Abstract
Epidermal growth factor receptor (EGFR) and its family members, ErbB2, ErB3 and ErB4, are receptor tyrosine kinases which send signals into the cell to regulate many critical processes including development, tissue homeostasis, and tumorigenesis. Central to the signaling of these receptors is their intracellular kinase domain, which is activated by ligand-induced dimerization of the receptor and phosphorylates several tyrosine residues in the C-terminal tail. The phosphorylated tail then recruits other signaling molecules and relays the signal to downstream pathways. A model of the autoinhibition, activation and feedback inhibition mechanisms for the ErbB kinase domain has emerged from a number of recent structural studies. Meanwhile, recent clinical studies have revealed the relationship between specific ErbB kinase mutations and the responsiveness to kinase inhibitor drugs. We will review these regulation mechanisms of the ErbB kinase domain, and discuss the binding specificity of kinase inhibitors and the effects of kinase domain mutations found in cancer patients from a structural perspective.
Keywords: Epidermal growth factor receptor, Receptor tyrosine kinase, Structural biology, Tyrosine kinase inhibitor, Lung cancer, Activation loop, Signal transduction, Growth factor receptor, Receptor Dimerization, Mig6
1. Background
The EGFR (epidermal growth factor receptor) family receptor tyrosine kinases (EGFR/ErbB1, ErbB2, ErB3 and ErB4) are cell surface receptors which, upon activation by their ligands, send signals to the cell to regulate many important processes [1–3]. The signaling is initiated by binding of a ligand (e.g. EGF) to the extracellular region of these receptors, which then transduce the signal to the intracellular region through the single transmembrane helix. The intracellular tyrosine kinase domain then phosphorylates several tyrosine residues in the C-terminal tail of the receptor, which recruits PTB and SH2 domains-containing signaling proteins and relay the signal to downstream signaling pathways.
Over two decades ago, Schlessinger and colleagues proposed the ligand-induced dimerization mechanism for EGFR activation [1, 4, 5]. Despite extensive investigation, how ligand engagement induced EGFR dimerization remained unclear. The breakthrough was made a few years ago by several groups, whose structural studies on the extracellular regions of the EGFR family members showed that the ligand induces a dramatic conformational change in the receptor which promotes dimerization by exposing elements of the dimer interface that are buried prior to ligand binding [6–11].
The remaining question was then how dimerization of the extracellular region induced activation of the intracellular portion of the receptor and downstream signaling pathways [2]. For other tyrosine kinases such as insulin receptor and Src, dimerization induces activation of the intracellular kinase domain by allowing trans-autophosphorylation of one or several tyrosine residues in the so-called activation loop, which normally cannot be performed in cis due to the geometric restraints [12, 13]. The activation loop is located at the mouth of the active site between the N-terminal (N-lobe) and the C-terminal lobes of the kinase domain. It usually serves as a switch in kinase regulation by converting between an unphosphorylated, closed conformation and a phosphorylated, open conformation that blocks and supports substrate binding, respectively. Phosphorylation of the corresponding tyrosine in EGFR, Tyr845, however does not seem to be critical for EGFR activation [14]. (Residue numbers used throughout this article are based on the mature EGFR protein sequence. This numbering scheme can be converted to the plus signaling peptide numbering convention by adding 24 to the residue number.) Consistent with this conclusion, the first crystal structure of the EGFR kinase domain adopts an active conformation, with Tyr845 in the activation loop unphosphorylated [15]. These observations led to the hypothesis that the kinase domain of EGFR is constitutively active. However, trans-autophosphorylation of the C-terminal tail can only take place when two receptor molecules are brought into close proximity by ligand-induced dimerization. This simplistic mechanism, however, fails to explain why the long (>200 residues) and presumably flexible C-terminal tail does not enter the active site of the kinase domain that is within the same receptor molecule and undergo phosphorylation in cis in the absence of the ligand.
A number of studies have shown that purified and unliganded EGFR possesses a low level of basal kinase activity towards substrate peptides, and EGF binding significantly stimulates the kinase activity [4, 5, 16–19]. The catalytic activity of the isolated EGFR kinase domain also increases upon treatment with reagents that induce oligomerization [20, 21]. These observations together point to a model in which the EGFR kinase domain is normally kept inactive, and an inter-molecular allosteric interaction is required for its activation.
The detailed structural basis for the autoinhibition and allosteric activation of the EGFR kinase domain has been revealed by a series of recent studies. In this review, we will summarize these important observations, and discuss the effects of the Mig6 inhibitory protein, small molecular kinase inhibitors, and EGFR mutations on kinase activity regulation, in the context of the structural studies.
2. Autoinhibition of the EGFR kinase domain
An important line of evidence for establishing the autoinhibited state of the EGFR kinase domain was the crystal structure that showed that the kinase domain adopts an inactive conformation similar to that first seen in the cyclin-dependent kinase 2 and the Src family kinases [22–25]. Two later crystal structures of the EGFR kinase domain, one bound to an ATP analogue, AMP-PNP, and the other bound to an allosteric inhibitory peptide (see below), show an essentially identical conformation [26, 27]. These three structures, obtained in different crystal forms with different ligands bound, strongly support that this CDK/Src-like conformation represents the preferred autoinhibited state of the EGFR kinase domain. Recent crystal structural studies show that the ErbB4 kinase domain also adopts the CDK/Src-like conformation, suggesting that ErbB4 also uses the same autoinhibition mechanism [28, 29]. In fact, the CDK/Src-like inactive conformation has now been observed in many divergent tyrosine and Ser/Thr kinases and may be a conserved mechanism for kinase autoinhibition in general [30–32].
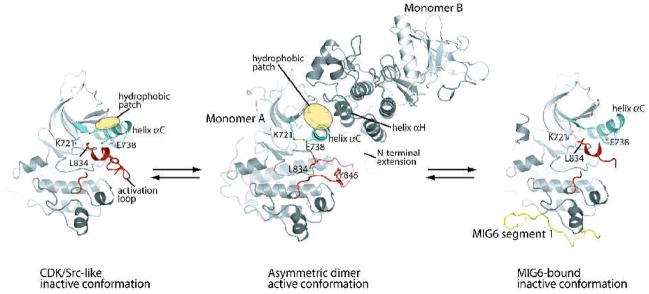
A pronounced structural feature of the CDK/Src-like conformation is the distinct orientation of an important α helix in the N-terminal lobe (N-lobe) of the kinase named helix αC (Figure 1). Compared to that in the active conformation, helix αC in the CDK/Src-like conformation is swung out in relation to the rest of the N-lobe of the kinase. The orientation of helix αC moves a conserved glutamate residue in the helix (Glu738) out of the active site and breaks the catalytically important salt bridge between the side chain of Glu738 and the side chain of a conserved Lysine in strand β3 (Lys721).
