Abstract
The ocelli are three simple photoreceptors on the vertex of the fruit fly head. We sought to identify the gene encoded by the classical ocellar mutant, reduced ocelli (rdo). Deficiency and inversion breakpoint mapping and P-element induced male recombination analyses were performed and Pray For Elves (PFE; CG15151; Fbgn0032661) emerged as a promising candidate for the rdo phenotype. The PFE locus maps to polytene region 36E on chromosome 2L between elfless (Fbgn0032660) and Arrestin 1 (Fbgn0000120). FlyBase annotation predicts that PFE encodes a serine/threonine kinase, yet protein prediction programs revealed no kinase domain. These analyses suggest that PFE simply encodes a leucine rich repeat molecule of unknown function, but presumably functions in nervous system protein-protein interaction. Two classical spontaneous alleles of rdo, rdo1 and rdo2, were characterized and the underlying mutations result from a small deletion spanning exon 1/intron 1 and a B104/roo insertion into the 3′UTR of PFE, respectively. Transposase-mediated excisions of several P-elements inserted into the PFE locus revert the rdo phenotype and a full-length PFE cDNA is sufficient to rescue rdo. A Gal4 enhancer trap reveals a broad adult neural expression pattern for PFE. Our identification and initial characterization of the rdo locus will contribute to the understanding of neurogenesis and neural development in the simple photoreceptors of the Drosophila visual system.
Keywords: Drosophila, ocelli, visual system, Pray For Elves, Leucine Rich Repeat
INTRODUCTION
The Drosophila adult visual system is composed of the two bilateral compound eyes and three triangularly arrayed simple eyes, the ocelli (Fig. 1A). The ocelli are located on the vertex of the fly head, one medial and two more posterior and lateral. The compound eyes and the ocelli derive from third instar larval eye-antennal imaginal discs (Fig. 1B). The two lateral ocelli each derive from a single disc while the medial ocellus arises from the fusion of both discs after puparium formation. Each ocellus possesses a dome-like corneal lens, ∼40 μm in diameter, that lies above a thin corneagenous cell layer and 75–95 photoreceptor cells.1,2 In Drosophila, six rhodopsins, Rh1-6, have been identified;3-11 Rh2 is specific to the ocellar photoreceptors and confers violet sensitivity.7,12,13 A layer of pigment cells surrounds the cluster of photoreceptor cells and each individual photoreceptor cell contains a single rhabdomere and pigment granules between the end of the rhabdomere and the nucleus. Photoreceptor cells are joined by belt desmosomes near the pigment granule layer. Photoreceptor cell axons, with associated glial cells, are found proximal to the photoreceptor cell nucleus.12,14 The axons of the photorecptor cells project to their corresponding ocellar ganglion and four giant ocellar interneurons from each ganglion project into the brain via the ocellar nerve.
Figure 1.
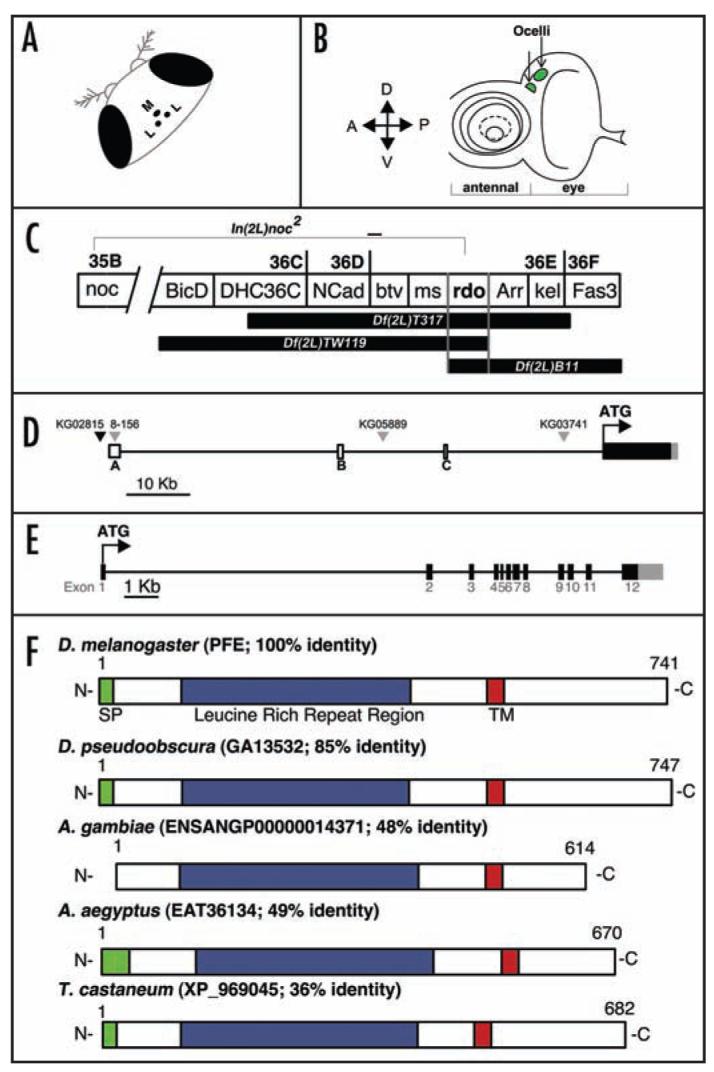
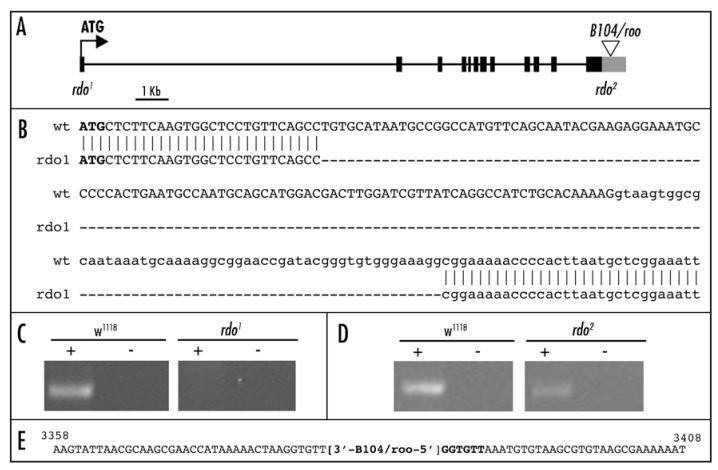
The rdo genetic region and the PFE locus. (A) Cartoon of the medial ocellus (M) and the two lateral ocelli (L) situated on the vertex of the Drosophila head between the compound eyes. (B) Cartoon of the third instar larval eye-antennal disc with the ocellar primordia shaded green. (C) The reduced ocelli region was mapped to 36DE using overlapping deficiencies Df(2L)TW119 and Df(2L)B11 and overlapping aberrations In(2L)noc2 and Df(2L)T317. (D) The PFE genomic region consists of a large 5′UTR with three non-coding exons (unshaded boxes) a coding region (black box) and a 3′UTR (grey box). Three P-elements (grey triangles), 8-156 (also referred to in the text as PFE-Gal4), KG05889, KG03741, inserted in the 5′UTR, are rdo- and one P-element (black triangle), KG02815, is inserted upstream of PFE and is rdo+. (E) The PFE coding region consists of twelve exons (black boxes) and 11 introns (thin line); the 3′UTR in exon 12 is also shown (grey box). (F) Diagram of the PFE protein and four insect orthologs (% identity based on output from BLAST 2 sequences is indicated). SP; signal peptide, TM; Transmembrane domain.
