Abstract
Virtually all cellular processes are carried out by dynamic molecular assemblies or multiprotein complexes, the compositions of which are largely undefined. They cannot be predicted solely from bioinformatics analyses nor are there well defined techniques currently available to unequivocally identify protein complexes (PCs). To address this issue, we attempted to directly determine the identity of PCs from native microbial biomass using Pyrococcus furiosus, a hyperthermophilic archaeon that grows optimally at 100 °C, as the model organism. Novel PCs were identified by large scale fractionation of the native proteome using non-denaturing, sequential column chromatography under anaerobic, reducing conditions. A total of 967 distinct P. furiosus proteins were identified by mass spectrometry (nano LC-ESI-MS/MS), representing ∼80% of the cytoplasmic proteins. Based on the co-fractionation of proteins that are encoded by adjacent genes on the chromosome, 106 potential heteromeric PCs containing 243 proteins were identified, only 20 of which were known or expected. In addition to those of unknown function, novel and uncharacterized PCs were identified that are proposed to be involved in the metabolism of amino acids (10), carbohydrates (four), lipids (two), vitamins and metals (three), and DNA and RNA (nine). A further 30 potential PCs were classified as tentative, and the remaining potential PCs (13) were classified as weakly interacting. Some major advantages of native biomass fractionation for PC identification are that it provides a road map for the (partial) purification of native forms of novel and uncharacterized PCs, and the results can be utilized for the recombinant production of low abundance PCs to provide enough material for detailed structural and biochemical analyses.
The repository of sequenced genomes is now above 850 (Genomes Online Database). Consequently there is a tremendous need to determine the function of gene products and identify groups of proteins that work together as complexes in distinct cellular processes. Genome-wide functional analyses suggest that there are 200–300 core biological functions that are essential to life (1). More often than not the functional units are assemblies composed of multiple proteins (2). Many biological processes involve multiprotein complexes that function as large and efficient machines, such as ribosomes (3, 4), flagella (5), and cellulosomes (6). In addition, the supramolecular organization of enzymes of partial or entire metabolic pathways as “metabolons” appears to provide certain advantages, such as substrate channeling (7–9). The identification of protein-protein interactions and functional, stable associations is extremely important in understanding the biology of a cell. However, predicting the nature of such complexes within a single genome, let alone for hundreds of genomes, remains a major challenge.
Although there are currently several methods available to study protein-protein interactions on a genome-wide scale (10–12), each has severe limitations. The in vivo two-hybrid system (13–15) requires tagged proteins, is limited to binary interactions, and is thought to generate a large percentage of false positives (16). The epitope tag affinity purification and tandem affinity purification methods have also been used extensively (17–21), but the tags can disrupt native protein-protein interactions, and the methods tend to be biased toward proteins that interact with high affinity and/or proteins of high abundance (10). The major limitation with all of these approaches is that they require genetic manipulation of the target organism, an ability limited to only a few well studied systems. Non-genetic techniques to identify protein-protein interactions include co-immunoaffinity precipitation to capture endogenous protein complexes, but this is not a genome-wide approach as it requires highly specific antibodies made against purified proteins (22). Two-dimensional blue native/SDS-PAGE and clear native-PAGE are also two widely used techniques that do not require genetic manipulation and allow for the analysis of protein complexes on a proteome-wide scale in a single experiment (23–26). However, they are limited in their dynamic range and typically identify only high abundance proteins (27).
The goal of this research is to develop a global method to identify novel protein complexes (PCs)1 independent of a genetic system and applicable to any organism with available native biomass. The approach involves multistep, non-denaturing column chromatography where the co-fractionation of proteins is used to identify potential complexes. As a model system we use the hyperthermophilic archaeon Pyrococcus furiosus, an anaerobe that grows optimally at 100 °C (28). Its genome sequence (29, 30) contains 2125 ORFs. A universal feature of prokaryotic genomes is the organization of genes into operons, which form basic transcriptional units (31) and are important in functional genomics. Using a neural network, we predicted that 1460 ORFs in the P. furiosus genome are contained within 470 operons (32), 349 of which were validated using DNA microarray data (33, 34). Operons typically encode functionally related proteins, which can include enzymes of the same pathway as well as heteromeric PCs. Herein heteromeric PCs encoded by two or more adjacent genes are referred to as Type 1 PCs, whereas heteromeric PCs encoded by two or more unlinked genes are referred to as Type 2 PCs. This pilot study focused on identifying stable, Type 1 heteromeric PCs in P. furiosus based on the co-fractionation of proteins during sequential column chromatography steps. In addition, a high throughput (HT) system was devised to allow protein identification by nano-LC-ESI-MS/MS. Our long term objective is to develop HT protocols for novel PC identification on a genome-wide basis using limited amounts of biomass.
EXPERIMENTAL PROCEDURES
Preparation of Anaerobic Cell-free Extract
P. furiosus (DSM 3638) was grown under anaerobic, reducing conditions at 90 °C in a 600-liter fermenter on maltose and peptides and harvested in the late log phase (35). The procedures for preparing anaerobic cell-free extract and for running the first chromatography step have been described previously (35). Briefly 300 g of frozen cells were gently lysed by osmotic shock anaerobically under a continuous flow of argon in 900 ml of 50 mm Tris-HCl (pH 8.0) containing 2 mm sodium dithionite as a reductant (Buffer A) and 0.5 μg/ml DNase I to reduce viscosity. The cell lysate was centrifuged at 100,000 × g for 2 h at 18 °C, and the clear supernatant, representing the cytoplasmic fraction, was immediately loaded onto the first column. This and all subsequent columns were run under anaerobic, reducing conditions where all buffers were degassed and maintained under a positive pressure of argon, and all liquid transfers were made using needles and syringes.
Large Scale Anaerobic Fractionation of Cytoplasmic Proteins
First Column (c1) Fractionation—
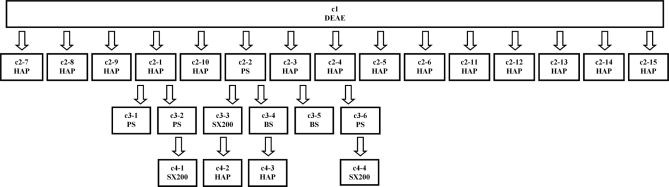
A column tree representing the complete fractionation procedure is shown in Fig. 1. All columns were run using an ÄKTA™ basic automated LC system (GE Healthcare). The P. furiosus cytoplasmic fraction was diluted 4-fold in Buffer A to reduce the ionic strength and was loaded onto a 10 × 20-cm (1.5-liter) column of DEAE-Sepharose Fast Flow (GE Healthcare) that had been equilibrated previously with the same buffer until the effluent was anaerobic and reducing. Unbound protein was washed off the column with Buffer A. Bound proteins were eluted using a linear gradient (15 liters) of 0–500 mm NaCl in Buffer A, and 125-ml anaerobic fractions were collected (120 fractions). Any remaining tightly bound proteins were eluted from the column using 1-liter step gradients of 1, 1.5, and 2 m NaCl, respectively, in Buffer A (high salt washes).
Fig. 1.
Column tree showing the four levels (c1–c4) of chromatography steps used to fractionate P. furiosus cytoplasmic proteins. The cytoplasmic proteins were fractionated using DEAE Fast Flow anion exchange chromatography as the first step (c1). Fractions from the first column were selected and combined to generate 15 pools that were fractionated through subsequent second (c2) and, for some control proteins, third (c3) and fourth (c4) level columns. The chromatography steps included hydroxyapatite (HAP), phenyl-Sepharose (PS), Superdex 200 (SX200), and blue Sepharose (BS), and these are indicated.
Second (c2) Level Column Fractionations—
A subset of 37 gradient fractions from the first column separation were selected using a combination of protein profiles and enzyme activities to create six fraction pools, c1-1 through c1-6. The control proteins and enzyme activities were as follows: c1-1, ferredoxin-NADPH oxidoreductase (FNOR) (36) and rubrerythrin (37); c1-2, 2-ketoisovalerate ferredoxin oxidoreductase (VOR) and pyruvate ferredoxin oxidoreductase (POR) (38); c1-3, aldehyde:ferredoxin oxidoreductase (39) and NADPH-rubredoxin oxidoreductase (40); c1-4, soluble hydrogenase 1 (SH1) (41); c1-5, rubredoxin (42); and c1-6, ferredoxin (Fd) (43). These pools, c1-1 through c1-6, were fractionated through six subsequent c2 columns. The remaining 83 gradient fractions were combined to generate nine additional pools, in this case based solely on SDS-PAGE and protein profiles. These pools, c1-7 through c1-15, were fractionated through nine subsequent c2 columns to give a total of 15 columns at the c2 level (Fig. 1). SDS-PAGE and native PAGE were carried out using precast 4–20% gradient gels (Criterion gel system, Bio-Rad). Protein concentrations were estimated by the method of Bradford (44) using bovine serum albumin as the standard.