Figure 1.
Autoinhibition, activation and feedback inhibition of the EGFR kinase domain. The EGFR kinase domain is autoinhibited by adopting the CDK/Src-like inactive conformation (left panel) [26]. The N-terminal portion of the activation loop (red) forms a short helix which packs against the N-lobe of the kinase and stabilizes the outwardly displaced conformation of helix αC (aqua). Upon ligand induced dimerization of the receptor, the kinase domain forms the asymmetric activating dimer (middle panel) [26]. In this dimer, monomer B activates monomer A by acting as a cyclin-like activator. The hydrophobic patch in the N-lobe of monomer A (yellow ellipse) is buried in the dimer interface. MIG6 is upregulated immediately after EGFR activation and serves as a feedback inhibitor. MIG6 segment 1 (yellow) binds the C-lobe face of the asymmetric dimer interface, sterically hindering the formation of the asymmetric dimer (right panel) [27].
The outward displacement of helix αC is sterically coupled to a closed conformation of the activation loop, which is incompatible with substrate peptide binding. Of particular note, the N-terminal portion of the activation loop immediately following the conserved Asp-Phe-Gly (DFG) motif forms a short helix. The first and fourth residues in this helix are two leucine residues (Leu834 and Leu837), which are conserved in many kinases. The sidechains of these two leucines point upwards and interact with the inner surface of the active site formed by hydrophobic residues from helix αC and strand β3, β4 and β5. These interactions stabilize the CDK/Src-like conformation by sterically hindering the formation of the catalytically important Lys721-Glu738 salt bridge and supporting the outwardly displaced orientation of helix αC. Leu834 and 837 are frequently mutated to arginine and glutamine respectively in human non-small cell lung cancer patients [33–36], highlighting their important role in stabilizing the inactive conformation of the EGFR kinase domain. These mutations will be discussed in more detail below. The activity of the isolated kinase domain bearing the Leu834Arg mutation is >15- fold higher than the wild type kinase domain, confirming that the activating effect of the mutation is exerted on the kinase domain itself [26, 37, 38]. In addition, mutations of some other residues involved in the hydrophobic interactions between the activation loop short helix and the N-lobe have also been shown to promote spontaneous activation of EGFR [39].
An analysis of these inactive structures provided insights into the intrinsic stability of the EGFR kinase domain in the CDK/Src-like inactive conformation. The top face of the N-lobe formed by helix αC and the rest of the N-lobe in the EGFR kinase domain contains a hydrophobic patch (Figure 1). In the CDK/Src-like conformation, some of the hydrophobic residues in this patch are either partially or completely buried in the groove between helix αC and strands β4 and β5. In contrast, these residues are mostly exposed in the active conformation, and consequently, the exposed hydrophobic area is much larger, which is energetically unfavorable. The kinase domain has to overcome this free energy penalty to convert to the active conformation, which is presumably driven by the ligand-induced dimerization of the receptor.
The ErbB4 kinase domain adopts the essentially identical inactive conformation, and most of the key residues involved in stabilizing the inactive conformation are conserved [28, 29]. While the structures of the ErbB2 and ErB3 kinase domains are not available at present, their high degree of sequence homology to EGFR and ErbB4 suggests that they also use the same mechanism for autoinhibition.
3. The allosteric activation mechanism
As mentioned above, some early studies pointed to a model in which the EGFR kinase domain is normally autoinhibited, but is activated by an inter-molecular allosteric interaction upon ligand-induced dimerization of the receptor [4, 5, 16–21]. More compelling evidence for this model came from recent studies using purified kinase domain of EGFR [26]. These experiments showed that the isolated kinase domain is monomeric in solution and possesses low catalytic activity. The activity dramatically increased when the local concentration of the kinase domain was increased by attaching it to the surface of lipid vesicles. These experiments strongly suggest that an intermolecular interaction between the isolated kinase domain induced by the high local concentration converts the kinase from the CDK/Src-like inactive conformation to the active conformation.
Three structures of the EGFR kinase domain, obtained under different crystallization conditions, showed the active conformation in the same crystal form [15, 26]. The inactive conformation was seen only in crystals either with ligands that prevent activation (lapatinib or Mig6, see below) or with a specific mutation that prevents the formation of the active crystal form (see below) [22, 26, 27]. The persistent active conformation in crystals appears to contradict the fact that the kinase domain is stable in the inactive state in solution. This apparent paradox can be explained by the existence of a specific intermolecular interaction between the kinase domain molecules in the crystal lattice which converts the kinase to the active conformation. This provides a link between the structural and biochemical data for understanding the kinase domain activating mechanism. In both cases, high local concentrations promote the specific intermolecular interaction that is required for activating the kinase.
Analyses of structures in the active conformation revealed two prominent dimers in the crystal lattice; one is symmetric and the other asymmetric [26]. The symmetric dimer is mediated by the interaction between a fragment in the C-terminal tail which is sandwiched between the N-lobe of its own kinase domain and the C-lobe of the dimer partner. Mutational analyses showed that this symmetric dimer is not required for ligand-induced activation of EGFR. However, it may contribute to fine tuning of EGFR activity by exerting an autoinhibitory effect, as suggested by Landau et al [40, 41].
The asymmetric dimer is formed between the bottom of the C-lobe of one kinase monomer (monomer B) and the top of the N-lobe of the other (monomer A) (Figure 1). It is worth pointing out that an earlier computational study, carried out in the absence of any direct structural information on EGFR, suggested several dimer models, one of which is an asymmetric dimer similar to this crystallographic dimer [42]. The interaction between monomer B and A resembles that between cyclin A and active cyclin dependent kinase 2, with the C-lobe of monomer B taking the position of cyclin A in engaging the N-lobe of the kinase partner, although the structure of the C-lobe of the EGFR kinase is completely unrelated to that of cyclin [26]. This asymmetric dimer interaction is incompatible with the CDK/Src-like inactive conformation of the kinase due to large conformational difference in the N-lobe, especially helix αC, of the kinase domain. Taken together, these observations led to a model for the activation of the EGFR kinase domain in which monomer B in the asymmetric dimer acts as a cyclin-like allosteric activator for monomer A.