Compared to the wealth of knowledge pertaining to visual transduction in the compound eyes relatively little is known about the function of the ocelli. Once thought to be important in the generation of endogenous rhythms, the ocelli, and indeed the compound eyes, are neither essential nor necessary for effectively wild-type circadian activity even though a major clock protein, PERIOD, is expressed in the photoreceptor cells of these structures.15 Indeed, no ocelli, ocelliless and reduced ocelli mutants all have effectively wild-type circadian locomotor rhythms.16 The ocelli are, however, involved in simple visual guidance and modulation of sensitivity of the compound eyes during phototaxis.17,18 In this work, a combination of molecular aberration breakpoint and P-element-mediated mapping, phenotype reversion and rescue, and molecular characterization of two extant rdo alleles were performed to demonstrate that leucine rich repeat protein PFE is encoded by the classical mutant reduced ocelli (rdo). Pray For Elves (PFE) was named by Suzanna Lewis to voice her growing frustration and feelings of isolation during the FlyBase release 3 genome annotation. She prayed for magic elves to help her finish her work (FBrf0151677).
RESULTS
Bioinformatics
Flies trans-heterozygous for the Df(2L)B11 and Df(2L)TW119 deficiencies and flies trans-heterozygous for Df(2L)T317 and In(2L)noc2 exhibit reduced ocelli defects. Based on the estimated breakpoints of the overlapping deficiencies and the reported polytene chromosome breakpoint of In(2L)noc2 (Fig. 1C), PFE was investigated as a candidate for the reduced ocelli (rdo) phenotype. The PFE genomic region is 57,843 basepairs due to the presence of 39,054 basepair-spanning 5′ UTR and the 10,767 basepair intron between exons 1 and 2 (Fig. 1D and E); no additional annotated genes are predicted within these large exons. Three non-coding exons are interspersed within the large 5′UTR and these total a mere 694 basepairs. Twelve coding exons are predicted for PFE and from the first ATG-containing exon to the predicted translation stop site, the PFE transcript is 2,226 basepairs; an 856 basepair 3′UTR is also predicted (Fig. 1E). A full-length, 3,792 basepair PFE cDNA clone, complete with 3′ and 5′ UTRs (Accession# BT004502; from the Berkeley Drosophila Genome Project Head cDNA Library clone #GH07373) was used for these analyses and the accuracy of the clone was first verified by sequencing.
PFE is predicted to encode a 741 amino acid protein with a signal peptide (amino acids 1–17), a leucine rich repeat (LRR) region (amino acids 105–416) and a single transmembrane domain (integral to the membrane amino acids 506–528) and regions of low complexity in the C-terminal end beyond the transmembrane region (Fig. 1F). PFE is predicted to localize to the plasma membrane and the N-terminal LRR region is predicted to be extracellular. Leucine rich repeats are 20-29 basepair motifs predicted to be involved in a great diversity of biological functions that require protein-protein interaction.19 The core LRR motif is LxxLxL,20 where x is any amino acid; PFE is predicted to encode 10 such repeats. There are no additional domains predicted for PFE and pairwise alignment of PFE with all LRR containing proteins encoded in the D. melanogaster genome21,22 reveals only 20–30% identity almost exclusively in the LRR region (data not shown). PFE therefore seems to simply encode an LRR protein, despite predictions inferred from genome annotation that this molecule has a serine/threonine kinase function.21
BLAST analysis of PFE revealed four similar molecules with no predicted function, from D. pseudoobscura (GA13532, 85% identity), A. gambiae (Q7Q1I6, ENSANGP00000014371, 51% identity), A. aegyptus (EAT36134; 49% identity) and T. castaneum (XP_969045; 36% identity). These molecules have the same general protein architecture as PFE although the number of LRRs varies slightly and the A. gambiae ortholog has no predicted signal peptide (Table 1). tBLASTn analysis identified PFE orthologs in all of the sequenced23 Drosophilid species (Table 1). A neighbor-joining tree was generated with the orthologs listed in Table 1, and as expected, the Drosophilid members are most closely related (Fig. 2). Very little is known about the function of the ocelli in these insect species and interestingly, unlike D. melanogaster, A. gambiae, A. aegyptus and T. castaneum all possess ocelli at larval stages, but not as adults. Indeed, in the yellow fever mosquito, A. aegyptus, the larvae possess a group of four lateral ocelli on each side of the head capsule.24 And the rhodopsin molecules in these simple eyes have a peak absorbance at a wavelength of 515 nm and a maximal spectral sensitivity at 520 nm (green);25,26 in D. melanogaster ocelli the peak spectral sensitivities are at 350–370 nm (ultra-violet) and 445 nm (blue).18
Table 1.
PFE orthologs
| Species | %Identity | Span | #LRRs | ID |
|---|---|---|---|---|
| D. melanogaster | 100 | 1–741 | 10 | PFE/CG15151 |
| D. simulans | 98 | 1–741 | 9 | wu050602 |
| D. sechellia | 98 | 1–741 | 10 | br051028 |
| D. yakuba | 98 | 1–741 | 8 | caf051213 |
| D. erecta | 95 | 4–741 | 8 | ca051209 |
| D. ananasse | 76 | 1–741 | 7 | caf051209 |
| D. pseudoobscura | 85 | 1–741 | 11 | GA13532 |
| D. persimilis | 80 | 1–741 | 8 | br051028 |
| D. willistoni | 61 | 4–741 | 6 | caf060213 |
| D. mojavensis | 57 | 1–741 | 6 | caf051209 |
| D. virilis | 57 | 4–741 | 4 | caf051209 |
| D. grimshawii | 57 | 1–741 | 6 | caf051209 |
| A. gambiae | 51 | 22–639 | 10 | ENSANG00000014371 |
| A. aegyptus | 49 | 18–647 | 10 | EAT36134 |
| T. castaneum | 36 | 5–539 | 9 | XP_969045 |
Orthologs of PFE in Drosophilid species, mosquito species and the red flour beetle. Single-pass transmembrane domains are predicted for all proteins listed and, with the exception of A. gambiae, all have predicted signal peptides. % identity and span (regions of shared identity) are based on BLAST 2 sequence output comparing protein x and PFE and #LRRs is the number of Leucine rich repeats predicted by SMART. See Experimental Procedures for details of bioinformatics.