Third (c3) and Fourth (c4) Level Column Fractionations—
Six c3 and four c4 level columns were run using fraction pools that were created for the further purification of the control proteins by the published procedures (Fig. 1). Detailed information about the individual c2, c3, and c4 column chromatography steps is summarized in supplemental Table 1.
Protein Identification Using Mass Spectrometry
Protein Digestion—
The steps for denaturation, reduction, alkylation, and digestion were automated for most samples using 96-well plates and a MassPrep robotic liquid handler (Waters, Milford, MA). To each well containing 50 μl of a fraction sample was added 50 μl of pure trifluoroethanol. The plate was heated to 60 °C for 30 min to complete the denaturation process. Disulfide bonds were reduced by adding 10 μl of dithiothreitol (200 mm in 50 mm ammonium bicarbonate) and alkylated by adding 14 μl of iodoacetamide (200 mm). After incubation for 30 min in the dark at 25 °C, the protein was digested for 18 h at 40 °C using 1.5 μg of trypsin (Trypsin Gold mass spectrometry, Promega, Madison, WI). Digestion was halted by lowering the pH to 4 by the addition of 5% formic acid.
Separation of Peptides and Mass Spectrometry—
Two methods were used for separation. A longer gradient was used for more complex samples, whereas a shorter gradient was used in a rapid HT LC-MS/MS analysis of less complex samples. A 96-well plate-based HT method was devised to rapidly separate peptides by reverse phase LC or MS/MS analysis using a two-pump setup. Using an Agilent 1100 microwell autosampler, 8 μl of the digested sample was injected and transported at 50 μl/min (using a quaternary pump) through the dead volume of a microwell autosampler to a C18 trap cartridge on a six-port switching valve. After 3 min, the flow was switched to the nanocapillary pump at a flow rate of 500 nl/min. An analytical column/nanoelectrospray tip (75-μm inner diameter, 6 cm in length) fabricated with a P-2000 (Sutter Instruments) laser puller and packed with C18 resin (5 μm; Agilent Zorbax SB) under high pressure was used to separate the peptides prior to their analysis with an LTQ mass spectrometer in data-dependent MS/MS mode. The reverse phase gradient separation was performed using water and acetonitrile (0.1% formic acid) as the mobile phases. For the HT LC-MS analysis, the gradient consisted of 20–35% acetonitrile over 20 min to 90% over 8 min prior to equilibration in 5% acetonitrile (0.1% formic acid) for 15 min before reinjection. For more complex c1 protein samples, a longer, conventional (CV) gradient elution was used consisting of 5–8% acetonitrile over 5 min to 35% over 113 min, to 55% over 12 min, and then to 98% maintained for 15 min (wash) before re-equilibration at 5% for 15 min.
MS/MS analysis was performed on the LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific Inc.) using 2 kV at the tip. One MS spectrum was followed by four MS/MS scans on the most abundant ions after the application of the dynamic exclusion list. The LTQ software utility extractmsn.exe (Thermo Fisher Scientific Inc., version 3, September 27, ©1997–2007) was used to peak pick and extract peak lists from the tandem mass spectra into peak list (.dta) files. Mascot Daemon was used to run this program externally and to submit the searches to Mascot (Matrix Science Ltd.) for protein identification. The Mascot searches were conducted at a 95% confidence level using tryptic peptides derived from the public National Center for Biotechnology Information (NCBI) annotation of the P. furiosus genome (NC_003413, December 2, 2007) (29, 30) containing a total of 2125 protein sequences. The false positive rate was estimated by searching 20 data sets of P. furiosus LC-MS/MS data against a decoy database containing reverse sequences from the P. furiosus genome. A Mascot ion score threshold of 35 for the 20 searches yielded an acceptable and low false positive rate of 0.398 ± 0.57%. Thus, the Mascot ion score threshold was set to 35 for all searches, including single peptide assignments, so that the false positive rate was consistently less than 1%. Furthermore there was little issue with the peptides matching multiple members of a protein family because of the use of a single small proteome. Mascot was searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 2.0 Da. Iodoacetamide derivative of cysteine and oxidation of methionine were specified as fixed and variable modifications, respectively. The enzyme was selected as trypsin with a maximum of one missed cleavage allowed for the search. The fragmentation pattern was optimized for the instrument used by selecting the Mascot search parameter as “ESI-trap.” The MS/MS data for all unique peptides identified for each protein are presented in supplemental Table 2.
Data Management
All MS/MS data were stored in a custom Structural Query Language (SQL) Server 2005 database that preserves the hierarchical (parent-child) nature of the multilevel fractionation process. All samples analyzed were assigned two-dimensional barcodes. A user interface developed in Microsoft Access 2003 shows the hierarchical fractionation steps and automatically generates a Microsoft Excel PivotTable for each column that displays the proteins identified by MS/MS along the rows, chromatography fractions along the columns, and the number of peptides detected where they intersect.
Protein Complex Identification
Identification of potential Type 1 PCs was accomplished by generating text-based PivotTables (described above) for each of the 26 column chromatography steps. The text files were processed by a custom script written in the R statistical language. For each column, this script created a number of tentative PCs consisting of proteins that (i) co-eluted in one or more column fractions and (ii) were encoded by adjacent genes on the same DNA strand (Type 1 PCs). These tentative PCs from each column that contained any proteins in common were merged to generate a final list of potential PCs and assigned unique PC identification numbers.
Protein Complex Evaluation
The quality of the predicted PCs (i.e. the association value) was evaluated using a custom algorithm applied to the column data contained in the PivotTable described above (restricted to the potential subunits identified in the prediction step). This algorithm may be used for evaluating both Type 1 and Type 2 PCs. For each predicted PC, the algorithm computes a per fraction subscore based on the proportion of potential subunits within the predicted PC that appear in that fraction and sums these subscores to produce an overall raw score. The assigned subscores range from −0.1 (when half of the potential subunits are observed in a fraction) to 1.0 (when all of the potential subunits are observed in a fraction) with a zero score assigned when none of the subunits are observed in a given fraction. Specifically the function used to assign the subscores is the piecewise linear function with one linear piece between the points (0, 0) and (0.5, −0.1) and another linear piece between (0.5, −0.1) and (1, 1). The points of the function were selected based on 14 known PCs by optimizing over piecewise linear functions (with points (0, 0), (0.5, y0), and (1, y1), −2 ≤ y0 ≤ 0 and 0 ≤ y1 ≤ 2) so that the sum of the positions of the known PCs within the resulting PC list, sorted by score (in descending order), was minimized. Raw scores for putative complexes (−13.6 to 43.1) were rescaled in a relative fashion to lie within the interval [0, 1].
Protein Annotations
The protein annotations are based on the public National Center for Biotechnology Information (NCBI) annotation of the genome (45) except where the protein or protein complex (a) is known and has been previously purified from P. furiosus (indicated by a reference), (b) shows sequence similarity to a known protein or protein complex purified from a closely related organism (also referenced), or (c) was listed as a hypothetical protein but has a known domain in the InterPro database (46).
RESULTS
Fractionation of Cytoplasmic Proteins Using Column Chromatography
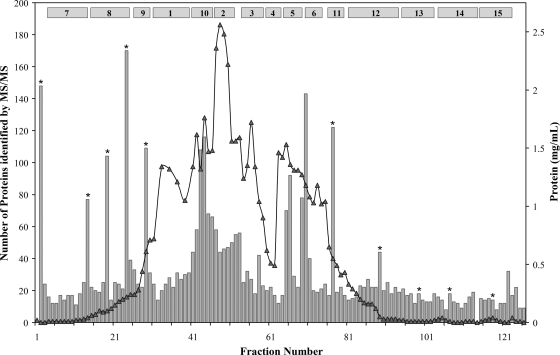
Fractionation was carried out under anaerobic and reducing conditions using sequential, non-denaturing, column chromatography to determine which proteins co-fractionated as potential multiprotein complexes. The cytoplasmic fraction was initially resolved by anion exchange chromatography (column level c1) into 126 fractions. The c1 fractions were analyzed by MS/MS, and the results are summarized in Fig. 2 along with the protein concentration profile. CV LC-ESI-MS/MS was used for c1 fractions containing the bulk of the protein as determined by protein concentrations and overall protein profile (fraction positions 31–75 of 126), whereas the new HT LC-ESI-MS/MS methodology, capable of handling 96-well format plates in a much shorter time and developed for this project, was used for the lower complexity fractions. Ten fractions were analyzed using both approaches to evaluate the efficacy of the HT method (Fig. 2). A total of 403 proteins were identified in these fractions, 387 by CV MS/MS and 111 by HT MS/MS (with 95 proteins in common). For more complex fractions containing at least 60 proteins identified by CV MS, typically less than 25% were identified by HT MS, whereas for less complex fractions containing 20 or so proteins (by CV MS) the two methods identified a similar number of proteins. Only the HT approach was used to analyze all c2, c3, and c4 fractions. A total of 696 proteins were identified in the c1 fractions by the two approaches.