Mutational analyses confirm the critical role for the asymmetric dimer in the activation of EGFR, both in the context of full length receptor in cells and the isolated kinase domain in the lipid vesicle-based assay [26]. For example, a Val924 to arginine mutation, which disrupts the C-lobe face of the asymmetric dimer interface but is far away from the kinase active site, abolishes both ligand-induced autophosphorylation of the full length receptor and lipid vesicle-induced activation of the isolated kinase domain [26, 43]. This Val924Arg mutant kinase domain has been crystallized with an ATP analogue, AMP-PNP, which shows the CDK/Src-like inactive conformation [26]. The fact that a single point mutation located far away from the active site leads to crystallization of the EGFR kinase in the CDK/Src-like inactive conformation strongly supports that the CDK/Src-like conformation is the preferred inactive state of the kinase domain, and the active conformation seen in the original crystal form is dependent on the asymmetric dimer interface.
The asymmetric dimer interface is dominated by a helix-helix packing interaction between helix αH of monomer B and helix αC of monomer A, which keeps helix C in the active conformation [26]. The interface buries a large hydrophobic surface area, the core of which is contributed mainly from the hydrophobic patch alongside of helix αC that is largely buried in the CDK/Src-like conformation but exposed in the active conformation (Figure 1). Therefore, the asymmetric dimer stabilizes the active conformation at least in part by compensating for the free energy penalty associated with the exposure of the hydrophobic patch in the active conformation.
Sequence analyses show that the asymmetric dimer interface is conserved in the two other catalytically active members in the family, ErbB2 and ErbB4, suggesting that ErbB2 and ErbB4 are likely to use the same activation mechanism. This is confirmed by a recent structural study showing that ErbB4 also forms an asymmetric dimer essentially identical to that of EGFR and the dimer is important for ErbB4 activation [28]. The conserved asymmetric dimer interface also underlies the ability of different members in the EGFR family to form heterodimers to activate one another [44]. An exception is ErbB3, which shows high sequence homology to other members in the family at the C-lobe face of the dimer interface but not at the N-lobe face. Unlike other members in the family, ErbB3 is a catalytically inactive kinase with several key residues in the active site mutated. The conserved C-lobe face allows ErbB3 to function as a cyclin-like activator for other members in the family through heterodimerization, nicely explaining the functional role of this catalytically dead kinase. The lack of conservation on the N-lobe face of ErbB3 is likely due to loss of selective pressure, since ErbB3 does not need to assume the position of monomer A (the kinase monomer that is activated).
4. Effect of the Activation Loop Conformation on the Kinase Domain
The centrally located activation loop is a common critical switch for protein kinase regulation [13]. The activation loops in active kinases all adopt the essentially identical open and extended conformation that is capable of supporting substrate peptide binding (Figure 1). The CDK/Src-like conformation of the activation loop is one of several different inactive conformations observed for kinases, which contribute to kinase inhibition by blocking substrate binding and/or stabilizing the inactive conformation. As mentioned above, phosphorylation of one or several tyrosine residues in the activation loop of kinases such as Src and Insulin receptor is required for conversion from the inactive to the active conformation [13, 45].
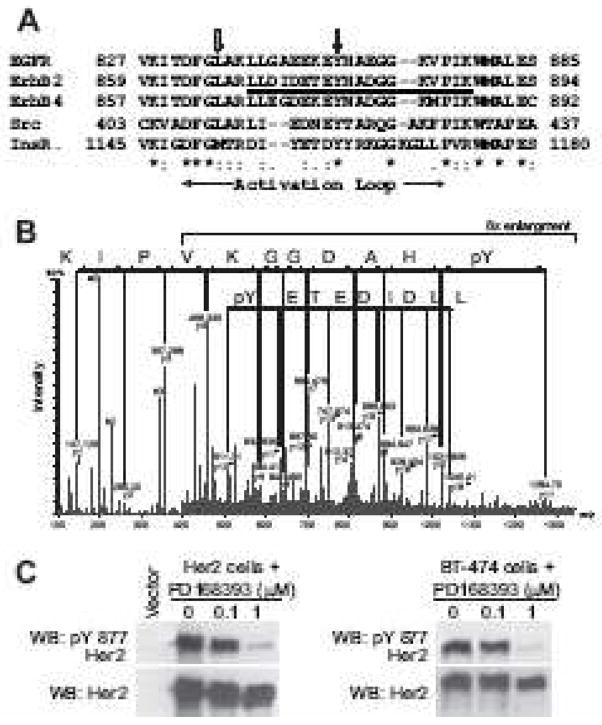
All four human ErbB kinases have a Tyr residue in their activation loop (Fig 2A), prompting the initial question of whether ErbB kinases could also use activation loop phosphorylation as a regulatory mechanism. Multiple mutational studies in transfected cell lines have been performed to address this question [14, 46–48]. These mutational studies have provided conflicting answers with two studies showing that the EGFR Tyr845Phe and ErbB2 Tyr877Phe mutations have a demonstrable phenotype and two studies stating that these mutations are without effect. A recent study using purified EGFR kinase domain found that there is only a subtle difference in the kinase activity of WT and Tyr845Phe kinase domains [26].
Figure 2.
Identification of Tyr877 phosphorylation site in ErbB2 kinase domain from [49]. (A) Multiple sequence alignment of the kinase domains of human EGFR, ErbB2, ErbB4, Src, and insulin receptor. The phosphopeptide identified by MS in (B) is underlined. Black and gray arrows mark the conserved tyrosine residue and EGFR Leu834, respectively. The activation loop is indicated by horizontal arrows. (B) MS/MS spectra of the peptide, LLDIDETE(pY)HADGGKVPIK, bearing the phosphotyrosine residue at Tyr877 of ErbB2. The charge state of the parent ion is +3, thus yielding both +1(upper line) and +2 charged daughter ions (lower line) on fragmentation. The mass difference corresponding to the phosphotyrosine residue, y11 minus y10 ion, is seen in both +1 and +2 charged fragment series. (C) ErbB2-transfected NIH3T3 fibroblasts or BT-474 breast cancer cells which overexpress endogenous ErbB2 were treated with the ErbB2 inhibitor, PD168393, for 1 hour and immunoblotting performed with phospho-Tyr877 specific ErbB2 antibody or anti-total ErbB2 antibody.
In contrast, three proteomic studies of the ErbB2 signaling pathway have all observed that the ErbB2 activation loop tyrosine residue (Tyr877) is phosphorylated and its phosphorylation status correlates with ErbB2 activity level in intact cells (Fig. 2) [49–51]. Evidence for ErbB2 activation loop phosphorylation on Tyr877 has been obtained using two independent methods: mass spectrometry (Fig. 2B) and immunoblotting with a phospho-specific antibody (Fig. 2C). ErbB2 Tyr877 phosphorylation has also been demonstrated in multiple cell lines [49–51]. Treatment of ErbB2 transfected fibroblasts with an ErbB2 kinase inhibitor markedly reduced the level of Tyr877 phosphorylation (Fig. 2C) [49]. Similarly, a rapid increase in the phosphorylation of ErbB2 Tyr877 was observed upon EGF treatment of a human mammary epithelial cell line that co-expressed EGFR and ErbB2 [51]. This EGF-induced phosphorylation of ErbB2 is likely the result of a heterodimer formation between EGFR and ErbB2.
