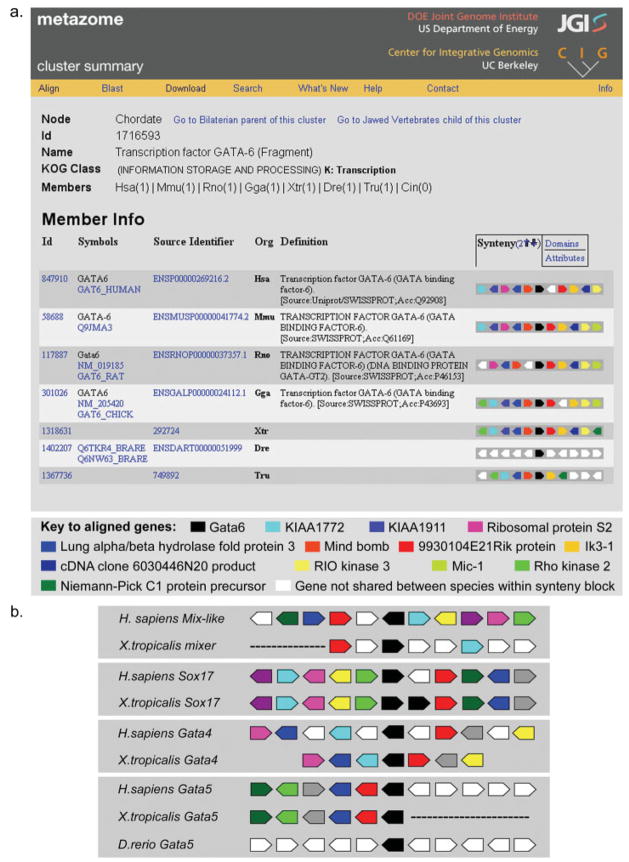
Through comparative genomics, the sequencing of the X. tropicalis genome may help overcome some of the problems described previously. To try to enrich for functional regulatory sequences, methods that scan genomes for transcription factor binding sequences have been combined with those that compare genome sequences from different species to find regions where sequence is evolutionarily conserved, perhaps as a result of functional constraints (“purifying selection”). These programs use a variety of alignment algorithms, coupled with programs that calculate and provide visual representations of sequence conservation. An example of this combined approach is rVISTA, which identifies binding sites in conserved regions of two locally aligned genomic sequences (Loots and Ovcharenko, 2004). Comparative genomics approaches of this kind are most successful when the two species in question are sufficiently distant from one another in evolutionary terms to allow sequences without function to diverge from one another. Humans and mice diverged from their last common ancestor ~75 million years ago and share between 66.7% and 75.9% identity in their non-coding regions (Waterston et al., 2002). This degree of conservation is close to the conventional criterion of >70% identity used to identify candidate sequences that may be conserved for functional reasons, and therefore significantly decreases the resolution with which regulatory elements can be detected by human-mouse genome comparisons. The greater evolutionary distance between the human and X. tropicalis genomes (360 million years) is likely to have resulted in a significantly greater degree of sequence divergence. While the same is true of the zebrafish (Danio rerio), the gene order on its chromosomes differs considerably from that of humans. This difference complicates the identification of orthologous sequences. The shared synteny and gene order between organisms is readily viewed using the powerful online tool Metazome (www.metazome.net). This provides a user-friendly visual representation of multi-species genome comparisons of regions around any gene of interest, together with links to the underlying genome sequence data and tools for generating and analyzing alignments. An example of the output of Metazome is shown in Figure 1a, in which synteny and gene order relationships are shown for ten genes flanking the Gata6 locus. Together with Mix-like/mixer, Sox17, Gata4, and Gata5, Gata6 functions downstream of nodal-related signaling in the developing endoderm of the early embryo as part of a nonlinear transcriptional network (Sinner et al., 2006). Inter-species alignments of the regions containing these genes show similar patterns of conserved synteny and gene order when human genes are compared with their X. tropicalis orthologues (Fig. 1a,b). However, where comparisons can be made with the corresponding regions of the zebrafish genome (Gata6, Fig. 1a; Gata5, Fig. 1b), synteny and gene order within the aligned regions are not found to be conserved. Inter-genome comparisons of other loci using Metazome support the notion that gene order in X. tropicalis is often similar to that of humans, and analysis of this similarity is ongoing. If this is found to be broadly true on a genome-wide scale it will make the alignment of genomes, identification of orthologous genes and comparison of corresponding regulatory elements simpler between humans and Xenopus than between humans and zebrafish.