Fig. 2.
Protein elution profile (line) of the c1 column showing the number of proteins identified in each fraction (bars) and the fraction pools (numbered boxes) generated for second level (c2) column fractionation. Fractions that were analyzed by both CV and HT MS/MS are indicated by an asterisk, and the total number of proteins identified by both methods is given.
To gain insight into which potential PCs remain associated after additional separation procedures, the c1 fractions were combined into 15 pools (see Fig. 2), and each was subjected to a c2 chromatography step. Pooling was based on the location of abundant proteins within the fractions as determined by SDS-PAGE and specific enzyme assays, minimizing the division of major proteins into multiple c2 columns and providing known, stable heteromeric PCs that should co-elute through multiple separation steps and serve as positive controls for detection by HT MS/MS. Four assayable heteromeric PCs (abbreviated FNOR, POR, VOR, and SH1) were chosen for this purpose. In addition, five known homomeric PCs served as negative controls for the co-fractionation analysis. These included three oxidoreductases, aldehyde:ferredoxin oxidoreductase, rubrerythrin, and NADPH-rubredoxin oxidoreductase, and two small redox proteins (rubredoxin and Fd) that are electron carriers for numerous oxidoreductase-type enzymes. None of these five proteins are known to form heteromeric PCs that are stable through multiple separation steps.
As shown in Fig. 1, the c1 fraction pools were fractionated through second level (c2) columns generating a total of 790 c2 fractions. To further purify the control proteins, six c2 fraction pools (∼5.3% of the total c2 fractions) were fractionated on c3 columns, and four c3 fraction pools (∼13% of the total c3 fractions) were fractionated on c4 columns. The c3 and c4 columns yielded 229 and 131 fractions, respectively. Overall the cytoplasmic extract was resolved into 1276 fractions, and an additional 280 proteins were identified in the c2–c4 fractions that were not found in the c1 fractions. The total number of proteins identified was 976, representing 46% of the predicted proteome (supplemental Tables 2–4). Given that the genome contains 2125 ORFs, 75% of which are expressed under a given growth condition (33) with ∼25% of those being membrane-associated (47), the upper limit for the number of different cytoplasmic proteins is about 1200. Approximately 80% of these were identified, showing the validity of the HT MS approach for less complex fractions.
Identification and Distribution of Potential Type 1 Heteromeric Protein Complexes
Identification of potential heteromeric PCs was carried out computationally using the MS/MS data generated for each fraction. Proteins that co-fractionated and were encoded by adjacent genes made up 243 or 25% of the total number of proteins identified. These proteins are predicted to form 106 potential Type 1 PCs (labeled PC-1 to PC-106; supplemental Table 5). Components from 71 of these were identified in c1 fractions as well as in the downstream c2, c3, and/or c4 fractions, whereas the remaining 35 were detected in downstream fractions only, either in fractions from a single column (26 PCs) or in fractions from multiple columns (nine PCs). A total of 32 of the 71 were identified as PCs only in c1 fractions.
Known PCs (15)
A total of 61 of the 243 proteins that constitute the 106 potential PCs are classified as known because they have been purified previously from P. furiosus. Of these, 43 are components of 15 potential PCs identified in the unbiased assessment based on the fractionation procedure (Tables I and II) and include the four PCs that were used as positive controls for the fractionation procedure (FNOR, POR, VOR, and SH1). One of these predicted complexes (PC-34; see Table I) actually represents two of the positive control PCs (POR and VOR) that are encoded by adjacent genes (PF0965–PF0971), one of which (PF0971) encodes a common subunit (61). Consequently the 43 proteins are actually components of 16 known PCs. The identification of these known PCs validates the criteria that were used to identify novel PCs.
Table I.
Known PCs (15)
Association values were calculated as described under “Protein Complex Identification” and “Protein Complex Evaluation” under “Experimental Procedures.” Fractions denote the number of chromatography fractions in which the protein was identified. The protein annotation differs from that in the public NCBI annotation if the protein component is known (as indicated by the reference), has similarity to a known complex from a closely related organism (also referenced), or has a known InterPro domain (indicated within angle brackets). The operon number is taken from Ref. 32. PC-34 represents two known PCs, making the total sixteen.
| Complex number | Gene | Annotation | pI | Association value | Fractions | Operon number | Refs. |
|---|---|---|---|---|---|---|---|
| Control PCs | |||||||
| PC-33 | PF0891 | Hydrogenase I β | 6.3 | 0.40 | 100 | 200 | 41 |
| PF0892 | Hydrogenase I γ | 9.0 | 45 | 200 | |||
| PF0893 | Hydrogenase I δ | 5.2 | 39 | 200 | |||
| PF0894 | Hydrogenase I α | 5.7 | 51 | 200 | |||
| PC-34 | PF0965 | Pyruvate ferredoxin oxidoreductase β | 9.1 | 0.42 | 112 | 215 | 48 |
| PF0966 | Pyruvate ferredoxin oxidoreductase α | 5.0 | 151 | 215 | |||
| PF0967 | Pyruvate ferredoxin oxidoreductase δ | 5.2 | 100 | 215 | |||
| PF0968 | 2-Ketoisovalerate ferredoxin oxidoreductase β | 9.1 | 42 | 215 | 38 | ||
| PF0969 | 2-Ketoisovalerate ferredoxin oxidoreductase α | 5.3 | 62 | 215 | |||
| PF0970 | 2-Ketoisovalerate ferredoxin oxidoreductase δ | 5.5 | 64 | 215 | |||
| PF0971 | Pyruvate/2-ketovalerate ferredoxin oxidoreductase γ | 5.4 | 240 | 215 | |||
| PC-49 | PF1327 | FNOR I α | 7.1 | 0.46 | 65 | 307 | 36 |
| PF1328 | FNOR I β | 7.2 | 20 | 307 | |||
| Known PCs | |||||||
| PC-2 | PF0018 | DNA polymerase II small subunit | 5.0 | 0.20 | 22 | 4 | 49 |
| PF0019 | DNA polymerase II large subunit | 6.2 | 27 | 4 | |||
| PF0020 | 〈β-Lactamase-like (lactamase_B)〉 | 5.5 | 57 | 5 | |||
| PC-5 | PF0092 | Replication factor C, large subunit | 5.8 | 0.26 | 8 | 22 | 50 |
| PF0093 | Replication factor C, small subunit | 6.7 | 37 | 22 | |||
| PF0094 | Protein-disulfide oxidoreductase | 4.9 | 25 | 51 | |||
| PC-10 | PF0181 | 〈ATPase, V1/A1 complex, subunit F〉 | 5.2 | 0.11 | 2 | 44 | 52 |
| PF0182 | ATPase subunit A | 6.1 | 72 | 44 | |||
| PF0183 | ATPase subunit B | 5.3 | 106 | 44 | |||
| PF0184 | ATPase subunit D | 9.6 | 19 | 44 | |||
| PC-22 | PF0533 | Indolepyruvate oxidoreductase α | 6.8 | 0.80 | 64 | 118 | 53 |
| PF0534 | Indolepyruvate ferredoxin oxidoreductase β | 6.4 | 37 | 118 | |||
| PC-26 | PF0598 | Aspartate carbamoyltransferase, regulatory subunit | 7.8 | 0.39 | 40 | 135 | 54 |
| PF0599 | Aspartate carbamoyltransferase, catalytic subunit | 6.1 | 34 | 135 | |||
| PC-46 | PF1245 | Proline dehydrogenase α | 8.1 | 0.46 | 48 | 285 | 55 |
| PF1246 | Proline dehydrogenase β | 7.2 | 25 | 285 | |||
| PC-50 | PF1331 | Hydrogenase II δ | 5.9 | 0.44 | 20 | 308 | 56 |
| PF1332 | Hydrogenase II α | 5.0 | 62 | 308 | |||
| PC-56 | PF1433 | mbh11 membrane-bound hydrogenase β | 5.1 | 0.31 | 8 | 332 | 57 |
| PF1434 | mbh12 membrane-bound hydrogenase α | 6.6 | 26 | 332 | |||
| PC-62 | PF1562 | DNA-directed RNA polymerase subunit a | 5.7 | 0.21 | 15 | 360 | 58 |
| PF1563 | DNA-directed RNA polymerase subunit a` | 6.4 | 51 | 360 | |||
| PF1564 | DNA-directed RNA polymerase subunit b | 6.3 | 99 | 360 | |||
| PF1565 | DNA-directed RNA polymerase subunit h | 8.8 | 2 | 360 | |||
| PC-65 | PF1577 | Hypothetical protein PF1577 | 9.3 | 0.23 | 14 | 363 | |
| PF1578 | DNA topoisomerase VI subunit A | 8.4 | 4 | 363 | 59 | ||
| PF1579 | DNA topoisomerase VI subunit B | 8.8 | 3 | 363 | |||
| PC-68 | PF1642 | DNA-directed RNA polymerase subunit k | 9.5 | 0.23 | 5 | 378 | 58 |
| PF1643 | DNA-directed RNA polymerase subunit n | 6.1 | 13 | 379 | |||
| PC-106 | PF2018 | Replication protein A32 | 5.1 | 0.25 | 15 | 457 | 60 |
| PF2019 | Replication protein A14 | 4.9 | 3 | 457 | |||
| PF2020 | Replication protein A41 | 4.9 | 18 | 457 |
Table II.