While structural studies have shown that the activation loops in both the EGFR and ErbB4 kinase domains are able to achieve the active conformation in the absence of phosphorylation, it cannot be ruled out that activation loop phosphorylation can stabilize the active conformation and contribute to the activation of kinase activity. Further functional studies remain to be performed to pinpoint the mechanistic role of activation loop phosphorylation in the regulation of the ErbB kinases.
5. Regulation of the kinase by the juxtamembrane region and the C-terminal tail
As with many receptor tyrosine kinases [52–54], the activity of the EGFR kinase domain is regulated by the intracellular juxtamembrane segment and the C-terminal tail. The C-terminal portion of the juxtamembrane region (residues 673–685, also referred to as the N-terminal extension of the kinase domain) is involved directly in the asymmetric activation dimer interface (Figure 1), and mutations at this region abolish dimerization-induced kinase activation [26]. The N-terminal portion of the juxtamembrane segment contains a highly positively charged segment (residues 645–657), which has been shown to be required for EGFR receptor dimerization and activation [55]. A recent study shows that this portion of the juxtamembrane region is involved directly in regulating the formation of the allosteric asymmetric kinase domain dimer [43]. The precise mechanism of how this is achieved awaits a structural study of this portion of the juxtamembrane region in the context of the asymmetric kinase dimer.
The regulatory role for the C-terminal tail of EGFR was suggested by various deletion mutations at the C-terminal tail, which can increase autophosphorylation and/or transforming activity of the receptor [56, 57]. A portion of the C-terminal tail (residues 961–998) immediately following the kinase domain has been included in the EGFR kinase domain proteins used for structural and biochemical studies [15, 22, 26, 27]. In vitro kinase assays show that removal of part of the tail (965–998) results in higher activation levels of the kinase domain by attaching to lipid vesicles, suggesting that the tail has a direct regulatory effect on the kinase domain [26].
Crystal structures show that this portion of the tail makes extensive interactions with the kinase domain [15, 22, 26, 27, 37]. The tail makes contacts with both the C-lobe and the N-lobe of the kinase domain in all these structures, although it appears to be rather flexible, and the details of the interactions differ in one structure from another. Of particular note, residues 971–979 in the tail in the lapatinib bound structure forms a short helix and blocks the kinase active site, which may partially explain the autoinhibitory effect of the tail [22]. Residues 982–994 wraps around the backside of the kinase N-lobe, and in the active crystal form, this part of the tail mediates the symmetric dimer mentioned above, which has been proposed to play an autoinhibitory role [40, 41]. However, given that the kinase domain in this dimer adopts the active conformation and the active site is available for substrate binding, it is not clear how this dimeric configuration causes autoinhibition at present.
6. MIG6-mediated feedback inhibition
Attenuation of the signal in a timely manner is a critical component of appropriate signal transduction. In addition to the well-characterized mechanisms for downregulation of EGFR signaling such as internalization, dephosphorylation and degradation, recent studies identified MIG6 (mitogen-induced gene 6; also named gene 33 and RALT, receptor associated late transducer) as a feedback inhibitor of EGFR family members that operates through a distinct mechanism [27, 58–63]. EGFR or ErbB2 activation induces the expression of MIG6, which then binds directly to the cytoplasmic region of the receptors and inhibits their activity. Studies using mouse models and cancer cells have shown that MIG6-mediated inhibition of EGFR is physiologically important, and loss of MIG6 promotes tumor formation [64–67].
MIG6 is a cytoplasmic protein of ~50 kDa, only the C-terminal portion of which is involved in binding EGFR [59, 61–63]. This region of MIG6 displays sequence similarity to the non-catalytic region of a non-receptor tyrosine kinase ACK1, which also binds to EGFR [68]. A recent crystallographic study of the EGFR kinase domain and peptides derived from the EGFR-binding region in MIG6 show that a 25-residues segment (residues 337–361, MIG6 segment 1) in MIG6 binds to the bottom of the kinase C-lobe, far away from the active site (Fig. 1) [27]. The EGFR kinase domain in this structure adopts the same CDK/Src-like inactive conformation, once again confirming that this is the preferred inactive state of the kinase. The footprint of MIG6 segment 1 on the EGFR kinase domain overlaps largely with the C-lobe face of the asymmetric activating dimer interface, suggesting that binding of MIG6 segment 1 can inhibit EGFR by preventing the formation of the asymmetric dimer. Biochemical experiments confirmed this binding mode, showing that MIG6 segment 1 only inhibits kinase activity of EGFR in the context of asymmetric dimer, but not the basal activity of the monomeric kinase in solution. This inhibition mechanism identifies a distinct approach towards highly specific inhibitors for the ErbB kinases, as it binds to a region that is conserved only in this family of kinases.
The EGFR-Mig6 structure, however, does not explain the preferential binding of MIG6 to EGFR in the active state [59, 60, 62], as the region on the kinase domain that binds to MIG6 segment 1 remains essentially identical in the inactive and active conformations of the kinase [27]. Further studies have shown that a segment C-terminal to segment 1 (segment 2, residues 362–412) confers EGFR activation state sensitivity on MIG6. This was initially suggested by two observations. The first is that the C-terminus of segment 1 is located close to a channel leading to the binding site of several peptidic kinase inhibitors that bind specifically to the kinase active site [69, 70]. The second is that segment 2 in MIG6 displays high sequence similarity to the corresponding region in ACK1, which also binds to EGFR in an activation sensitive manner [68]. In vitro kinase assays show that a longer peptide spanning both segment 1 and 2 displays a significantly higher inhibitory potency to a constitutively active form of the EGFR kinase domain Leu834Arg, but not the autoinhibited wild type kinase. The structural details for the EGFR kinase domain/MIG6 segment 2 interactions are not known at the present. It likely binds to the active site or the N-lobe of the EGFR kinase which shows a large conformational difference between the inactive and active conformations.