Figure 2.
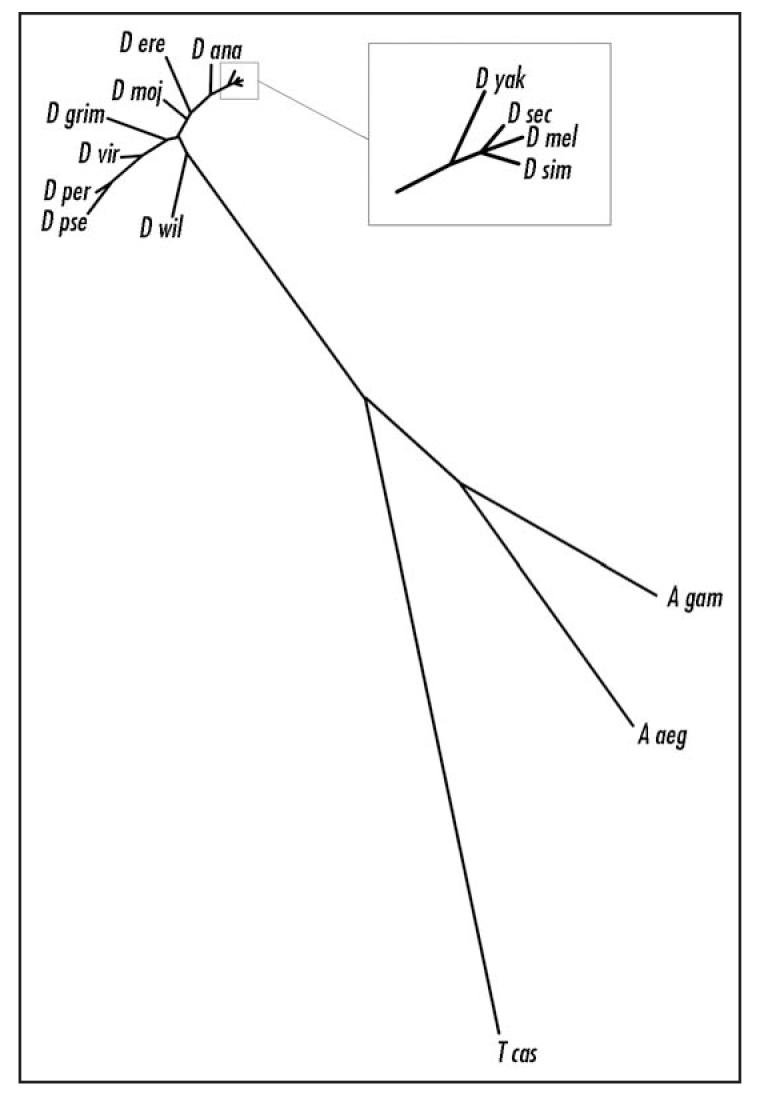
Neighbor-joining tree of PFE orthologs. Comparison of the Drosophilid, mosquito and beetle PFE orthologs. See Experimental Procedures for construction of tree.
P-element induced male recombination (PIMR) mapping of rdo
As mentioned previously, we had some evidence, based on breakpoint mapping to suggest that rdo encodes PFE. PIMR27 on the P-element P{SuporP}KG02815 (henceforth KG02815) inserted to the left of the 5′UTR of PFE (Fig. 1D) was performed in order to map rdo2 relative to the P-element insertion. Recombinants were analyzed in trans with Df(2L)TW119 to map the rdo2 mutation relative to the KG02815 P-element cross-over event. In trans with Df(2L)TW119, four Sp+ pr class recombinants were all rdo- while in the Sp pr+ class, one recombinant was rdo+ and another, designated KG02815-D, was rdo-. Two overall conclusions can be made from analysis of these male recombinant lines: 1) rdo2 maps to the right of P-element KG02815, as expected if rdo encodes PFE, and 2) the rdo phenotype in KG02815-D is likely the result of a flanking deletion into the 5′UTR of PFE during P-element mobilization.28 However, further molecular analyses of this line have not been performed.
P-element insertions at PFE
Three P-element inserts into different locations along the ∼40 kilobase PFE 5′UTR (pre-ATG) (Fig. 1D) were investigated for these analyses including: P{SuporP}KG03741, P{SuporP}KG05889 and the Gal4 enhancer trap line P{GawB}8-156 (henceforth, KG03741, KG05889 and PFE-Gal4, respectively).29,30 The rdo phenotype resulting from insertion of the KG elements is somewhat unexpected since the transposons do not insert into the predicted non-coding exons. The defects seen in these lines are likely due to the suppressor of Hairy wing [su(Hw)] chromatin insulator sites flanking the mini-white gene on the P-element. The su(Hw) insulators and the propensity for P-elements to insert at the 5′ end of genes, often results in altered levels of gene expression by disruption of the typical enhancer/promoter dynamic.30-32 These three P-element lines are all rdo-; the KG05889 insertion chromosome is homozygous lethal and therefore was tested for reduced ocelli in trans with Df(2L)TW119. Mobilizations of KG03741, KG05889 and PFE-Gal4 were also performed using standard transposase mediated techniques and excisions of these elements reverted the rdo phenotype (data not shown).