Fractionation grid for control and known PCs
This table shows the proteins (indicated by their PF number) identified by LC-MS/MS along rows and the chromatography step along the columns. The squares generated where they intersect are color-coded dark gray if the identified PC components co-eluted in the same fractions from a particular column and light gray if a PC component eluted separately in the fractions of a particular column. Blank squares denote that the protein was not identified in any fractions of that column.
Thirteen of the known PCs have been purified previously from P. furiosus biomass, whereas three (PC-5, PC-26, and PC-46) have been characterized using recombinant proteins. Two PCs represent soluble subcomplexes of known membrane-bound PCs, ATP synthase (A1 subcomplex: PC-10 (52)) and the membrane-bound hydrogenase (MBH: PC-56; (57)). In one instance, only two component proteins of a known heterotetrameric PC, hydrogenase II (PC-50 (56)), were detected in any fraction at any level, whereas three PCs were each predicted to contain an additional subunit (PC-5: replication factor C complex (50), PC-2: DNA polymerase II (49), and PC-65: DNA topoisomerase (59)). The third component of one of them (PC-5) has also been purified (protein-disulfide oxidoreductase (51, 62)). It is possible that all three potential PCs are functional complexes that dissociate upon purification. All the known PCs are predicted to be encoded by operons (32) with the exceptions of the third components in the trimeric PCs noted above (PC-2 and PC-5), supporting the notion that they are dimeric PCs (see Table I). Association scores for all the known complexes were above 0.2 with the exception of PC-10, the subcomplex of the ATPase. Excluding one member of the complex (PF0181) results in a much higher score (0.48), suggesting that PF0181 is weakly associated.
Expected PCs (5)
Five potential Type 1 PCs containing 12 proteins were categorized as expected PCs because homologous complexes have been characterized from related organisms (Table III and supplemental Table 5). These PCs are involved in protein biosynthesis (PC-37 (63) and PC-58 (64, 65)), RNA processing (PC-63 (66)), central carbon metabolism (PC-74 (67)), and amino acid metabolism (PC-76 (68)). The expected PCs had association scores in the range of 0.2–0.3 and, with one exception (PC-58), are predicted to be encoded by operons.
Table III.
| Complex number | Gene | Annotation | pI | Association value | Fractions | Operon number | Refs. |
|---|---|---|---|---|---|---|---|
| PC-37 | PF0989 | Phenylalanyl-tRNA synthetase α subunit | 9.2 | 0.29 | 13 | 220 | 63 |
| PF0990 | Phenylalanyl-tRNA synthetase β subunit | 4.9 | 9 | 220 | |||
| PC-58 | PF1461 | Glutamyl-tRNAGln amidotransferase subunit D | 5.7 | 0.27 | 36 | 64, 65 | |
| PF1462 | Glutamyl-tRNAGln amidotransferase subunit E | 5.6 | 21 | ||||
| PC-63 | PF1568 | Exosome complex exonuclease 1 | 8.9 | 0.23 | 1 | 361 | 66 |
| PF1569 | Exosome complex RNA-binding protein 1 | 8.9 | 6 | 361 | |||
| PC-74 | PF1767 | 2-Ketoglutarate:ferredoxin oxidoreductase δ | 6.9 | 0.23 | 4 | 401 | 67 |
| PF1768 | 2-Ketoglutarate:ferredoxin oxidoreductase α | 6.1 | 6 | 401 | |||
| PC-76 | PF1795 | Sarcosine oxidase, α subunit | 9.1 | 0.23 | 15 | 407 | 68 |
| PF1796 | Polyferredoxin | 6.9 | 2 | 407 | |||
| PF1797 | l-Proline dehydrogenase α subunit | 7.9 | 3 | 407 | |||
| PF1798 | l-Proline dehydrogenase β subunit | 6.1 | 5 | 407 |
Novel Potential PCs (86)
The remaining 86 potential Type 1 PCs have been categorized as novel PCs in that they were not anticipated and/or have not been characterized previously. Thirteen of them have orthologous interactions in the Escherichia coli and/or Saccharomyces cerevisiae (European Bioinformatics Institute IntAct database (Refs. 69 and 70; see supplemental Table 6). These were analyzed using additional criteria that included homology, the extent of overlap between component proteins, abundance of protein partners (number of fractions in which they were identified), operon structure, and evidence for co-regulation from DNA microarray data. Based on this information, the novel potential PCs were categorized as strong (43 PCs), tentative (30 PCs), or weak (13 PCs) (Tables IV, V, and VI, respectively). Some of the biological implications of the identified strong PCs are discussed below.
Table IV.
Strong novel PCs (43)
For details, see the legend to Table I. Association values <0.2 are in parentheses. HypC, hydrogenase expression/formation protein C; HD, predicted hydrolase; TLDD, tolerance for the letD; Tneap, Thermotoga neopolitana; RAMP, receptor activity modifying protein; NUDIX, NUcleoside DIphosphate linked to some other moiety X.