7. Small Molecule Tyrosine Kinase Inhibitors
Small molecule kinase inhibitors targeting the ErbB family have been the subject of intensive drug development and clinical trials in cancer therapy [71]. Three drugs have received FDA approval for use in cancer patients and numerous more are moving through the clinical trials pipeline. The FDA-approved drugs include gefitinib (Iressa) and erlotinib (Tarceva), which are selective for EGFR, and lapatinib (Tykerb), which targets both EGFR and ErbB2. Crystal structures of more than seven inhibitor–ErbB kinase complexes have been solved [15, 22, 28, 29, 37, 72] and we will focus on three of them. These three structures, shown in Fig.3, represent the active and CDK/Src-like inactive conformations of EGFR with a reversible inhibitor bound and the inactive conformation with an irreversible inhibitor bound. All three inhibitors are ATP-mimetics and bind in the ATP-binding site.
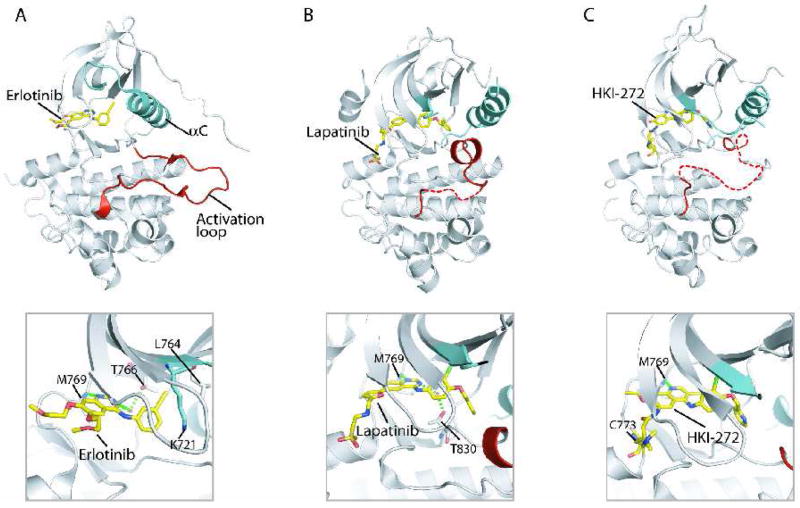
Figure 3.
Structures of EGFR kinase domain bound to small molecule kinase inhibitors. (A) Erlotinib - EGFR structure [15]. (B) Lapatinib - EGFR structure [22]. (A) HKI-272 - EGFR structure [72]. The upper panels show the entire EGFR kinase domain with inhibitor bound, revealing the active (A) and inactive (B and C) conformations of EGFR. The lower panels show close-up views of the bound inhibitors.
The first inhibitor-kinase structure solved was of erlotinib bound to EGFR [15]. Erlotinib binds to the ATP binding site of EGFR with the kinase domain adopting an active structure (Fig. 3A). The two aromatic rings of erlotinib form an interplanar angle of 42°, which directs the acetylene group into a pocket created by Thr766, Lys721, and Leu764. Nitrogen N1 of the quinazoline ring of erlotinib makes a hydrogen bond with Met769 amide nitrogen and N3 of the quinazoline ring makes a water-mediated hydrogen bond with Thr766 side chain. Binding of gefitinib to EGFR is essentially the same as erlotinib [37].
In contrast, lapatinib binds to the EGFR kinase in the CDK/Src-like inactive conformation (Fig. 3B) [22]. Like erlotinib, the N1 of the quinazoline ring of lapatinib makes a hydrogen bond with Met769 amide nitrogen but N3 of the quinazoline ring makes a water-mediated hydrogen bond with a different Thr residue (Thr830). Lapatinib contains a bulky 3-fluorobenzyl-oxy group which is accommodated by an extra hydrophobic pocket created by the outwardly displayed helix αC in the CDK/Src-like conformation [22, 25]. Simple modeling shows that binding of lapatinib to the active kinase would result in steric clash, likely explaining why lapatinib binds preferentially to the inactive structure.
HKI-272 is an irreversible inhibitor of the ErbB kinases. It possesses an acrylamide-like or crotonamide moiety, which can form a covalent bond with nucleophiles such as thiol groups. When bound in the ATP binding pocket of EGFR, the crotonamide group forms a bond with Cys 773 located in the hinge region of the kinase (Fig. 3C) [72]. Lung cancer cases bearing the gefitinib-sensitizing EGFR mutations can become gefitinib-resistant through the acquisition of a second mutation Thr766Met. Even in the presence of this mutation, these tumors remain sensitive to HKI-272 [73, 74]. The crystal structure of HKI-272 bound to EGFR Thr766Met shows the kinase domain in an inactive structure with helix αC displaced outward, similar to the lapatinib-bound structure [72]. The covalent bond between Cys773 and the crotonamide group of HKI-272 was seen, and hydrogen bond interactions of the quinazoline ring of HKI-272 with hinge region residues on EGFR were similar to other inhibitor bound structures.
Despite the high degree of sequence similarity in the active site of different members in the ErbB family, the ErbB kinases show strikingly different sensitivity to these kinase inhibitors (Table 1). Erlotinib and gefitinib are potent inhibitors of EGFR, but are much weaker inhibitors of ErbB2 and ErbB4. In contrast, lapatinib has similar Ki’s for EGFR and ErbB2 but is 100-fold less potent inhibitor of ErbB4 [22]. Extensive structure-activity relationship studies have been conducted on the chemical structure of the small molecule inhibitors to tailor specificity towards various kinases (reviewed in [75]), but the amino acid residues in the ErbB kinase domains which cause these differences in inhibitor specificity remain unclear. Comparison of the lapatinib-EGFR structure with that of lapatinib-ErbB4 shows that the residues that contact lapatinib are conserved between EGFR and ErbB4 [22, 28].
Table 1.
Inhibition Constants (Ki) for Erlotinib, Gefitinib, and Lapatinib (from reference [22]).
| EGFR | ErbB2 | ErbB4 | |
|---|---|---|---|
| Erlotinib | 0.7 nM | 1.0 μM | 1.5 μM |
| Gefitinib | 0.4 nM | 870 nM | 1.1 μM |
| Lapatinib | 3 nM | 13 nM | 347 nM |
An alternative mechanism that may lead to differential inhibitor specificity is the dynamic equilibrium of the kinase between different conformations. This mechanism has been used to explain the strong preference of imatinib (Gleevec) for the Abl kinase over the closely related Src family kinases [76]. Considering the different conformational preference of the kinase inhibitors discussed above, the dynamic equilibrium between the active, CDK/Src-like and possibly other inactive conformations of the ErbB kinase domains may have an important role in determining their sensitivity to these inhibitors. This notion is supported by recent studies showing that certain activation mutations found in lung cancers are more sensitive toward gefitinib, which prefers the active conformation (see below).