Extant alleles rdo1 and rdo2 disrupt PFE
Two extant spontaneous rdo alleles, rdo1 and rdo2,33 were characterized for this study. It has been previously shown that rdo mutations result in variable reductions in size of the ocellar lens2, although rdo1 is generally more penetrant than rdo2 (personal observations), and the number of ocellar receptor cells is reduced by about half in rdo homozygotes or rdo/Df(2L)TW119 heterozygotes.2
We sequenced the PFE coding regions of these alleles to identify the mutations. rdo1 is a small 158 basepair deletion of a region extending from exon 1 into intron 1 (Fig. 3A and B). RT-PCR analysis reveals that no PFE transcript is detectable in rdo1 mutants (Fig. 3C). The 3′UTR of rdo2 consistently failed to PCR amplify and iPCR analysis of rdo2 indicated that a B104/roo retrotransposon34 is inserted into the 3′UTR of PFE; a 6 bp insertion site duplication (GGTGTT) is also evident (Fig. 3E). RT-PCR analysis of rdo2 was performed and PFE transcript is detectable in these mutants (Fig. 3D). To determine if the level of PFE transcript is reduced in rdo2 quantitative real time PCR was performed. Total mRNA pools were isolated from adults and PFE was weakly expressed in both w1118 and rdo2; an approximate 2-fold difference in transcript copy number is evident in rdo2 (no significant difference, ANOVA p = 0.1686, Table 2). Since rdo2 is a B104/roo insertion into the 3′UTR it is possible that mRNA stability is variable—consistent with the weaker penetrance of this allele—and, as a result, a significant difference is masked. It is therefore possible that the level of PFE protein is reduced, but in the absence of a PFE antibody this possibility cannot be presently addressed. Alternatively, the effects on transcript level may not be uniform throughout the adult tissues. Finally, we PCR amplified the PFE coding regions in In(2L)noc2/Df(2L)T317 flies but were unable to find the breakpoints of the inversion, suggesting that the breakpoint of In(2L)noc2 is likely to be in the 40 Kb 5′UTR of PFE.
Figure 3.
Molecular identification of the rdo1 and rdo2 mutations. (A) Schematic of the PFE coding region (black boxes) and 3′UTR (grey box) shown with B104/roo insertion (unfilled triangle). (B) Sequence alignment of exon 1 (UPPERCASE) and a portion of intron 1 (lowercase) between wild-type and rdo1. ATG start is indicated in bold, deleted region of rdo1 indicated by ‘-’. (C) RT-PCR analysis of PFE in control and rdo1 cDNA pools reveals no detectable PFE transcript in rdo1. DmCa1D primers were used as a positive control (data not shown). (D) RT-PCR analysis of PFE in control and rdo2 cDNA pools indicates PFE transcript is detectable in both strains. DmCa1D primers were used as a positive control (data not shown). Real time PCR was conducted to analyze possible differences in PFE transcript level (see Table 2) (E) The rdo2 mutation is a B104/roo retrotransposon insertion (Accession #AY180917) into the 3′UTR of PFE with a flanking insertion site duplication (GGTGTT; bold). Numbering of wild-type sequence (normal font weight) is based on NCBI accession BT004502.
Table 2.
Real time PCR analysis of PFE in rdo2
| Drosophila Strain |
Biological Samples |
Transcripts (Fold-Change) |
P (ANOVA) | Ct (Gapdh2) | Ct (PFE) |
|---|---|---|---|---|---|
| w1118 | 3 | 1.0 | 15.57 ± 0.29 | 25.37 ± 1.23 | |
| rdo2 | 3 | -1.741 ± 0.37 | 0.1686 | 15.72 ± 0.24 | 27.11 ± 0.86 |
The level of wild-type (w1118) transcript is defined as 1.0. Data (Mean ± SD) are fold-change in level of PFE transcript. ‘-’ indicates a reduction in amount of PFE transcript relative to control (no significant difference). Data are normalized based on difference in average cycle threshold (Ct) for Gapdh2 and PFE.
The PFE-Gal4 enhancer trap driving UAS-PFE rescues rdo
A PFE-Gal4 enhancer trap insertion (P{GawB}8-156) into noncoding exon A (Fig. 1D) of PFE causes the rdo phenotype. A full-length PFE cDNA was cloned into the pUAST vector (UAS-PFE) and, in w; PFE-Gal4; UAS-PFE/TM3 flies, rdo appears to be fully rescued to wild-type, as determined by observing the ocelli under a dissecting scope (n = 30) and with SEM (n = 3). These data indicate that UAS-PFE is sufficient to rescue, and is encoded by, rdo (Fig. 4). Likewise, with the ubiquitous driver spaghetti-squash-Gal435, UAS-PFE is also sufficient to rescue rdo in Df(2L)TW119/Df(2L)B11 flies (data not shown). Heterozygous PFE-Gal4 (rdo+) driven expression of UAS-nls-GFP (nls; nuclear localization signal) is evident in all third instar larval imaginal discs in proneural cluster cells (Fig. 5A; A. Christiansen, personal communication) and this expression pattern is identical to previously reported in situ data using a full length PFE cDNA probe.36 In adults, GFP expression is evident in much of the nervous system including, but not limited to, the femoral chordotonal organ neurons and bristle neurons, the wing vein campaniform sensilla and wing margin bristle neurons, the compound eye facets, the lateral and medial ocelli and the olfactory chemoreceptors of the third antennal segment (Fig. 6). One caveat from these data is that the enhancer trap may not respond to any negative regulatory elements. In other words, in adults, PFE expression may be limited to the ocelli—consistent with the mutant phenotype—but, in the absence of adequate silencers, expression in the nervous system is robust. On the other hand, PFE may be functionally redundant in all tissues except the ocelli and therefore rdo only manifests in the ocelli.
Figure 4.
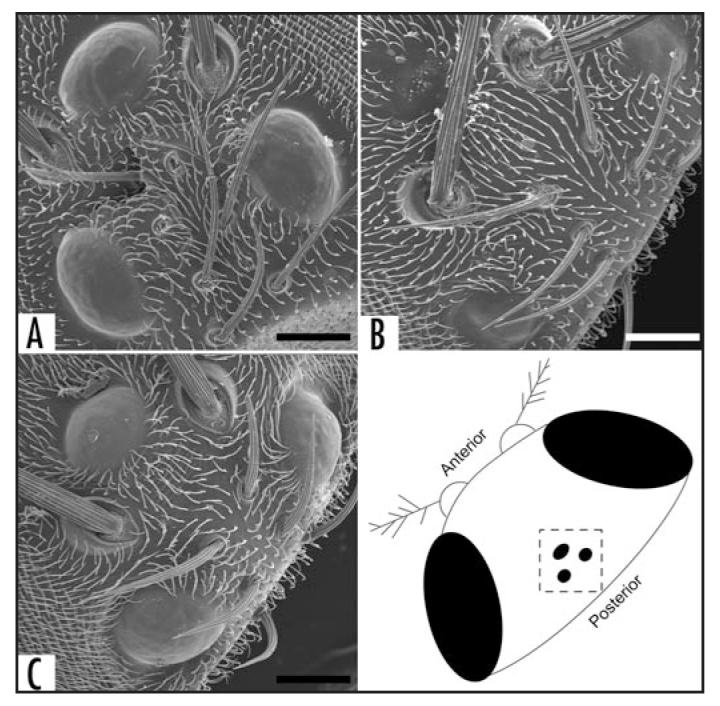
Rescue of rdo with UAS-PFE. (A) w1118 control line with fully formed ocelli with normal scalloping evident on the ocellar lenses. (B) w; PFE-Gal4 (rdo) line with reduced ocellar lenses. The lateral lenses exhibit relatively normal scalloping while the medial ocellus exhibits corneal nipples (after Stark and Sapp, 1989). (C) w; PFE-Gal4; UAS-PFE/TM3 flies have restored ocelli and scalloped lenses. Scale bars = 25 μm.