| Complex number | Gene | Annotation | pI | Association value | Fractions | Operon number | Refs. |
|---|---|---|---|---|---|---|---|
| PC-1 | PF0014 | Hypothetical protein PF0014 | 6.4 | 0.25 | 2 | 3 | |
| PF0015 | Hypothetical protein PF0015 | 5.0 | 9 | 3 | |||
| PC-6 | PF0096 | 〈4Fe-4S ferredoxin, iron-sulfur binding (4FE4S_FERREDOXIN)〉 | 8.2 | 0.24 | 2 | 23 | |
| PF0097 | 42-kDa subunit bacteriochlorophyll synthase-like protein | 6.1 | 13 | 23 | |||
| PC-7 | PF0138 | Ferritin/ribonucleotide reductase-like (ferritin/RR_like) | 5.4 | 0.26 | 37 | ||
| PF0139 | 2-Keto acid ferredoxin oxidoreductase 3γ | 8.4 | 28 | 38 | |||
| PC-8 | PF0146 | Potassium channel, putative | 4.7 | 0.31 | 13 | 35 | |
| PF0147 | Potassium channel related protein | 4.8 | 24 | 35 | |||
| PC-9 | PF0160 | TLDD-like protein | 5.1 | 0.27 | 17 | 41 | |
| PF0161 | 〈Peptidase U62, modulator of DNA gyrase (PmbA_TldD)〉 | 6.7 | 4 | 41 | |||
| PC-11 | PF0198 | Phosphoribosylformylglycinamidine synthase subunit I | 5.9 | 0.24 | 1 | 48 | |
| PF0199 | Phosphoribosylformylglycinamidine synthase subunit II | 5.2 | 3 | 48 | |||
| PC-12 | PF0202 | Isocitrate dehydrogenase | 6.5 | 0.36 | 53 | 49 | 71 |
| PF0203 | Citrate synthase | 7.2 | 15 | 49 | 72 | ||
| PC-17 | PF0390 | Hypothetical protein PF0390 | 5.3 | 1.00 | 86 | 87 | |
| PF0391 | Hypothetical protein PF0391 | 8.9 | 47 | 87 | |||
| PC-24 | PF0548 | 〈Hydrogenase expression/formation protein (HYPC)〉 | 4.9 | 0.21 | 7 | 122 | |
| PF0549 | 〈Hydrogenase formation HypD protein (Hydrgn_mat_hypD)〉 | 6.8 | 13 | 122 | |||
| PC-28 | PF0639 | 〈CRISPR-associated HD (cas3_HD)/CH〉 | 8.3 | 0.25 | 21 | 146 | |
| PF0640 | ATP-dependent RNA helicase, putative | 8.9 | 38 | 146 | |||
| PF0641 | 〈CRISPR-associated protein, MJ0382 (cas_cas5a)〉 | 9.4 | 16 | 146 | |||
| PC-29 | PF0642 | 〈CRISPR-associated regulatory Csa2 (DUF73)〉 | 6.4 | 0.39 | 24 | 146 | |
| PF0643 | Hypothetical protein PF0643 | 6.6 | 38 | 146 | |||
| PC-30 | PF0753 | 2-Keto acid:ferredoxin oxidoreductase subunit β | 8.8 | 0.73 | 41 | 170 | 38 |
| PF0754 | 2-Keto acid:ferredoxin oxidoreductase subunit α | 5.5 | 40 | 170 | |||
| PC-32 | PF0872 | 〈kaiA binding (KaiA-binding)〉 | 6.2 | 0.23 | 9 | 195 | |
| PF0873 | Hypothetical protein PF0873 | 8.1 | 32 | ||||
| PC-35 | PF0972 | 〈Hydroxymethylglutaryl-coenzyme A synthase (HMG_coA_syn)〉 | 7.9 | 0.31 | 8 | 216 | |
| PF0973 | Acetyl-CoA acetyltransferase | 4.8 | 18 | 216 | |||
| PF0974 | Hypothetical protein PF0974 | 8.5 | 18 | ||||
| PC-38 | PF1033 | Putative peroxiredoxin | 5.5 | 0.20 | 8 | ||
| PF1034 | Orotate phosphoribosyltransferase | 5.5 | 15 | ||||
| PC-40 | PF1121 | 〈CRISPR-associated protein Cas5, Tneap type (cas_Cas5t)〉 | 9.3 | 0.21 | 1 | 254 | |
| PF1122 | 〈CRISPR-associated regulatory DevR (cas_Cst2_DevR)〉 | 6.1 | 6 | 255 | |||
| PF1123 | 〈CRISPR-associated protein, CXXC_CXXC region〉 | 6.7 | 17 | 255 | |||
| PC-41 | PF1124 | 〈CRISPR-associated protein, TM1791 (cas_TM1791_cmr6)〉 | 8.9 | 0.23 | 2 | 256 | |
| PF1125 | 〈CRISPR-associated protein, TM1791.1 (cas_Cmr5)〉 | 8.6 | 2 | 256 | |||
| PF1126 | 〈CRISPR-associated RAMP Cmr4 (cas_RAMP_Cmr4)〉 | 5.6 | 14 | 256 | |||
| PC-42 | PF1186 | NADH oxidase | 5.7 | 0.21 | 12 | ||
| PF1187 | Hypothetical protein PF1187 | 5.1 | 7 | ||||
| PC-47 | PF1268 | 〈Cobalamin-independent methionine synthase MetE, N-terminal〉 | 5.2 | 0.23 | 2 | 291 | |
| PF1269 | Methionine synthase | 5.9 | 4 | 291 | |||
| PC-48 | PF1283 | Rubrerythrin | 5.7 | (0.16) | 92 | 294 | 37 |
| PF1284 | 〈Histone fold (histone-fold)〉 | 9.1 | 29 | ||||
| PF1285 | 〈FeS assembly protein SufD (sufD)〉 | 7.1 | 0.35 | 18 | 295 | ||
| PF1286 | 〈FeS assembly protein SufB (sufB)〉 | 6.0 | 9 | 295 | |||
| PF1287 | 〈FeS assembly ATPase SufC (sufC)〉 | 6.1 | 11 | 295 | |||
| PC-51 | PF1339 | Hypothetical protein PF1339 | 5.3 | 0.48 | 17 | 310 | |
| PF1340 | 〈von Willebrand factor, type A (VWFADOMAIN)〉 | 8.5 | 18 | 310 | |||
| PC-52 | PF1356 | Glucose-1-phosphate uridylyltransferase | 5.3 | 0.21 | 28 | 313 | |
| PF1357 | UDP- or dTTP-glucose 4-epimerase or 4–6-dehydratase | 5.8 | 3 | 313 | |||
| PC-60 | PF1529 | Pyridoxine biosynthesis protein | 5.8 | 0.20 | 53 | 351 | |
| PF1530 | Putative thiazole biosynthetic enzyme | 5.3 | 7 | ||||
| PC-64 | PF1573 | Hypothetical TLDD protein | 5.6 | 0.25 | 7 | 362 | |
| PF1574 | 〈Peptidase U62, modulator of DNA gyrase (PmbA_TldD)〉 | 5.3 | 13 | 362 | |||
| PC-69 | PF1713 | Carbamoyl-phosphate synthase small subunit | 5.9 | 0.34 | 8 | 388 | |
| PF1714 | Carbamoyl-phosphate synthase large subunit | 5.7 | 21 | 388 | |||
| PC-72 | PF1728 | NDP-sugar synthase | 5.4 | 0.24 | 32 | 393 | |
| PF1729 | Phospho-sugar mutase | 5.6 | 31 | 393 | |||
| PC-75 | PF1771 | 2-Keto acid:ferredoxin oxidoreductase α | 5.3 | 0.40 | 14 | 401 | 38 |
| PF1772 | 2-Keto acid:ferredoxin oxidoreductase β | 9.2 | 16 | 401 | |||
| PF1773 | 2-Keto acid:ferredoxin oxidoreductase γ | 6.0 | 20 | 401 | |||
| PC-77 | PF1807 | 50 S ribosomal protein L32e | 11.2 | 0.22 | 9 | 408 | |
| PF1808 | 50 S ribosomal protein L6 | 8.7 | 13 | 408 | |||
| PF1809 | 30 S ribosomal protein S8 | 9.6 | 4 | 408 | |||
| PC-79 | PF1824 | 50 S ribosomal protein L4 | 10.6 | 0.25 | 17 | 408 | |
| PF1825 | 50 S ribosomal protein L3 | 10.1 | 6 | 408 | |||
| PC-81 | PF1837 | Acetyl-CoA synthetase II β | 5.6 | 0.25 | 69 | 410 | 73 |
| PF1838 | Acetyl-CoA synthetase NTB α | 5.6 | 14 | 410 | |||
| PC-82 | PF1842 | 〈Prokaryotic chromosome segregation and condensation protein〉 | 4.7 | 0.23 | 1 | 412 | |
| PF1843 | Chromosome segregation protein smc | 8.7 | 4 | 412 | |||
| PC-83 | PF1857 | Cobalt transport ATP-binding protein cbio | 6.4 | 0.24 | 1 | ||
| PF1858 | Cysteine synthase | 8.8 | 2 | ||||
| PC-85 | PF1909 | Ferredoxin | 3.9 | (0.10) | 71 | 43 | |
| PF1910 | Sulfide dehydrogenase α SudX | 8.0 | 0.22 | 67 | 428 | 36 | |
| PF1911 | Sulfide dehydrogenase SudY | 5.7 | 6 | 428 | |||
| PC-91 | PF1999 | Glycine dehydrogenase subunit 1 | 5.5 | 0.25 | 2 | 452 | |
| PF2000 | Glycine dehydrogenase subunit 2 | 6.0 | 5 | 452 | |||
| PC-92 | PF2005 | Glycerol-3-phosphate dehydrogenase | 5.9 | 0.25 | 7 | 454 | |
| PF2006 | NADH oxidase | 6.3 | 4 | 454 | |||
| PF2007 | Hypothetical protein PF2007 | 9.2 | 3 | 454 | |||
| PC-94 | PF0008 | Hit family protein | 6.2 | 0.20 | 21 | 2 | |
| PF0009 | 〈Molybdenum cofactor biosynthesis (MoeB)〉 | 5.6 | 5 | 2 | |||
| PC-95 | PF0070 | 〈Prefoldin (prefoldin)〉 | 9.3 | 0.22 | 4 | 15 | |
| PF0071 | 〈Pseudouridine synthase and archaeosine transglycosylase domain〉 | 8.2 | 7 | 15 | |||
| PC-98 | PF0838 | Hypothetical protein PF0838 | 9.6 | 0.22 | 12 | 186 | |
| PF0839 | 〈PilT protein, N-terminal (PIN)〉 | 4.7 | 1 | 186 | |||
| PC-100 | PF1558 | 30 S ribosomal protein S7 | 10.0 | 0.28 | 9 | 360 | |
| PF1559 | 30 S ribosomal protein S12 | 10.6 | 19 | 360 | |||
| PC-101 | PF1589 | 〈PfkB (PfkB)〉 | 6.9 | 0.22 | 7 | ||
| PF1590 | 〈NUDIX (NUDIX_hydrolase)〉 | 6.0 | 8 | 365 | |||
| PC-102 | PF1648 | 30 S ribosomal protein S11 | 10.4 | 0.25 | 23 | 380 | |
| PF1649 | 30 S ribosomal protein S4 | 10.0 | 22 | 380 | |||
| PF1650 | 30 S ribosomal protein S13 | 10.9 | 11 | 380 | |||
| PC-103 | PF1679 | 3-Isopropylmalate dehydratase large subunit (LeuC) | 5.5 | 0.26 | 9 | 383 | |
| PF1680 | 3-Isopropylmalate dehydratase small subunit (LeuD) | 6.6 | 4 | 383 | |||
| PC-104 | PF1803 | 50 S ribosomal protein L30 | 10.2 | 0.21 | 7 | 408 | |
| PF1804 | 30 S ribosomal protein S5P | 9.6 | 35 | 408 |
Table V.