8. ErbB mutations in Lung Cancer
Mutations in the EGFR kinase domain have been identified in approximately 10% of human non-small cell lung cancer cases and produce cancers that are very sensitive to treatment with gefitinib and erlotinib [34–36]. The most common mutations observed in the EGFR kinase domain are 1) Leu834Arg and Leu837Gln point mutations in the activation loop, 2) small in-frame deletions in exon 19 which commonly delete Leu723-Ala726 but can affect a variable number of codons ranging between residues 722 to 735, 3) Gly695X point mutations in the P-loop (X=Cys, Ser, or Ala), and 4) small in-frame insertions or duplications in exon 20 [33]. The deletions in exon 19 and the insertions in exon 20 fall on either side of the αC helix. These mutations are activating and cause oncogenic transformation of cell lines [37, 77]. Similar exon 20 in-frame insertion mutations in ErbB2 have been found in 1.6% of non-small cell lung cancer cases and these insertions add amino acids to the C-terminal end of the ErbB2 αC helix [78]. In cell culture, the EGFR Leu834Arg, Gly695X, and exon 19 deletion mutations result in enhanced sensitivity to gefitinib and erlotinib, but the EGFR or ErbB2 exon 20 insertions result in resistance to these kinase inhibitors [34, 79, 80].
As described above, Leu834 plays an important role in stabilizing the CDK/Src-like inactive conformation of the EGFR kinase domain. The Leu834Arg mutation likely destabilizes the CDK/Src-like conformation and shifts the equilibrium toward the active conformation, leading to kinase activation as well as higher sensitivity to gefitinib [26, 37]. This is supported by the fact that the Leu834Arg EGFR kinase domain shows >15-fold more activity than wild-type EGFR in in vitro kinase assays [26, 37, 38].
Similarly, the Gly695Ser mutation activates the kinase by ~10 fold [37]. Gly695 is located at the P-loop of the kinase, which adopts a conformation that favors a glycine at the 695 position and plays a role in stabilizing the CDK/Src-like conformation of the kinase. The Gly695Ser mutation destabilizes this conformation of the P-loop, and in turn destabilizes the CDK/Src-like conformation and favors the active conformation, [37]. Consistently, crystal structures of the Leu834Arg and Gly695Ser mutations demonstrate that both mutations are easily accommodated in the active conformation of EGFR.
The in-frame deletions in EGFR exon 19 involve residues that are located in the loop between strand β3 and helix αC. The activating effects of these mutations are predicted to be due to destabilization of the CDK/Src-like conformation, as the β3-αC loop packs against and stabilizes the short helix formed by the activation loop [26]. Thus, by disrupting the inactive conformation, these oncogenic mutations are able to drive cell proliferation in the lungs to produce human lung cancers.
9. Concluding Remarks
The past 5–7 years have significantly enhanced our knowledge of the ErbB kinase domain. Important questions that remain to be addressed include the structural relationship between the kinase domain and the juxtamembrane domain and the C-terminal tail, and how exactly they participate in regulation of the kinase domain. Further understanding of the role of the activation loop in kinase regulation is also required. Finally, the allosteric activation and inhibition mechanisms revealed by the asymmetric dimer and EGFR-Mig6 structures pave the way to new modes of targeting the ErbB family. The potential of developing peptidomimetic or allosteric drugs will offer significant treatment advances for cancer and other diseases that involve dysregulation of the ErbB tyrosine kinase family.
Acknowledgments
We are grateful for critical reading by Linda Pike, Daniel Leahy, and Philip Cole. RB is supported by the NIH (K22 Transition Career Development Award CA128951). XZ is a Virginia Murchison Linthicum Scholar in Medical Research at University of Texas Southwestern Medical Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schreiber AB, Libermann TA, Lax I, Yarden Y, Schlessinger J. Biological role of epidermal growth factor-receptor clustering. Investigation with monoclonal anti-receptor antibodies. J Biol Chem. 1983;258:846–853. [PubMed] [Google Scholar]
- 2.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 3.Ushiro H, Cohen S. Identification of phosphotyrosine as a product of epidermal growth factor-activated protein kinase in A-431 cell membranes. J Biol Chem. 1980;255:8363–8365. [PubMed] [Google Scholar]
- 4.Yarden Y, Schlessinger J. Self-phosphorylation of epidermal growth factor receptor: evidence for a model of intermolecular allosteric activation. Biochemistry. 1987;26:1434–1442. doi: 10.1021/bi00379a034. [DOI] [PubMed] [Google Scholar]
- 5.Yarden Y, Schlessinger J. Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry. 1987;26:1443–1451. doi: 10.1021/bi00379a035. [DOI] [PubMed] [Google Scholar]
- 6.Cho HS, Leahy DJ. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 2002;297:1330–1333. doi: 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- 7.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, Jorissen RN, Nice EC, Burgess AW, Ward CW. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 8.Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 9.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr, Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 11.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Kofler M, Jorissen RN, Nice EC, Burgess AW, Ward CW. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell. 2003;11:495–505. doi: 10.1016/s1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 12.Hubbard SR. Structural analysis of receptor tyrosine kinases. Prog Biophys Mol Biol. 1999;71:343–358. doi: 10.1016/s0079-6107(98)00047-9. [DOI] [PubMed] [Google Scholar]
- 13.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 14.Gotoh N, Tojo A, Hino M, Yazaki Y, Shibuya M. A highly conserved tyrosine residue at codon 845 within the kinase domain is not required for the transforming activity of human epidermal growth factor receptor. Biochem Biophys Res Commun. 1992;186:768–774. doi: 10.1016/0006-291x(92)90812-y. [DOI] [PubMed] [Google Scholar]
- 15.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 16.Canals F. Signal transmission by epidermal growth factor receptor: coincidence of activation and dimerization. Biochemistry. 1992;31:4493–4501. doi: 10.1021/bi00133a016. [DOI] [PubMed] [Google Scholar]