Figure 5.
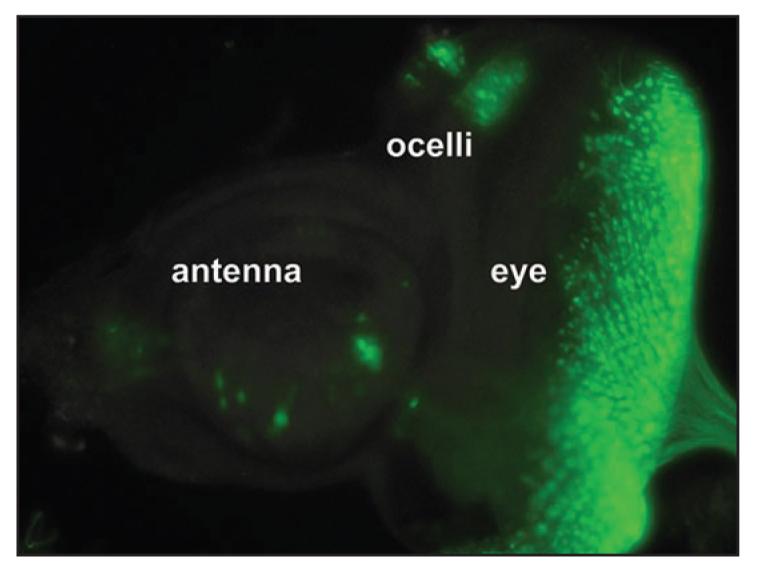
The PFE-Gal4 enhancer trap drives expression of UAS-nls-GFP in the eye-antennal disc proneural cluster cells UAS-nls-GFP expression pattern was observed in PFE-Gal4 heterozygotes (rdo+). GFP expression is evident in third instar larval eye-antennal imaginal disc proneural cluster cells including the ocellar, eye and antennal primordia. Photoshop was used to reduce the background over the entire image. The color range was selected to include only the intensely green areas and applied as a duplicated masking layer over the original image layer. The original image layer was then converted to black and white with a gradient map (black to white).
Figure 6.
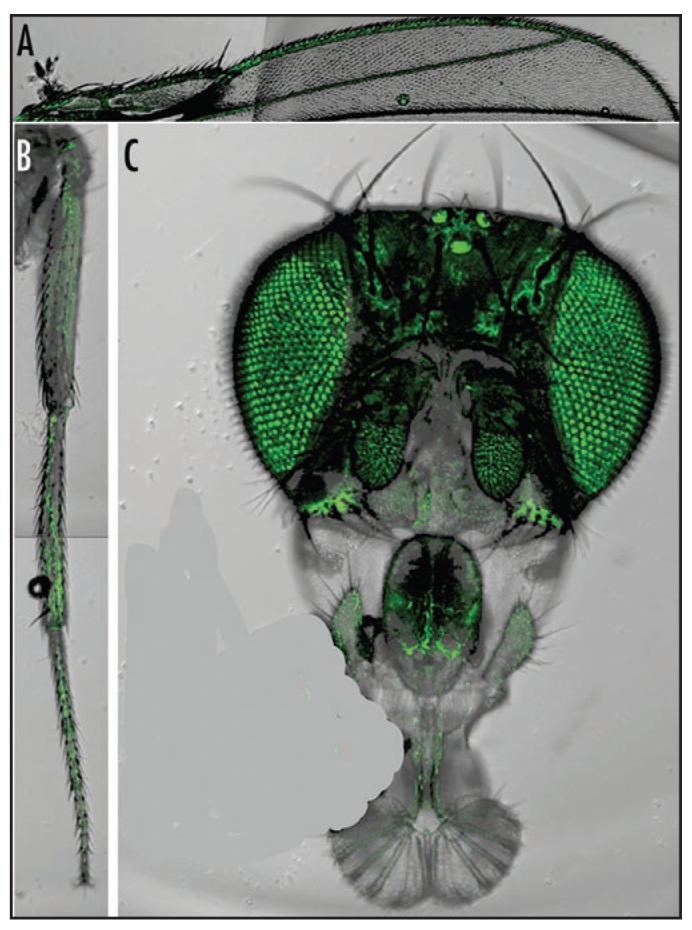
PFE-Gal4 drives expression of UAS-GFP in the adult nervous system. UAS-GFP expression pattern was observed in PFE-Gal4 heterozygotes (rdo+). (A) GFP expression is evident in the bristle neurons along the L1 wing vein margin and the campaniform sensilla of L2 wing vein. Wing vein L3 and the wing chordotonal organ cluster are not shown in this view but GFP expression is evident in these regions. (B) Expression in the bristle neurons of the tibia and tarsi is evident. (C) Expression in the adult head is evident in olfacatory chemoreceptors in the third antennal segment and palps, in the compound eyes and in the ocelli. An air bubble was airbrushed out of (C) using Photoshop.
CONCLUSIONS
We have presented several pieces of evidence that indicate PFE is encoded by rdo. Initially our genetic mapping with overlapping deficiencies and other chromosomal aberrations refined the rdo region. P-element induced male recombination indicated that rdo2 was to the right of the KG02815 P-element insert. Furthermore, several P-elements inserted into the PFE 5′UTR resulted in rdo and this phenotype was reverted when the elements were precisely excised. A UAS-PFE construct, when driven by a PFE-specific Gal4 enhancer trap, reverts the rdo phenotype, indicating that PFE is sufficient to rescue rdo. We also identified the mutations in PFE in two extant alleles of rdo. rdo1 results from a small deletion and lacks detectable PFE transcript and is therefore a null allele. rdo2 is a B104/roo retrotransposon insertion into the 3′UTR of PFE and transcripts are detectable by real time PCR and not significantly reduced compared to controls.