FIG. 1.
Comparative genomic alignment of endoderm specification factors (a) Synteny and gene order relationships amongst genes flanking the chordate Gata6 cluster, as seen in the online analysis tool Metazome. Note. This screenshot was edited to focus on the aligned genes, omitting the Functional Analysis section relating to Gata6. Orthologous genes in aligned genomes are color-coded as described. Abbreviations: Hsa, Homo sapiens; Mmu, Mus musculus; Rno, Rattus norvegicus; Gga, Gallus gallus; Xtr, Xenopus tropicalis; Dre, Danio rerio; Tru, Takifugu rubripes. (b) Synteny and gene order relationships amongst genes flanking the Mix-like/mixer, Sox17/Sox17beta, Gata4, and Gata5 genes. Corresponding regions of the human genome and X. tropicalis genomic scaffolds are shown in each case. For the Gata5 cluster, the corresponding region of the D. rerio genome is also aligned, as for Gata6 in (a). Alignments were based on Metazome output and on annotation of the available X. tropicalis genomic scaffolds (assembly 4.1). Orthologous genes are color-coded, with clustered genes shown in black. Dotted lines indicate regions not included in the currently available genomic scaffolds.
Although synteny and gene order are often poorly conserved between the genomes of mammals and teleost fish, sequence comparisons have been made. In particular, attention has been focused on comparing mammalian genomes with that of the pufferfish Takifugu (fugu) rubripes. The primary reason for such comparisons is the extensive loss of large extraneous regions of non-coding sequence from the fugu genome, which has eliminated much of its “junk” DNA while leaving important regulatory sequences intact. However, these comparisons also benefit from the large evolutionary distance and associated sequence divergence between the species. The divergence that has occurred over more than 400 million years between mammals and pufferfish allows larger shared genomic regions to be narrowed down to smaller stretches of sequence (<100 bp) with regulatory functions (Baroukh et al., 2005). This approach has been used successfully to identify regulatory elements associated with several important developmental genes, including Hoxb4, Pax6, and sonic hedgehog (Aparicio et al., 1995; Woolfe et al., 2005), along with genes that have additional non-developmental functions such as the regulation of genes involved in lipid metabolism (COUP-TFII; [Baroukh et al., 2005]). These studies support the suggestion that similar comparisons between mammalian and Xenopus genomes might provide greater resolution in the detection of functionally constrained sequence elements than is achieved through human-rodent comparisons.
VALIDATING GENE NETWORKS IN VIVO
The sequencing of the X. tropicalis genome and the application of available bioinformatics tools will inevitably lead to the identification of conserved non-coding sequences. Transgenesis has been used in both the mouse and the zebrafish to test the function of conserved elements from vertebrate genomes. However, there are significant technical disadvantages to conducting transgenesis in these model organisms. Transgenesis in the mouse is inefficient, costly, and initially produces mosaic animals in which only a subset of cells carry the transgene. Although it is faster and cheaper in zebrafish, transgenesis by DNA microinjection, I-SceI meganuclease, or transposon-based methods similarly results in mosaic animals in which it can be difficult to interpret the activity of a regulatory element. Xenopus is an alternative model for these assays. A technique for the production of transgenic frogs by restriction enzyme-mediated integration (REMI) was developed by Kristen Kroll and Enrique Amaya and has since become a well-established technique within the Xenopus research community (Kroll and Amaya, 1996). The technique involves incubating linearized transgene DNA with sperm nuclei in the presence of a restriction enzyme followed by transplantation of the nuclei into unfertilized eggs by microinjection, resulting in fertilization. Unlike transgenesis in the mouse, REMI in Xenopus can generate hundreds of transgenic embryos in a single experiment and is effective in both X. laevis and X. tropicalis (Hirsch et al., 2002; Offield et al., 2000). More recently, microinjection of DNA linearized with the yeast meganuclease I-SceI (also known as omega meganuclease) has been found to be a highly efficient method of transgenesis in both X. tropicalis and X. laevis (Ogino et al., 2006; Pan et al., 2006). A Tol2 transposon-based method has also been used with success (Hamlet et al., 2006), as has the integrase ϕC31 (Allen and Weeks, 2005). In Xenopus, both REMI and I-SceI meganuclease transgenesis commonly result in the integration of the transgene before the first nuclear division, resulting in non-mosaic transgenic animals that transmit the transgene through the germline. In X. tropicalis, REMI generates non-mosaic embryos from ~2%–5% of those injected (Hirsch et al., 2002), while 10% of injected embryos are non-mosaic using the I-SceI meganuclease method (Ogino et al., 2006).