- 17.Posner I, Engel M, Levitzki A. Kinetic model of the epidermal growth factor (EGF) receptor tyrosine kinase and a possible mechanism of its activation by EGF. J Biol Chem. 1992;267:20638–20647. [PubMed] [Google Scholar]
- 18.Erneux C, Cohen S, Garbers DL. The kinetics of tyrosine phosphorylation by the purified epidermal growth factor receptor kinase of A-431 cells. J Biol Chem. 1983;258:4137–4142. [PubMed] [Google Scholar]
- 19.Yarden Y, Schlessinger J. The EGF receptor kinase: evidence for allosteric activation and intramolecular self-phosphorylation. Ciba Found Symp. 1985;116:23–45. doi: 10.1002/9780470720974.ch3. [DOI] [PubMed] [Google Scholar]
- 20.Mohammadi M, Honegger A, Sorokin A, Ullrich A, Schlessinger J, Hurwitz DR. Aggregation-induced activation of the epidermal growth factor receptor protein tyrosine kinase. Biochemistry. 1993;32:8742–8748. doi: 10.1021/bi00085a004. [DOI] [PubMed] [Google Scholar]
- 21.Wedegaertner PB, Gill GN. Activation of the purified protein tyrosine kinase domain of the epidermal growth factor receptor. J Biol Chem. 1989;264:11346–11353. [PubMed] [Google Scholar]
- 22.Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, Alligood KJ, Rusnak DW, Gilmer TM, Shewchuk L. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 23.De Bondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim SH. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- 24.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 25.Schindler T, Sicheri F, Pico A, Gazit A, Levitzki A, Kuriyan J. Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol Cell. 1999;3:639–648. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–744. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu C, Tarrant MK, Choi SH, Sathyamurthy A, Bose R, Banjade S, Pal A, Bornmann WG, Lemmon MA, Cole PA, Leahy DJ. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16:460–467. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood ER, Shewchuk LM, Ellis B, Brignola P, Brashear RL, Caferro TR, Dickerson SH, Dickson HD, Donaldson KH, Gaul M, Griffin RJ, Hassell AM, Keith B, Mullin R, Petrov KG, Reno MJ, Rusnak DW, Tadepalli SM, Ulrich JC, Wagner CD, Vanderwall DE, Waterson AG, Williams JD, White WL, Uehling DE. 6-Ethynylthieno[3,2-d]- and 6-ethynylthieno[2,3-d]pyrimidin-4-anilines as tunable covalent modifiers of ErbB kinases. Proc Natl Acad Sci U S A. 2008;105:2773–2778. doi: 10.1073/pnas.0708281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levinson NM, Kuchment O, Shen K, Young MA, Koldobskiy M, Karplus M, Cole PA, Kuriyan J. A Src-like inactive conformation in the abl tyrosine kinase domain. PLoS Biol. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deindl S, Kadlecek TA, Brdicka T, Cao X, Weiss A, Kuriyan J. Structural basis for the inhibition of tyrosine kinase activity of ZAP-70. Cell. 2007;129:735–746. doi: 10.1016/j.cell.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 32.Min X, Lee BH, Cobb MH, Goldsmith EJ. Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure. 2004;12:1303–1311. doi: 10.1016/j.str.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 33.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006;118:257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 34.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 35.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 36.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carey KD, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, Park F, Haley JD, Gibson N, Sliwkowski MX. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 39.Choi SH, Mendrola JM, Lemmon MA. EGF-independent activation of cell-surface EGF receptors harboring mutations found in gefitinib-sensitive lung cancer. Oncogene. 2007;26:1567–1576. doi: 10.1038/sj.onc.1209957. [DOI] [PubMed] [Google Scholar]
- 40.Landau M, Ben-Tal N. Dynamic equilibrium between multiple active and inactive conformations explains regulation and oncogenic mutations in ErbB receptors. Biochim Biophys Acta. 2008;1785:12–31. doi: 10.1016/j.bbcan.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Landau M, Fleishman SJ, Ben-Tal N. A putative mechanism for downregulation of the catalytic activity of the EGF receptor via direct contact between its kinase and C-terminal domains. Structure. 2004;12:2265–2275. doi: 10.1016/j.str.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Groenen LC, Walker F, Burgess AW, Treutlein HR. A model for the activation of the epidermal growth factor receptor kinase involvement of an asymmetric dimer? Biochemistry. 1997;36:3826–3836. doi: 10.1021/bi9614141. [DOI] [PubMed] [Google Scholar]
- 43.Thiel KW, Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc Natl Acad Sci U S A. 2007;104:19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 45.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. Embo J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segatto O, Lonardo F, Pierce JH, Bottaro DP, Di Fiore PP. The role of autophosphorylation in modulation of erbB-2 transforming function. New Biol. 1990;2:187–195. [PubMed] [Google Scholar]
- 47.Zhang HT, O’Rourke DM, Zhao H, Murali R, Mikami Y, Davis JG, Greene MI, Qian X. Absence of autophosphorylation site Y882 in the p185neu oncogene product correlates with a reduction of transforming potential. Oncogene. 1998;16:2835–2842. doi: 10.1038/sj.onc.1201820. [DOI] [PubMed] [Google Scholar]
- 48.Biscardi JS, Maa MC, Tice DA, Cox ME, Leu TH, Parsons SJ. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem. 1999;274:8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 49.Bose R, Molina H, Patterson AS, Bitok JK, Periaswamy B, Bader JS, Pandey A, Cole PA. Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc Natl Acad Sci U S A. 2006;103:9773–9778. doi: 10.1073/pnas.0603948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukherji M, Brill LM, Ficarro SB, Hampton GM, Schultz PG. A phosphoproteomic analysis of the ErbB2 receptor tyrosine kinase signaling pathways. Biochemistry. 2006;45:15529–15540. doi: 10.1021/bi060971c. [DOI] [PubMed] [Google Scholar]
- 51.Wolf-Yadlin A, Kumar N, Zhang Y, Hautaniemi S, Zaman M, Kim HD, Grantcharova V, Lauffenburger DA, White FM. Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol Syst Biol. 2006;2:54. doi: 10.1038/msb4100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiara F, Bishayee S, Heldin CH, Demoulin JB. Autoinhibition of the Platelet-derived Growth Factor {beta}-Receptor Tyrosine Kinase by Its C-terminal Tail. J Biol Chem. 2004;279:19732–19738. doi: 10.1074/jbc.M314070200. [DOI] [PubMed] [Google Scholar]