As demonstrated by Gal4-enhancer trap analysis and in situ hybridization with a full-length PFE cDNA, a broad neural expression pattern is evident for PFE although the only observed mutant phenotype is reduced ocelli; the most parsimonious explanation for this difference is that PFE may simply be expressed in non-essential regions. On the other hand, these data suggest that PFE mutants may not exhibit a more severe sensory phenotype due to functional redun-dancy with other LRR proteins. Furthermore, PFE is predicted to be post-transcriptionally regulated by microRNAs and this regulation may be spatially active in all tissues other than the ocelli. PFE may also be translationally or post-translationally regulated and, as a result, the protein, or putative active version of the protein, may only be present in the ocelli. Perhaps a more enticing explanation is that additional phenotypes have not been uncovered due to the purely morphological approach to our experimental design; in our laboratory strains, PFE may be involved in a presently unidentified, non-essential sensory process. rdo mutants have normal sound-evoked potentials in the antennal nerve (D. Eberl, unpublished). No electroretinograms have been performed on rdo. Nevertheless, we were able to definitively attribute rdo to PFE and future studies will focus on investigating the role of this LRR protein in ocellar system development. We are particularly interested in determining if PFE is redundant to other LRR molecules and the role of the LRR motif in protein-protein interactions necessary for proper formation of the ocelli.
EXPERIMENTAL PROCEDURES
Animals
Genetic strains of Drosophila melanogaster included seven mutant strains, four P-element lines and one normal control. The control strain was the w1118 (FBal0018186) strain; transformants were generated in the w1118 background. Stock of spontaneous reduced ocelli alleles of the genotypes rdo1 (FBal0014506) and rdo2 pr (FBal0014507) were obtained from the Bloomington Stock Center. Chromosomal deficiencies used to map rdo were Df(2L)TW119, cn bw/CyO37 (Bloomington; FBab0001642), Df(2L)B11, nub bpr/CyO (Bob Hardy, UCSD; FBab0040370), In(2L)noc2/CyO38 (John Roote, Cambridge; FBab0004671), and Df(2L)T317, b pr cn sca/CyO39 (Bloomington; FBab0001563). The w; TW12, Tft pr/CyO pr (Bloomington; FBab0001643) strain was used for PIMR. P-element lines KG03741 (FBst0013966), KG05889 (FBst0014132) and KG02815 (FBst0012989) were isolated from the Berkeley Drosophila Genome Project KG Enhancer Trap screen (http://www.fruitfly.org/p_disrupt/) and obtained from the Bloomington Stock Center. The 8-156 (PFE-Gal4) enhancer trap line was a generous gift from Audrey Christiansen and was originally obtained from the private enhancer trap collection in the Heberlein lab.
Bioinformatics
Protein architectures for PFE and orthologs were analyzed using the following servers and default parameters: SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/), TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/) and SMART (http://smart.embl-heidelberg.de/). Subcellular localization of PFE was predicted with PSORT II (http://psort.hgc.jp). BLAST analyses were performed with the predicted full-length PFE protein sequence as input. D. pseudoobscura, mosquito and red flour beetle PFE orthologs were identified by BLASTp (http://www.ncbi.nlm.nih.gov/BLAST/) with no filter for low complexity. PFE orthologs in other Drosophilid species were identified with tBLASTn at DroSpeGe (http://insects.eugenes.org/species/blast/). DNA sequences from the DroSpeGe search output were six-frame translated at the BCM Search Launcher (http://searchlauncher.bcm.tmc.edu/) and aligned with PFE with BLAST 2 Sequences (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi) to determine % identity. Sequences were aligned with ClustalW (with slow/accurate parameters) at http://align.genome.jp/ to generate an unrooted Neighbor-Joining Tree.
PIMR
Prior to performing PIMR, rdo2 was crossed into a background with several 2nd chromosome markers and a source of transposase—the resulting strain had the following genotype: w+; Sp rdo2 pr rl cn/CyO; Dr,Δ2-3/TM6, Ubx. Females of this strain were crossed to males of the P-element carrying strain, w/Y; KG02815. w+/Y; Sp rdo2 pr rl cn/KG02815; Dr, Δ2-3/+ males were collected and crossed to w; TW12 Tft pr/CyO pr females. Sp+ pr or Sp pr+ recombinants were collected over CyO or the Tft chromosome.
Reverse transcriptase PCR and real time PCR
For reverse transcriptase PCR, total RNA was isolated from tissues using the RNeasy mini kit (Qiagen 74104) with DNaseI treatment (Qiagen 79254) per kit instructions. For first strand synthesis, 5 μl of isolated total RNA was added to 1.25 μl poly-d(T)12-18 primer (Amersham 27-7858-02), 1 μl dNTPs and 1.25 μl DEPC-H2O and this mixture was heated to 65°C for 5 minutes and then cooled on ice for 5 minutes. Ten μl of 5x reaction buffer (250 μM Tris-HCl pH 8.3, 375 μM KCl, 15 μM MgCl2; Invitrogen 18064-022) and 5 μl 0.1M DTT (Invitrogen 18064-022) and 2.5 μl of Recombinant RNasin ribonuclease inhibitor (Promega N251A) were added and the mixture was heated to 42°C for 2 minutes. Then reverse transcriptase (Invitrogen 18064-022) or DEPC treated water was added for Rt+ or Rt-, respectively, and then incubated for 42°C for 50 minutes. The first strand synthesis reaction was inactivated at 70°C for 15 minutes. The cDNA was then used in a standard PCR reaction with DmCa1D (FBgn0001991) control primers DmCa1DF [5′-CAACCGGATGTGAAGTGCG-3′] and DmCa1DR [5′-CTT GGCACTTCGCCTGAAGG-3′] (data not shown), as well as, PFE primers directed against exons 1 and 3, PFE-RT1F [5′-GCTCTTCAAGTGGCTCCTG-3′] and PFE-RT3R [5′-GAAACGTTGAACTGGCCATG-3′] primers and the program [{95°C/30 s, 54Δ7°C/30 s, 72°C/30 s} × 30; {72°C/5 m} × 1].