Tentative novel PCs (30)
For details, see the legend to Table I. PINc, nucleotide-binding protein; DOD, dodecapeptide; SAP, DNA binding domain named after SAF-A/B, Acinus, and PIAS; PUA, PseudoUridine and Archaeosine transglycosylase; CBS, cystathionine-beta synthase; SAM, S-adenosylmethionine.
| Complex number | Gene | Annotation | pI | Association value | Fractions | Operon number | Ref. |
|---|---|---|---|---|---|---|---|
| PC-3 | PF0037 | Hypothetical protein PF0037 | 8.5 | 0.19 | 6 | 9 | |
| PF0038 | Glyoxalase II family member | 6.0 | 48 | 9 | |||
| PC-14 | PF0232 | Transcription regulatory protein, arsR family | 6.2 | 0.15 | 5 | 57 | |
| PF0233 | Acetyl-CoA synthetase, α subunit | 6.0 | 45 | 73 | |||
| PC-15 | PF0378 | 50 S ribosomal protein L31 | 10.1 | 0.19 | 14 | 85 | |
| PF0379 | 50 S ribosomal protein L39e | 12.4 | 25 | 85 | |||
| PC-16 | PF0382 | Prefoldin, β subunit | 9.5 | 0.18 | 16 | 86 | 74 |
| PF0383 | Hypothetical protein PF0383 | 4.6 | 17 | 86 | |||
| PC-18 | PF0431 | Phosphoribosylaminoimidazole synthetase | 5.3 | 0.23 | 2 | 98 | |
| PF0432 | Putative sugar-catabolism phosphotransferase | 5.2 | 4 | 98 | |||
| PC-19 | PF0440 | Ribonucleotide reductase | 6.1 | 0.15 | 44 | 75 | |
| PF0441 | Galactose-1-phosphate uridylyltransferase | 8.6 | 6 | ||||
| PC-20 | PF0491 | Transcription initiation factor E subunit α | 8.8 | 0.20 | 7 | 110 | |
| PF0492 | Hypothetical protein PF0492 | 5.6 | 18 | ||||
| PC-23 | PF0540 | Thymidylate synthase | 8.9 | 0.23 | 15 | ||
| PF0541 | Methionine aminopeptidase | 6.7 | 2 | 76 | |||
| PC-27 | PF0637 | 〈CRISPR-associated protein, MJ0385 (cas_Csa4)〉 | 6.3 | 0.23 | 7 | 146 | |
| PF0638 | 〈CRISPR-associated protein, MJ0385 (cas_Csa4)〉 | 6.8 | 2 | 146 | |||
| PC-36 | PF0982 | Hypothetical protein PF0982 | 4.8 | 0.05 | 1 | 218 | |
| PF0983 | PCNA sliding clamp (proliferating cell nuclear antigen) | 4.5 | 104 | 218 | 77 | ||
| PC-39 | PF1115 | 〈PUA (PUA)〉 | 8.0 | 0.20 | 11 | 253 | |
| PF1116 | Glycerol-3-phosphate cytidyltransferase | 8.6 | 24 | ||||
| PC-43 | PF1189 | Hypothetical protein PF1189 | 5.5 | 0.14 | 2 | 268 | |
| PF1190 | 〈Rubrerythrin (rubrerythrin)〉 | 5.2 | 66 | 268 | |||
| PC-45 | PF1242 | Dehydrogenase subunit α | 8.5 | 0.23 | 16 | 284 | |
| PF1243 | Hypothetical protein PF1243 | 5.0 | 2 | 284 | |||
| PC-53 | PF1375 | Elongation factor Tu | 8.7 | 0.39 | 79 | 318 | |
| PF1376 | 30 S ribosomal protein S10 | 10.2 | 19 | 318 | |||
| PC-54 | PF1379 | Cell division protein pelota | 7.2 | 0.22 | 4 | 319 | |
| PF1380 | Arginyl-tRNA synthetase | 6.2 | 33 | 319 | |||
| PC-55 | PF1385 | DNA polymerase, bacteriophage type | 9.0 | 0.25 | 5 | 321 | |
| PF1386 | 〈Helix-turn-helix type 3 (HTH_CROC1)〉 | 9.3 | 4 | 321 | |||
| PC-57 | PF1454 | 〈Radical SAM (Radical_SAM)〉 | 6.3 | 0.20 | 33 | 334 | |
| PF1455 | 〈von Willebrand factor, type A (VWFADOMAIN)〉 | 4.9 | 2 | 334 | 78 | ||
| PC-59 | PF1482 | 〈β-Grasp fold, ferredoxin-type (ferredoxin_fold)〉 | 8.2 | 0.21 | 12 | 338 | |
| PF1483 | 〈DNA-binding SAP (SAP)〉 | 5.3 | 19 | ||||
| PC-61 | PF1556 | Peptidyl-tRNA hydrolase | 9.5 | 0.19 | 8 | 359 | |
| PF1557 | 〈Rossmann-like α/β/α sandwich fold〉 | 5.8 | 31 | 359 | |||
| PC-66 | PF1614 | Hypothetical protein PF1614 | 8.6 | 0.14 | 1 | 372 | |
| PF1615 | 〈Intein DOD homing endonuclease (INTEIN_ENDONUCLEASE)〉 | 9.0 | 57 | 372 | |||
| PC-70 | PF1716 | 〈Nucleotide-binding protein, PINc (PINc)〉 | 9.0 | 0.19 | 1 | 389 | |
| PF1717 | 〈Translation initiation factor IF-2 γ subunit〉 | 8.2 | 37 | 389 | |||
| PF1718 | 〈Wyosine base formation (Wyosine_form)〉 | 6.9 | 20 | ||||
| PC-71 | PF1721 | 〈Triphosphoribosyl-dephospho-CoA protein (CitG)〉 | 7.8 | 0.15 | 1 | 390 | |
| PF1722 | Archaeal histone a2 | 9.6 | 50 | ||||
| PC-73 | PF1730 | Thymidylate kinase | 9.2 | 0.23 | 1 | 393 | |
| PF1731 | Signal recognition particle SRP54 | 9.4 | 5 | 393 | 79 | ||
| PC-78 | PF1813 | 50 S ribosomal protein L24 | 10.4 | 0.25 | 2 | 408 | |
| PF1814 | 50 S ribosomal protein L14 | 11.3 | 8 | 408 | |||
| PC-80 | PF1827 | 〈Uncharacterized conserved protein, 2×CBS, MJ1225 type〉 | 8.5 | 0.18 | 37 | ||
| PF1828 | Hypothetical protein PF1828 | 8.9 | 11 | ||||
| PC-84 | PF1874 | Glyceraldehyde-3-phosphate dehydrogenase | 6.0 | 0.18 | 18 | ||
| PF1875 | Hypothetical protein PF1875 | 4.4 | 15 | ||||
| PC-86 | PF1926 | Recombinase (RadA) | 6.2 | 0.21 | 14 | 80 | |
| PF1927 | 〈Uncharacterized conserved protein UCP015877, zinc finger〉 | 7.0 | 2 | ||||
| PC-88 | PF1974 | Thermosome, single subunit | 4.9 | 0.08 | 99 | 81 | |
| PF1975 | 〈Phosphoenolpyruvate carboxylase, Archaea (DUF557)〉 | 5.9 | 4 | ||||
| PC-89 | PF1990 | Transcription antitermination protein NusG | 8.2 | 0.26 | 6 | 450 | |
| PF1991 | 50 S ribosomal protein L11 | 5.4 | 7 | 450 | |||
| PC-93 | PF2039 | Hypothetical protein PF2039 | 5.5 | 0.23 | 2 | 463 | |
| PF2040 | 〈Profilin/allergen (PROFILIN)〉 | 9.1 | 3 | 463 |
Table VI.