The ease and efficiency with which non-mosaic transgenic Xenopus can be generated by these techniques not only means that regulatory elements identified by bioinformatic analysis can be tested for function in vivo. It also makes unbiased, non-sequence based approaches to promoter and enhancer analysis feasible. Non-coding sequences flanking genes of interest can be fragmented and tested in large numbers to determine their ability to direct the expression of reporter genes, and the resulting expression patterns can be unequivocally interpreted in the absence of the mosaicism inherent in other model systems. This type of “promoter bashing” has been done with great success in invertebrate models such as the sea urchin (Yuh et al., 1994), but Xenopus represents the only equivalent vertebrate model. Importantly, the testing of conserved human regulatory elements in zebrafish (Fisher et al., 2006; Shin et al., 2005), and fugu elements in transgenic mice (Aparicio et al., 1995), has shown that the conservation of these elements at the sequence level is truly associated with conserved biological function, highlighting the relevance of the use of distantly related model organisms to gain insight into gene regulation in higher organisms, including humans.
FROM GENE SEQUENCE TO GENE FUNCTION
Although morpholinos have proved to be invaluable tools for blocking gene function in Xenopus, there is a need to develop truly genetic techniques for comprehensively analyzing the functions of genes. This is a challenge facing geneticists studying any organism. The long and successful history of forward genetic screening in Drosophila and the mouse, together with the study of the genetic basis of inherited human disease, has provided geneticists with informative mutations in many important genes and forms the basis of our understanding of the genetic control of development, normal biology and disease. However, this has relied on the chance mutation of genes involved in some readily observed feature, followed by an often lengthy process of positional cloning to characterize the mutated gene. In the age of genome sequencing, there is a need to work in the opposite direction—from gene to phenotype. Morpholinos that inhibit either the translation or splicing of specific mRNAs go some way towards this, because they can be designed to target any gene of interest and their effectiveness can be determined using antibodies against the targeted protein or, in the case of splice morpholinos, RT-PCR (Draper et al., 2001; Heasman et al., 2000; Sivak et al., 2005). Rana et al. (2006) have recently investigated the utility of morpholinos as tools for large-scale reverse screening in X. tropicalis, testing morpholinos designed to target mRNAs corresponding to 202 genes. For ~65% of the targeted genes, abnormal developmental phenotypes were observed and could be broadly categorized into synphenotype groups. The specificity of these effects was demonstrated in many cases by the use of a second morpholino targeting a distinct region of the mRNA, which typically resulted in a similar phenotype. The success of this approach suggests that in the near future it may be a valuable large-scale screening tool for identifying novel genes regulating development.
With DNA sequence as a starting point, morpholino “knockdown” experiments can provide a broad indication of the biological requirement for certain genes in a similar way to gene knockout studies in the mouse. The problem with these techniques is that they are relatively crude when compared with the analysis of more subtle point mutations. They tell us very little about the relationship between gene sequence and gene function. Genetic analyses of allelic series in Drosophila have shown how important this kind of analysis can be in uncovering distinct biochemical pathways and functions associated with particular regions of genes and their encoded proteins. Studies of human genetic diseases also highlight the importance of this often overlooked aspect of gene biology. Different mutations in the same gene can cause surprisingly diverse mutation-specific diseases. For example, mutations in different regions of the protein phosphatase SHP-2 cause diseases as diverse as juvenile leukaemia and severe cardiac malformations (Tartaglia et al., 2001, 2003). Reverse genetic techniques are clearly needed to fully understand gene function in vertebrate model systems.