For real time PCR, first strand synthesis was performed on isolated total RNA using the Superscript III First Strand Synthesis System (Invitrogen 18080-051) and oligo(dT)20 primer per kit instructions. Real time PCR was carried out with isolated cDNAs and the SYBR Green PCR Master Mix (Applied Biosystems 4309155) with Gapdh2 control primers Gapdh2-f [5′-AGCGCTGGTGCCGAATAC-3′] and Gapdh2-r [5′-AGTGAGTGGATGCCTTGTCGAT-3′] and PFE primers (described in reverse transcriptase section above). To control for pipetting errors, cDNAs from a biological sample were amplified in triplicate for both control and PFE primers; three biological samples per strain type were analyzed. NoRT controls for Gapdh2 and PFE primer sets were also included in the real time PCR reaction for each biological sample; no template controls were also performed for both primer sets. Real time PCR was performed using the following cycling conditions: [{95°C/10 m} × 1; {95°C/15 s, 60°C/1 m} × 40] and a single dissociation protocol {60°C + 35°C/20 minutes}. In the dissociation step, a single peak was obtained for Gapdh2 and PFE primers indicating only a single, specific product was amplified.
Inverse PCR
30 flies were collected and placed on ice briefly. Flies were homogenized in 200 μl Buffer A {100 μM Tris-HCl, pH 7.5, 100 μM EDTA, 100 μM NaCl, 0.5% SDS}. Another 200 μl of Buffer A was added and tissues were homogenized again and placed at 65°C for 30 minutes. Eight hundred microliters of LiCl/KAc Solution {1 part 5 M KAc, 2 parts 6 M LiCl} were added and the sample was incubated on ice for 10 minutes. Tubes were centrifuged for 15 minutes at max speed. One milliliter of supernatant was removed to a new tube and 600 μl isopropanol were added. Tubes were centifuged for 15 minutes at max speed. The supernatant was discarded and washed with 70% EtOH. Pellets were resuspsended in 150 μl TE. 10 μl of the sample in TE with 2.5 μl Sau3AI with 2.5 μl BSA, 2 μl 100 μg/ml RNAseA, 2.5 μl Sau3AI buffer and 5.5 μl H2O was digested for 2.5 hours at 37°C and heat inactivated 20 minutes at 65°C. 10 μl of the Digested Genomic DNA was ligated in a large volume (12 μl 10X ligation buffer, 1 μl 400,000 U/μl Ligase and 97 μl H2O) overnight at 4°C. 360 μl QG buffer from QIAquick Gel Extraction Kit (Qiagen 28704) and 120 μl isopropanol was added. Contents were added to gel extraction column and centrifuged at 13,000 rpm for 1 minute. The flow through was discarded and 750 μl PE buffer from QIAquick Gel Extraction Kit (Qiagen 28704) was added to the column for a 2 minute incubation; the tube was centrifuged briefly and the flow through discarded, followed by a 1 minute spin to dry column. Ligated products were eluted with 50 μl EB from QIAquick Gel Extraction Kit (Qiagen 28704) and a 1 minute spin. PCR was performed with 10 μl of template, 1 μl of Forward primer, 1 μl Reverse Primer, 1 μl dNTPs (50X), 5 μl 10X buffer, 1.5 μl MgCl2, 0.5 μl Taq and 30 μl H2O. The left side of the break was determined using PFE-iLeft-inF [5′-GTACACACGATTCATTTCGACAC-3′] and PFE-iLeft-inR [5′-GTGAAACACTGGTAAAGTTCG-3′] and the right side of the break was determined with PFE-iRight-outF [5′-GGTAAGGTCTTCTTGCGAGC-3′] and PFE-iRight-outR [5′-GAACTAGCATGTAAGGCATGTC-3′].
Rescue construct
A full-length (including 5′ and 3′ UTRs) cDNA clone of PFE was PCR amplified from the BDGP EST clone GH07373, using primers POT2F [5′-AATGCAGGTTAACCTGGCTTATCG-3′] and POT2R [5′-AACGCGGCTACAATTAATACATAACC-3′], and the program [{95°C/30 s, 56.5°C/30 s, 72°C/2 m} × 30; {72°C/10 m} × 1]. The PCR product was cloned into TOPO-XL and confirmed by sequencing. Full-length PFE was directionally subcloned into pUAST40 and injected by the Duke University Model System Genomics Facility.
Scanning electron microscopy
Heads were isolated and fixed in 2.5% gluteraldehyde for 24 hours at 4°C, postfixed in 1% OsO4/Phosphate Buffer for 1 hour, dehydrated in an ethanol series, critical point dried and finally sputter coated with a layer of Gold/Palladium. Images were taken on a Hitachi S-4000.
Statistical analysis
Real time PCR data were analyzed by ANOVA using SAS version 6.12 (Sas Institute Inc., Cary, NC, USA).
ACKNOWLEDGEMENTS
We would like to thank Audrey Christiansen and the Bloomington Drosophila stock center for providing fly stocks, the Elena Sivan Loukianova and the UI Central Microscopy Research Facility for SEMs, Jill Pottratz for deficiency mapping and the Duke University Model Systems Genomics Facility for injection of the UAS-PFE construct.