Weak or unlikely novel PCs (13)
For details, see the legend to Table I. PINc, nucleotide-binding protein; PRC, photosynthetic reaction centre.
| Complex number | Gene | Annotation | pI | Association value | Fractions | Operon number | Ref. |
|---|---|---|---|---|---|---|---|
| PC-4 | PF0042 | Metal-dependent hydrolase | 6.0 | 0.00 | 6 | 9 | |
| PF0043 | Phosphoenolpyruvate synthetase | 5.2 | 153 | 82 | |||
| PC-13 | PF0207 | Argininosuccinate synthase | 5.3 | 0.14 | 84 | 51 | |
| PF0208 | Argininosuccinate lyase | 5.6 | 7 | 51 | |||
| PC-21 | PF0495 | Reverse gyrase | 7.5 | 0.18 | 19 | 111 | 83 |
| PF0496 | 〈Transcriptional regulator winged helix (Wing_hlx_tran_reg)〉 | 5.6 | 55 | ||||
| PC-25 | PF0588 | Phospho-sugar mutase | 5.7 | 0.14 | 18 | 133 | |
| PF0589 | Mannose-6-phosphate isomerase | 5.8 | 53 | 133 | |||
| PC-31 | PF0868 | NDP-sugar synthase | 5.1 | 0.08 | 147 | ||
| PF0869 | Aspartyl-tRNA synthetase | 5.3 | 15 | ||||
| PC-44 | PF1203 | Formaldehyde:ferredoxin oxidoreductase | 6.0 | 0.11 | 112 | 272 | 84 |
| PF1204 | Seryl-tRNA synthetase | 6.7 | 89 | 273 | |||
| PF1205 | 〈Nucleotide-binding protein, PINc (PINc)〉 | 8.9 | 3 | 273 | |||
| PC-67 | PF1640 | 30 S ribosomal protein S2 | 9.3 | 0.07 | 99 | 378 | |
| PF1641 | 〈Enolase (enolase)〉 | 4.4 | 11 | 378 | |||
| PC-87 | PF1931 | 〈Circadian clock protein KaiC (KAIC)〉 | 8.6 | 0.13 | 48 | 434 | |
| PF1932 | 〈Transcriptional regulator winged helix (Wing_hlx_tran_reg)〉 | 5.2 | 16 | 434 | |||
| PC-90 | PF1993 | 50 S ribosomal protein L10 | 4.9 | 0.16 | 10 | 450 | |
| PF1994 | 50 S ribosomal protein L12 | 4.3 | 97 | 450 | |||
| PC-96 | PF0220 | d-Arabino-3-hexulose-6-phosphate formaldehyde lyase | 6.3 | 0.15 | 52 | ||
| PF0221 | Hypothetical protein PF0221 | 5.0 | 2 | 54 | |||
| PC-97 | PF0463 | Hydrolase related to 2-haloalkanoic acid dehalogenase | 5.1 | 0.07 | 27 | 105 | |
| PF0464 | Glyceraldehyde-3-phosphate ferredoxin oxidoreductase | 6.3 | 68 | 105 | 85 | ||
| PC-99 | PF1499 | 30 S ribosomal protein S19e | 9.7 | 0.18 | 22 | 341 | |
| PF1500 | 〈PRC-barrel-like (PRCH_cytoplasmic)〉 | 4.6 | 50 | 342 | |||
| PC-105 | PF1956 | Fructose-1,6-bisphosphate aldolase class I (Fba1) | 6.6 | 0.17 | 17 | 440 | 86 |
| PF1957 | Agmatinase | 5.0 | 23 | 440 |
Strong, Novel Potential PCs and Their Biological Implications—
Properties of the 43 strong, novel potential PCs are summarized in Table IV. All have association scores of ≥0.2 and the majority (39 PCs) are predicted to be encoded by operons.
P. furiosus utilizes peptides as a carbon source (28), and a pathway for amino acid catabolism has been proposed (Fig. 3; Refs. 61, 87). The various 2-ketoacids produced from amino acids by aminotransferases are converted to the CoA derivative by four heteromeric Fd-linked 2-keto acid oxidoreductases (KORs) that were all identified in this study. These are specific for pyruvate (POR: PC-34 (48)), aromatic (indolepyruvate ferredoxin oxidoreductase: PC-22 (53)), branched chain 2-keto acids (VOR (αβγδ): PC-34 (88)), and 2-ketoglutarate:ferredoxin oxidoreductase (αβγδ: PC-74 (expected) (67)). The acyl-CoA derivatives generated are used for ATP synthesis by two heteromeric (α2β2) ADP-dependent acyl-CoA synthetases (ACSs), ACS I (PF1540/PF1787) and ACS II (PF0532/PF1837) (73); however, these are Type 2 PCs (encoded by unlinked genes) and would not be identified as complexes by the present methodology.
Fig. 3.
Pathway of amino acid catabolism in P. furiosus and the proposed roles of five novel Type 1 PCs. The PCs are PC-30, PC-35, PC-75, PC-81, and PC-85. IOR, indolepyruvate oxidoreductase; KGOR, 2-ketoglutarate:ferredoxin oxidoreductase; red, reduced; ox, oxidized.
The P. furiosus genome contains genes that encode two additional KOR paralogs (38) and three paralogs of the ACS α subunits, all of unknown function. We have now identified PCs corresponding to the two unknown KOR paralogs (Fig. 3, PC-30 and PC-75), which presumably extend the repertoire of amino acid derivatives that are utilized by the KORs. We also identified an additional ACS complex (PC-81; PF1837/PF1838) that shares a subunit (PF1837) with ACS II. Both ACS β subunits and all five ACS α subunits were identified in the fractionation procedure (supplemental Table 3). The fact that at least one β subunit (PF1837) can form two distinct PCs with two α subunits (PF0532 and PF1838) indicates that the two β subunit paralogs may assemble with two or more α subunits to give a variety of Type 1 and Type 2 PCs. An additional PC (PC-35), composed of two proteins that are annotated as an acyl carrier protein and acetyl-CoA synthase, has been identified that may convert the acyl-CoAs generated by amino acid oxidation to as yet unknown products (see Fig. 3). In support of this, the expression of the genes encoding both components of PC-35 (PF0972/PF0973) are up-regulated when P. furiosus is grown on peptides (33, 38, 89). Another potential PC (PC-7) contains a component that is a paralog of the KOR γ-subunit, although its function is unknown.
As shown in Fig. 3, the reductant generated from amino acid oxidation reduces the redox protein Fd, which is subsequently used to reduce NAD(P) by the known PC, FNOR I (PC-49 (36)). We have now identified a paralog of FNOR I, PC-85, that is termed FNOR II. DNA microarray studies have shown that the expression of FNOR I and FNOR II is reciprocally regulated when P. furiosus is grown on peptides and carbohydrates (33), indicating that they catalyze the same reaction but have different physiological roles. PC-85 includes the known electron carrier protein Fd (43). Although Fd is not predicted to be part of the FNOR II operon, it does take part in the FNOR I reaction and would be predicted to interact with FNOR II. The association score for PC-85 was found to increase from 0.1 to 0.22 on removing Fd from the complex, indicating that it may be a weakly interacting component.