- 53.Shewchuk LM, Hassell AM, Ellis B, Holmes WD, Davis R, Horne EL, Kadwell SH, McKee DD, Moore JT. Structure of the Tie2 RTK domain: self-inhibition by the nucleotide binding loop, activation loop, and C-terminal tail. Structure Fold Des. 2000;8:1105–1113. doi: 10.1016/s0969-2126(00)00516-5. [DOI] [PubMed] [Google Scholar]
- 54.Hubbard SR. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5:464–471. doi: 10.1038/nrm1399. [DOI] [PubMed] [Google Scholar]
- 55.Aifa S, Aydin J, Nordvall G, Lundstrom I, Svensson SP, Hermanson O. A basic peptide within the juxtamembrane region is required for EGF receptor dimerization. Exp Cell Res. 2005;302:108–114. doi: 10.1016/j.yexcr.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 56.Chang CM, Shu HK, Ravi L, Pelley RJ, Shu H, Kung HJ. A minor tyrosine phosphorylation site located within the CAIN domain plays a critical role in regulating tissue-specific transformation by erbB kinase. J Virol. 1995;69:1172–1180. doi: 10.1128/jvi.69.2.1172-1180.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frederick L, Wang XY, Eley G, James CD. Diversity and Frequency of Epidermal Growth Factor Receptor Mutations in Human Glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 58.Wick M, Burger C, Funk M, Muller R. Identification of a novel mitogen-inducible gene (mig-6): regulation during G1 progression and differentiation. Exp Cell Res. 1995;219:527–535. doi: 10.1006/excr.1995.1261. [DOI] [PubMed] [Google Scholar]
- 59.Fiorentino L, Pertica C, Fiorini M, Talora C, Crescenzi M, Castellani L, Alema S, Benedetti P, Segatto O. Inhibition of ErbB-2 mitogenic and transforming activity by RALT, a mitogen-induced signal transducer which binds to the ErbB-2 kinase domain. Mol Cell Biol. 2000;20:7735–7750. doi: 10.1128/mcb.20.20.7735-7750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hackel PO, Gishizky M, Ullrich A. Mig-6 is a negative regulator of the epidermal growth factor receptor signal. Biol Chem. 2001;382:1649–1662. doi: 10.1515/BC.2001.200. [DOI] [PubMed] [Google Scholar]
- 61.Anastasi S, Baietti MF, Frosi Y, Alema S, Segatto O. The evolutionarily conserved EBR module of RALT/MIG6 mediates suppression of the EGFR catalytic activity. Oncogene. 2007;26:7833–7846. doi: 10.1038/sj.onc.1210590. [DOI] [PubMed] [Google Scholar]
- 62.Anastasi S, Fiorentino L, Fiorini M, Fraioli R, Sala G, Castellani L, Alema S, Alimandi M, Segatto O. Feedback inhibition by RALT controls signal output by the ErbB network. Oncogene. 2003;22:4221–4234. doi: 10.1038/sj.onc.1206516. [DOI] [PubMed] [Google Scholar]
- 63.Xu D, Makkinje A, Kyriakis JM. Gene 33 Is an Endogenous Inhibitor of Epidermal Growth Factor (EGF) Receptor Signaling and Mediates Dexamethasone-induced Suppression of EGF Function. J Biol Chem. 2005;280:2924–2933. doi: 10.1074/jbc.M408907200. [DOI] [PubMed] [Google Scholar]
- 64.Anastasi S, Sala G, Huiping C, Caprini E, Russo G, Iacovelli S, Lucini F, Ingvarsson S, Segatto O. Loss of RALT/MIG-6 expression in ERBB2-amplified breast carcinomas enhances ErbB-2 oncogenic potency and favors resistance to Herceptin. Oncogene. 2005;24:4540–4548. doi: 10.1038/sj.onc.1208658. [DOI] [PubMed] [Google Scholar]
- 65.Ferby I, Reschke M, Kudlacek O, Knyazev P, Pante G, Amann K, Sommergruber W, Kraut N, Ullrich A, Fassler R, Klein R. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med. 2006;12:568–573. doi: 10.1038/nm1401. [DOI] [PubMed] [Google Scholar]
- 66.Zhang YW, Staal B, Su Y, Swiatek P, Zhao P, Cao B, Resau J, Sigler R, Bronson R, Vande Woude GF. Evidence that MIG-6 is a tumor-suppressor gene. Oncogene. 2007;26:269–276. doi: 10.1038/sj.onc.1209790. [DOI] [PubMed] [Google Scholar]
- 67.Ballaro C, Ceccarelli S, Tiveron C, Tatangelo L, Salvatore AM, Segatto O, Alema S. Targeted expression of RALT in mouse skin inhibits epidermal growth factor receptor signalling and generates a Waved-like phenotype. EMBO Rep. 2005;6:755–761. doi: 10.1038/sj.embor.7400458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen F, Lin Q, Gu Y, Childress C, Yang W. Activated Cdc42-associated Kinase 1 Is a Component of EGF Receptor Signaling Complex and Regulates EGF Receptor Degradation. Mol Biol Cell. 2007;18:732–742. doi: 10.1091/mbc.E06-02-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lei M, Robinson MA, Harrison SC. The active conformation of the PAK1 kinase domain. Structure. 2005;13:769–778. doi: 10.1016/j.str.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 70.Depetris RS, Hu J, Gimpelevich I, Holt LJ, Daly RJ, Hubbard SR. Structural basis for inhibition of the insulin receptor by the adaptor protein Grb14. Mol Cell. 2005;20:325–333. doi: 10.1016/j.molcel.2005.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baselga J. Targeting tyrosine kinases in cancer: the second wave. Science. 2006;312:1175–1178. doi: 10.1126/science.1125951. [DOI] [PubMed] [Google Scholar]
- 72.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kwak EL, Sordella R, Bell DW, Godin-Heymann N, Okimoto RA, Brannigan BW, Harris PL, Driscoll DR, Fidias P, Lynch TJ, Rabindran SK, McGinnis JP, Wissner A, Sharma SV, Isselbacher KJ, Settleman J, Haber DA. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fry DW. Mechanism of action of erbB tyrosine kinase inhibitors. Exp Cell Res. 2003;284:131–139. doi: 10.1016/s0014-4827(02)00095-2. [DOI] [PubMed] [Google Scholar]
- 76.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 77.Greulich H, Chen TH, Feng W, Janne PA, Alvarez JV, Zappaterra M, Bulmer SE, Frank DA, Hahn WC, Sellers WR, Meyerson M. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shigematsu H, Takahashi T, Nomura M, Majmudar K, Suzuki M, Lee H, Wistuba, Fong KM, Toyooka S, Shimizu N, Fujisawa T, Minna JD, Gazdar AF. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 79.Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, Carpenter G, Gazdar AF, Muthuswamy SK, Arteaga CL. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 80.Minami Y, Shimamura T, Shah K, LaFramboise T, Glatt KA, Liniker E, Borgman CL, Haringsma HJ, Feng W, Weir BA, Lowell AM, Lee JC, Wolf J, Shapiro GI, Wong KK, Meyerson M, Thomas RK. The major lung cancer-derived mutants of ERBB2 are oncogenic and are associated with sensitivity to the irreversible EGFR/ERBB2 inhibitor HKI-272. Oncogene. 2007;26:5023–5027. doi: 10.1038/sj.onc.1210292. [DOI] [PubMed] [Google Scholar]