One approach to meeting this need is a technique known as TILLING (Targeting Induced Local Lesions in Genomes) (Stemple, 2004). Following mutagenesis, selected genes are amplified by PCR and then screened by direct sequencing, digestion with a mismatch-targeting endonuclease such as CelI, or by liquid chromatography (dHPLC) to detect induced mutations. This type of technique has successfully isolated mutations in specific genes in plants, Drosophila, zebrafish and mice. So, why has it not had a greater impact on our understanding of gene function? In part, this is because of inherent problems with the established model organisms. Such screens in the mouse are carried out on libraries of mutagenized embryonic stem (ES) cells and often identify numerous mutant cell lines for the targeted gene (Chen et al., 2000). However, it is expensive and laborious to produce mice from these lines by blastocyst injection and breeding and so it is often necessary to choose those lines with mutations that are thought likely to affect gene function. This introduces a bias against uncovering novel functional domains and therefore undermines the usefulness of the approach. While in zebrafish the technique screens heterozygous mutation carriers (Wienholds et al., 2002), all of which can be raised and used as founders for mutant lines, the presence of duplicated genes in the zebrafish genome poses a potential problem for mutant analysis. The presence of these duplicated genes is thought to stem from a fish-specific genome duplication event in an ancestor of teleost fishes (reviewed in Meyer and Van de Peer, 2005). While many duplicated genes have been lost in other teleost species, such as fugu, duplicate copies have been retained in zebrafish. This complicates the interpretation of the effects of mutations in some genes as a result of the potential for functional redundancy between duplicated copies.
X. tropicalis is free of many of the problems associated with other vertebrates used for reverse genetic screens, while retaining their advantages. It is closely related to mammals in terms of both the copy number and arrangement of genes in its genome (see earlier discussion) and, as with zebrafish, TILLING techniques can be applied to large numbers of animals to screen for those that are heterozygous carriers of mutations in specific genes. Efforts are currently underway to screen for mutations in many important developmental genes by direct sequencing and by dHPLC. An early report has described the successful isolation of animals carrying a nonsense mutation in the NFATC3 gene (Goda et al., 2006) and improvements to the screening approaches are likely to increase the yield of such mutants.
THE FUTURE FOR THE FROG
The sequencing of the X. tropicalis genome was not driven simply by a desire to compare its genome with that of other species. As we have described, the genome sequence is already driving Xenopus research in new directions that can illuminate human biology. Although there are currently no Xenopus models of human disease, it seems inevitable that these will arrive. When they do, the accessibility of Xenopus throughout its life cycle will open up new avenues for research into potential treatments through the large-scale screening of drug libraries (Tomlinson et al., 2005). Much still needs to be done to characterize the effects of a variety of mutagens in order to optimize the efficiency of both forward and reverse genetics in the frog but this work is underway and such screens, together with the genome sequence, transgenesis and other tools such as the UAS-Gal4 system (Chae et al., 2002) promise to turn Xenopus tropicalis into a valuable new genetic system for studying development and its control by complex genetic networks.
Acknowledgments
We thank David M. Goodstein and the Computational Genomics Group (Joint Genome Institute, USDOE-Lawrence Berkeley Laboratory) for permission to reproduce the Metazome output shown in Figure 1a.