FUNDING
Daniel F. Eberl: NIH grant DC04848
ABBREVIATIONS
- rdo
reduced ocelli
- PFE
Pray For Elves
- LRR
leucine rich repeat
- PIMR
P element induced male recombination
Reference
- 1.García-Alonso L, Fetter RD, Goodman CS. Genetic analysis of Laminin A in Drosophila: Extracellular matrix containing laminin A is required for ocellar axon pathfinding. Development. 1996;122:2611–21. doi: 10.1242/dev.122.9.2611. [DOI] [PubMed] [Google Scholar]
- 2.Stark WS, Sapp R. Ultrastructure of the ocellar visual system in normal and mutant Drosophila melanogaster. J Neurogenetics. 1989;5:127–53. doi: 10.3109/01677068909066203. [DOI] [PubMed] [Google Scholar]
- 3.Zuker CS, Montell C, Jones K, Laverty T, Rubin GM. A rhodopsin gene expressed in photoreceptor cell R7 of the Drosophila eye: Homologies with other signal-transducing molecules. J Neurosci. 1987;7:1550–7. doi: 10.1523/JNEUROSCI.07-05-01550.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuker CS, Mismer D, Hardy R, Rubin GM. Ectopic expression of a minor Drosophila opsin in the major photoreceptor cell class: Distinguishing the role of primary receptor and cellular context. Cell. 1988;53:475–82. doi: 10.1016/0092-8674(88)90167-5. [DOI] [PubMed] [Google Scholar]
- 5.Zuker CS, Cowman AF, Rubin GM. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985;40:851–8. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]
- 6.O’Tousa JE, Baehr W, Martin RL, Hirsh J, Pak WL, Applebury ML. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40:839–50. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- 7.Cowman AF, Zuker CS, Rubin GM. An opsin gene expressed in only one photoreceptor cell type of the Drosophila eye. Cell. 1986;44:705–10. doi: 10.1016/0092-8674(86)90836-6. [DOI] [PubMed] [Google Scholar]
- 8.Montell C, Jones K, Zuker C, Rubin G. A second opsin gene expressed in the ultraviolet-sen-sisite R7 photoreceptor cells of Drosophlia melanogaster. J Neurosci. 1987;7:1558–66. doi: 10.1523/JNEUROSCI.07-05-01558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papatsenko D, Sheng G, Desplan C. A new rhodopsin in R8 photoreceptors of Drosophila: Evidence for coordinate epxression with Rh3 in R7 cells. Development. 1997;124:1665–73. doi: 10.1242/dev.124.9.1665. [DOI] [PubMed] [Google Scholar]
- 10.Chou WH, Hall KJ, Wilson DB, Wideman CL, Townson SM, Chadwell LV, Britt SG. Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron. 1996;17:1101–15. doi: 10.1016/s0896-6273(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 11.Huber A, Schulz S, Bentrop J, Groell C, Wolfrum U, Paulsen R. Molecular cloning of Drosophila Rh6 rhodopsin: The visual pigment of a subset of R8 photoreceptor cells. FEBS Lett. 1997;406:6–10. doi: 10.1016/s0014-5793(97)00210-x. [DOI] [PubMed] [Google Scholar]
- 12.Pollock JA, Benzer S. Transcript localization of four opsin genes in the three visual organs of Drosophila; RH2 is ocellus specific. Nature. 1988;333:779–82. doi: 10.1038/333779a0. [DOI] [PubMed] [Google Scholar]
- 13.Mismer D, Michael WM, Laverty TR, Rubin GM. Analysis of the promoter of the Rh2 opsin gene in Drosophila melanogaster. Genetics. 1988;120:173–80. doi: 10.1093/genetics/120.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon CS, Hirosawa K, Suzuki E. Studies on the structure of ocellar photoreceptor cells of Drosphila melanogaster with special reference to subrhabdomeric cisternae. Cell Tisues Res. 1996;284:77–85. doi: 10.1007/s004410050568. [DOI] [PubMed] [Google Scholar]
- 15.Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J Neurosci. 1992;12:3321–49. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vosshall LB, Young MW. Circadian rhythms in Drosophila can be driven by period expression in a restricted group of central brain cells. Neuron. 1995;15:345–60. doi: 10.1016/0896-6273(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 17.Fischbach K, Reichert H. Interactions of visual subsystems in Drosophila melanogaster: A behavioural genetic analysis. Biol Behav. 1978;2:305–17. [Google Scholar]
- 18.Hu KG, Stark WS. The roles of Drosophila ocelli and compound eyes in phototaxis. J Comp Physiol A. 1980;135:85–95. [Google Scholar]
- 19.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–32. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 20.Kajava AV, Kobe B. Assessment of the ability to model proteins with leucine-rich repeats in light of the lastest strutural information. Protein Sci. 2002;11:1082–90. doi: 10.1110/ps.4010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grumbling G, Strelets V. The Flybase Consortium. FlyBase: Anatomical data, images and queries. Nucleic Acids Res. 2006;34:D484–8. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hynes RO, Zhao Q. The evolution of cell adhesion. J Cell Biol. 2000;150:F89–F95. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert D. DroSpeGe, a public database of Drosophila species genomes. doi: 10.1093/nar/gkl997. http://insectseu- genesorg/DroSpeGe/2005 [DOI] [PMC free article] [PubMed]
- 24.Brown P, White R. Rhodopsin of the larval mosquito. J Gen Physiol. 1972;59:401–14. doi: 10.1085/jgp.59.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seldin E, White R, Brown P. Spectral sensitivity of larval mosquito ocelli. J Gen Physiol. 1972;59:415–20. doi: 10.1085/jgp.59.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White R. Analysis of the development of the compound eye in the mosquito, Aedes aegypti. J Exp Zool. 1961;148:223–39. doi: 10.1002/jez.1401480305. [DOI] [PubMed] [Google Scholar]
- 27.Chen B, Chu T, Harms E, Gergen JP, Strickland S. Mapping of Drosophila mutations using site-specific male recombination. Genetics. 1998;149:157–63. doi: 10.1093/genetics/149.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preston CR, Sved JA, Engels WR. Flanking duplications and deletions associated with P-inuced male recombination in Drosophila. Genetics. 1996;144:1623–38. doi: 10.1093/genetics/144.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bier E, Baessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, Jan LY, Jan YN. Searching for a pattern and mutation in the Drosophila with a P-lacZ vector. Genes Dev. 1989;3:1273–87. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- 30.Roseman RR, Johnson EA, Rodesch CK, Bjerke M, Nagoshi RN, Geyer PK. A P element containing suppressor of Hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics. 1995;141:1061–74. doi: 10.1093/genetics/141.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellen HJ. Ten years of enhancer detection: Lessons from the fly. Plant Cell. 1999;11:2271–81. doi: 10.1105/tpc.11.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao GC, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci. 2000;97:3347–51. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindsley D, Zimm GG. The Genome of Drosophila melanogaster. Academic Press; San Diego, California: 1992. [Google Scholar]
- 34.Scherer G, Tschudi C, Perera J, Delius H. B104, a new dispersed repeated gene family in Drosophila melanogaster and its analogies with retroviruses. J Mol Biol. 1982;157:435–51. doi: 10.1016/0022-2836(82)90470-3. [DOI] [PubMed] [Google Scholar]
- 35.Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J Cell Biol. 2000;149:471–90. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reeves N, Posakony JW. Genetic programs activated by proneural proteins in the developing Drosophila PNS. Dev Cell. 2005;8:413–25. doi: 10.1016/j.devcel.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 37.Wright TRF, Hodgetts RB, Sherald AF. The genetics of dopa decarboxylase in Drosophila melanogaster. I. Isolation and characterization of deficiencies of the structural locus and the a-methyldopa hypersensitive locus. Genetics. 1976;84:267–85. doi: 10.1093/genetics/84.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashburner M, Aaron C, Tsubota S. The genetics of a small autosomal region of Drosophila melanogaster, including the structural gene for alcohol dehydrogenase. V. Characterization of x-ray-induced Adh null mutations. Genetics. 1981;102:421–35. doi: 10.1093/genetics/102.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steward R, Nusslein-Volhard C. The genetics of the Dorsal-Bicaudal-D region of Drosophila melanogaster. Genetics. 1986;113:665–78. doi: 10.1093/genetics/113.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]