APPENDIX
General Xenopus tropicalis Resource Sites
Xenbase
A general Xenopus web resource and excellent jump site http://www.xenbase.org/
JGI
The Joint Genome Institute Xenopus tropicalis genome sequencing site http://genome.jgi-psf.org/Xentr4/Xentr4.home.html
The Xenopus tropicalis genetic map (from the University of Houston) http://tropmap.biology.uh.edu/
Trans-NIH Initiative
Reports, publications and funding opportunities from the NIH for Xenopus tropicalis http://www.nih.gov/science/models/xenopus/
Xenopus tropicalis EST and BAC Data Bases and Sources
IMAGE Consortium
Xenopus cDNA library web site and resource http://image.llnl.gov/image/html/xenopuslib_info.shtml
NIH Xenopus tropicalis EST database http://xgc.nci.nih.gov/
Sanger Xenopus tropicalis EST data base http://www.sanger.ac.uk/Projects/X_tropicalis/
The Japanese Government sponsored Xenopus tropicalis EST database http://xenopus.nibb.ac.jp/
The British Government (MRC) sponsored Xenopus tropicalis EST database http://www.geneservice.co.uk/products/cdna/XtropEST.jsp
The Wellcome Xenopus tropicalis full-length cDNA database http://informatics.gurdon.cam.ac.uk/online/xt-fl-db.html
CHORI BAC Resources
Information and source for Xenopus tropicalis BAC and BAC library http://bacpac.chori.org/xenopus216.htm
Phyolgenetic Genome Comparison Sites
NCBI genome viewer and BLAST site http://www.ncbi.nlm.nih.gov/mapview/
Phylofoot
Phylogenetic jump page and general resource http://www.phylofoot.org/
Metazome
Comparative genomic analysis and synteny database http://www.metazome.net/
DCODE
Phylogenetic tool based website and jump page at Lawrence Livermore National Laboratory http://www.dcode.org/
ECR (from DCODE)
A very user friendly phylogenetic analysis tool http://ecrbrowser.dcode.org/
MAVID
Multiple genomic alignment server http://baboon.math.berkeley.edu/mavid/
VISTA
Comparative genomic server http://pipeline.lbl.gov/cgi-bin/gateway2
Sequence Formats Conversions Site
As its title says, a useful site for format conversions between most of the phylogenetic footprinting sites http://bioweb.pasteur.fr/seqanal/formats-uk.html
Prediction of Cis-Regulatory and Transcription Factor Binding Sites
TRED
Genome-wide cis-regulatory element prediction and annotation tool http://rulai.cshl.edu/TRED
TRANSFAC
Free online computational program for the identification of transcription factor binding sites http://www.gene-regulation.com/
JASPAR
Free online computational program for the identification of transcription factor binding sites http://mordor.cgb.ki.se/cgi-bin/jaspar2005/jaspar_db.pl
MAPPER
Multi-data based computational tool for the identification of transcription factor binding sites http://bio.chip.org/mapper
ABS
Public database of known binding sites identified in promoters of orthologous vertebrate genes http://genome.imim.es/datasets/abs2005/index.html
EEL
Free software for the identification of regulatory elements based on the identification of clustered transcription factor binding sites http://www.cs.helsinki.fi/u/kpalin/EEL/
Stubb
Free online program allowing long genomic sequences to be searched for clusters of transcription factor binding sites and has replaced its fore-runner, Ahab. http://stubb.rockefeller.edu/
Windowfit
Free online program for analysis of smaller, enhancer-length sequences to identify binding sites and predict their strength. http://stubb.rockefeller.edu/
LITERATURE CITED
- Allen BG, Weeks DL. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat Methods. 2005;2:975–979. doi: 10.1038/nmeth814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, Rigby P, Krumlauf R, Brenner S. Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc Natl Acad Sci USA. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroukh N, Ahituv N, Chang J, Shoukry M, Afzal V, Rubin EM, Pennacchio LA. Comparative genomic analysis reveals a distant liver enhancer upstream of the COUP-TFII gene. Mamm Genome. 2005;16:91–95. doi: 10.1007/s00335-004-2442-9. [DOI] [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci USA. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Bataille AR, Chen X, Poitras C, Laganiere J, Lefebvre C, Deblois G, Giguere V, Ferretti V, Bergeron D, Coulombe B, Robert F. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae J, Zimmerman LB, Grainger RM. Inducible control of tissue-specific transgene expression in Xenopus tropicalis transgenic lines. Mech Dev. 2002;117:235–241. doi: 10.1016/s0925-4773(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yee D, Dains K, Chatterjee A, Cavalcoli J, Schneider E, Om J, Woychik RP, Magnuson T. Genotype-based screen for ENU-induced mutations in mouse embryonic stem cells. Nat Genet. 2000;24:314–317. doi: 10.1038/73557. [DOI] [PubMed] [Google Scholar]
- Denissov S, van Driel M, Voit R, Hekkelman M, Hulsen T, Hernandez N, Grummt I, Wehrens R, Stunnenberg H. Identification of novel functional TBP-binding sites and general factor repertoires. Embo J. 2007;26(4):944–954. doi: 10.1038/sj.emboj.7601550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: A quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- Goda T, Abu-Daya A, Carruthers S, Clark MD, Stemple DL, Zimmerman LB. Genetic screens for mutations affecting development of Xenopus tropicalis. PLoS Genet. 2006;2:e91. doi: 10.1371/journal.pgen.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammer TC, Khokha MK, Lane MA, Lam K, Harland RM. Identification of mutants in inbred Xenopus tropicalis. Mech Dev. 2005;122:263–272. doi: 10.1016/j.mod.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Halfon MS. (Re)modeling the transcriptional enhancer. Nature Genetics. 2006;38(10):1102–1103. doi: 10.1038/ng1006-1102. [DOI] [PubMed] [Google Scholar]
- Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Hamlet MR, Yergeau DA, Kuliyev E, Takeda M, Taira M, Kawakami K, Mead PE. Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis. 2006;44:438–445. doi: 10.1002/dvg.20234. [DOI] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: A novel antisense approach. Dev Biol. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Hirsch N, Zimmerman LB, Gray J, Chae J, Curran KL, Fisher M, Ogino H, Grainger RM. Xenopus tropicalis transgenic lines and their use in the study of embryonic induction. Dev Dyn. 2002;225:522–535. doi: 10.1002/dvdy.10188. [DOI] [PubMed] [Google Scholar]
- Janssens H, Hou S, Jaeger J, Kim AR, Myasnikova E, Sharp D, Reinitz J. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nat Genet. 2006;38:1159–1165. doi: 10.1038/ng1886. [DOI] [PubMed] [Google Scholar]
- Kenwrick S, Amaya E, Papalopulu N. Pilot morpholino screen in Xenopus tropicalis identifies a novel gene involved in head development. Dev Dyn. 2004;229:289–299. doi: 10.1002/dvdy.10440. [DOI] [PubMed] [Google Scholar]
- Kolm PJ, Sive HL. Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol. 1995;171:267–272. doi: 10.1006/dbio.1995.1279. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Horikoshi T, Heaney SJ, van Baren MJ, van der Linde HC, Breedveld GJ, Joosse M, Akarsu N, Oostra BA, Endo N, Shibata M, Suzuki M, Takahashi E, Shinka T, Nakahori Y, Ayusawa D, Nakabayashi K, Scherer SW, Heutink P, Hill RE, Noji S. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci USA. 2002;99:7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots GG, Ovcharenko I. rVISTA 2.0: Evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANS-FAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Noramly S, Zimmerman L, Cox A, Aloise R, Fisher M, Grainger RM. A gynogenetic screen to isolate naturally occurring recessive mutations in Xenopus tropicalis. Mech Dev. 2005;122:273–287. doi: 10.1016/j.mod.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Offield MF, Hirsch N, Grainger RM. The development of Xenopus tropicalis transgenic lines and their use in studying lens developmental timing in living embryos. Development. 2000;127:1789–1797. doi: 10.1242/dev.127.9.1789. [DOI] [PubMed] [Google Scholar]
- Ogino H, McConnell WB, Grainger RM. Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mech Dev. 2006;123:103–113. doi: 10.1016/j.mod.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Palin KTJ, Ukkonen E. Locating potential enhancer elements by comparative genomics using the EEL software. Nature Protocols. 2006;1:368–374. doi: 10.1038/nprot.2006.56. [DOI] [PubMed] [Google Scholar]
- Pan FC, Chen Y, Loeber J, Henningfeld K, Pieler T. I-SceI meganuclease-mediated transgenesis in Xenopus. Dev Dyn. 2006;235:247–252. doi: 10.1002/dvdy.20608. [DOI] [PubMed] [Google Scholar]
- Rajewsky N, Vergassola M, Gaul U, Siggia ED. Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo. BMC Bioinformatics. 2002;3:30. doi: 10.1186/1471-2105-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana AA, Collart C, Gilchrist MJ, Smith JC. Defining synphenotype groups in Xenopus tropicalis by use of antisense morpholino oligonucleotides. PLoS Genet. 2006;2:e193. doi: 10.1371/journal.pgen.0020193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:e271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JT, Priest JR, Ovcharenko I, Ronco A, Moore RK, Burns CG, MacRae CA. Human-zebrafish non-coding conserved elements act in vivo to regulate transcription. Nucleic Acids Res. 2005;33:5437–5445. doi: 10.1093/nar/gki853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Liang Y, Siggia E. Stubb: A program for discovery and analysis of cis-regulatory modules. Nucleic Acids Res. 2006;34:W555–559. doi: 10.1093/nar/gkl224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinner D, Kirilenko P, Rankin S, Wei E, Howard L, Kofron M, Heasman J, Woodland HR, Zorn AM. Global analysis of the transcriptional network controlling Xenopus endoderm formation. Development. 2006;133:1955–1966. doi: 10.1242/dev.02358. [DOI] [PubMed] [Google Scholar]
- Sivak JM, Petersen LF, Amaya E. FGF signal interpretation is directed by Sprouty and Spred proteins during mesoderm formation. Dev Cell. 2005;8:689–701. doi: 10.1016/j.devcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Stemple DL. TILLING—a high-throughput harvest for functional genomics. Nat Rev Genet. 2004;5:145–150. doi: 10.1038/nrg1273. [DOI] [PubMed] [Google Scholar]
- Stewart D, Tomita A, Shi YB, Wong J. Chromatin immunoprecipitation for studying transcriptional regulation in Xenopus oocytes and tadpoles. Methods Mol Biol. 2006;322:165–181. doi: 10.1007/978-1-59745-000-3_12. [DOI] [PubMed] [Google Scholar]
- Tada M, O’Reilly MA, Smith JC. Analysis of competence and of Brachyury autoinduction by use of hormone-inducible Xbra. Development. 1997;124:2225–2234. doi: 10.1242/dev.124.11.2225. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Niemeyer CM, Fragale A, Song X, Buechner J, Jung A, Hahlen K, Hasle H, Licht JD, Gelb BD. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34:148–150. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- Taverner NV, Kofron M, Shin Y, Kabitschke C, Gilchrist MJ, Wylie C, Cho KW, Heasman J, Smith JC. Microarray-based identification of VegT targets in Xenopus. Mech Dev. 2005;122:333–354. doi: 10.1016/j.mod.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Tomlinson ML, Field RA, Wheeler GN. Xenopus as a model organism in developmental chemical genetic screens. Mol Biosyst. 2005;1:223–228. doi: 10.1039/b506103b. [DOI] [PubMed] [Google Scholar]
- Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle FC, Odom DT, Bell GW, Yuan B, Danford TW, Wiellette EL, Herbolsheimer E, Sive HL, Young RA, Smith JC. Zebrafish promoter microarrays identify actively transcribed embryonic genes. Genome Biol. 2006;7:R71. doi: 10.1186/gb-2006-7-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Schulte-Merker S, Walderich B, Plasterk RH. Target-selected inactivation of the zebrafish rag1 gene. Science. 2002;297:99–102. doi: 10.1126/science.1071762. [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, Abnizova I, Gilks W, Edwards YJ, Cooke JE, Elgar G. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Ransick A, Martinez P, Britten RJ, Davidson EH. Complexity and organization of DNA-protein interactions in the 5′-regulatory region of an endoderm-specific marker gene in the sea urchin embryo. Mech Dev. 1994;47:165–186. doi: 10.1016/0925-4773(94)90088-4. [DOI] [PubMed] [Google Scholar]