Based on their annotations, four PCs appear to be directly involved in the metabolism of amino acids: PC-47 (methionine), PC-91 (glycine), PC-69 (arginine), and PC-103 (leucine) (Table IV). The components of PC-103 (PF1679/PF1680) are annotated as subunits of a putative 3-isopropylmalate dehydratase and have paralogs (PF0938/PF0939) within the leucine biosynthesis operon that are more likely to encode the true 3-isopropylmalate dehydratase. Based on a study with the closely related species Pyrococcus horikoshii (90), PC-103 is more likely to be involved in lysine biosynthesis. PC-69 is annotated as carbamoyl-phosphate synthetase, and we have previously presented evidence indicating that it is involved in arginine biosynthesis (33). This heterodimeric carbamoyl-phosphate synthetase is distinct from the (monomeric) carbamate kinase-like enzyme (PF0676) previously purified from P. furiosus that was incorrectly termed a carbamoyl-phosphate synthetase (91).
Four PCs appear to be involved in various aspects of carbohydrate metabolism, one in the tricarboxylic acid cycle (PC-12) and three concerned with monosaccharide phosphate derivatives (PC-52, PC-72, and PC-101). All of these PCs are predicted to be encoded by operons with the exception of PC-101, which is proposed to encode a carbohydrate kinase and a NUDIX protein. The latter is a type of NDP hydrolase, and the crystal structure of a recombinantly produced ortholog (expressed using a single gene) from a hyperthermophilic archaeon is known (92). Given the unknown function of these proteins in hyperthermophiles such as P. furiosus, a physiological heteromeric complex is a possibility.
PC-92 is a heterotrimeric complex with two components that indicate an involvement in the oxidation of glycerol phosphate. The third component (PF2007) is a basic hypothetical protein, but it appears to be a real component as the three proteins co-purify to the c3 level and are encoded by an operon. PC-6 appears to be a redox-active complex involved in isoprenoid biosynthesis based on the annotations of its two components.
PC-60 appears to be involved in the biosynthesis of the B-type vitamins pyridoxal and thiamine, whereas two potential PCs (PC-48 and PC-24) are proposed to be involved in the metabolism of iron and nickel (Table IV). PC-48, a five-component complex, contains three proteins homologous to those involved in iron-sulfur cluster biosynthesis that have been previously characterized individually from bacteria (SufCBD (93)). The other two components of PC-48, a histone-like protein (PF1284) and ruberythyrin (PF1283), an enzyme previously purified from P. furiosus as a homomeric protein (37), are unlikely to be true components because they only co-fractionate with the SufCBD complex at the c1 level and are not part of the predicted SufCBD operon (Table IV). Indeed the association score for PC-48 increases from 0.16 to 0.35 when PF1283 and PF1284 are excluded.
PC-24 is proposed to be involved in nickel metabolism. P. furiosus evolves hydrogen gas and contains three distinct hydrogenases, all of which contain a nickel-iron ([NiFe]) catalytic site (94). The genome encodes homologs for proteins that are known to be required for the biosynthesis of the [NiFe] site, and two of them, HypC and HypD, are components of PC-24 (Table IV). These two proteins appear to form a weak/transient complex in bacteria (Refs. 95 and 96; supplemental Table 6) but in P. furiosus remain together after four levels of chromatography, indicating that PC-24 forms a strong complex. PC-24 is found in c4 fractions containing PC-33, the cytoplasmic hydrogenase I (SH1), suggesting that it may form a stable complex with this enzyme (supplemental Table 5). This aspect is currently under further study.
Several potential PCs are involved in various aspects of nucleotide and polynucleotide metabolism. PC-11, a heterodimeric complex, is involved in purine biosynthesis, whereas two PCs are paralogous complexes (PC-9 and PC-64) that appear to be involved in modulating the activity of DNA gyrase. In P. furiosus, this is presumably the unusual DNA topoisomerase or reverse gyrase that is unique to hyperthermophilic organisms (97). Coincidentally another potential PC that may be involved in DNA binding (PC-82) is annotated as a chromosome segregation protein (98). It appears to be a low abundance PC because the component proteins were only found in a total of four fractions (Table IV). PC-95 might bind RNA and also contains a chaperone-type protein.
Another group of potential PC components are homologs of the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (cas) system. CRISPRs are a distinctive feature of the genomes of most prokaryotes and, along with cas genes, are thought to be involved in resistance to bacteriophages and constitute a bacterial immune system similar to that of RNA interference in higher organisms (99–101). P. furiosus has two identified CRISPR/cas regions containing 23 potential cas genes in the genome (99). Seventeen putative Cas proteins were identified in this fractionation, providing the first evidence for the presence of Cas proteins in the native proteome (supplemental Table 3). Twelve were found in four Type 1 PCs (Table IV). The genes for two of these (sub)complexes (PC-28 and PC-29) are predicted to form a single operon (32), and transcriptional analysis indicates that the genes are down-regulated in cold-adapted cells (34). Also down-regulated are two more genes within this operon whose products form PC-27, which is in the tentative PC category (discussed below; see Table V). The genes for the remaining two (sub)complexes, PC-40 and PC-41, are in three contiguous operons and appear to be up-regulated in cells grown on carbohydrates relative to peptides (33). Whether these proteins play a role as an archaeal immune system remains to be seen.
A total of 44 ribosomal proteins were identified in the native fractionation (supplemental Table 3). Twenty-two of these formed five strong, four tentative, and three weak potential PCs (Tables IV, V, and VI, respectively). The multiple (sub)complexes are likely a consequence of not detecting many ribosomal proteins by the HT MS/MS technique. For example, as shown in supplemental Table 5, PC-77, PC-79, and PC-104 were identified in c2 fractions, but all seven component proteins are predicted to be part of the same operon.
For many of the strong PCs, there is no indication of their true function as their components are classified as “hypothetical” and/or are poorly annotated. For example, PC-17 is composed of two very abundant (found in at least 47 fractions) hypothetical proteins encoded by a predicted operon (with an association score of 1.0) that are highly likely to be a stable complex. In some cases, the general function of one subunit is known, e.g. PC-42 where one component is a flavoprotein and the other is a hypothetical protein. Some PC components are annotated as homologs of proteins in higher eukaryotes, such as PC-51 (a von Willebrand factor homolog), whereas in others annotations are hard to rationalize. For example, PC-98 contains an acidic pilin-related protein and a very basic conserved hypothetical protein.
Tentative Novel PCs (30)—
Thirty potential PCs containing 61 proteins were identified that are defined as tentative because they were detected as complexes only in c1 fractions (Table V and supplemental Table 5). Some are likely to be true PCs whose partners were simply not identified by HT MS/MS in the c2–c4 fractions. These include 11 PCs that are predicted to be encoded by operons and have association scores of ≥0.2. Many are also likely to be in low abundance PCs; in fact, six complexes were found only in c1 fractions that were analyzed by CV MS/MS (PC-18, -27, -55, -73, -78, and -93) and were not detected by HT MS/MS. Several PCs have protein components with predicted basic pI values (>8.0) suggesting that they would not bind to an anion exchange column (c1) unless they were part of a true (acidic) multiprotein complex
Weak or Unlikely Novel PCs (13)—
Thirteen PCs containing 27 proteins fall into this category where one or both of their components are abundant proteins found in many fractions but typically not in the same one(s), i.e. they appear to fortuitously co-elute with their proposed partner in a few fractions (Table VI). This is reflected in their association scores (<0.2) and includes five proteins that have been previously characterized from P. furiosus as homomeric proteins. For example, phosphoenolpyruvate synthase (82) was found in over 150 fractions, whereas its proposed partner in PC-4, a seemingly unrelated metallohydrolase, was found in only six fractions. Moreover the genes encoding the two proteins are not predicted to be in an operon, and the association score is 0. Conversely argininosuccinate synthase and argininosuccinate lyase (PC-13) are enzymes of the same pathway and potentially could form a weakly interacting complex. Additional experimentation will be necessary to test the validity and/or strength of this and other PCs listed in Table VI.
DISCUSSION
Experimental Approach for Identification of PCs—
In this pilot study, the cytoplasmic extract of native biomass, which contains soluble PCs not removed (sedimented) by ultracentrifugation, was fractionated using sequential, non-denaturing chromatography steps. These included anion exchange, hydroxyapatite, hydrophobic interaction, affinity, and size exclusion. The fractionation was carried out using anaerobic conditions to maintain the integrity of any oxygen-sensitive PCs, e.g. those containing redox-sensitive metal cofactors, that might dissociate if cofactors are lost (35). Use of the native proteome also eliminated the problems associated with tagged and two-hybrid approaches for PC identification. Proteins were identified using a combination of CV and HT MS/MS on individual fractions generated at each chromatography level, whereas the identification of potential PCs utilized the MS/MS data in combination with co-fractionation data and bioinformatics analysis. This took advantage of the idea that gene clustering in prokaryotes can imply functional coupling (102, 103). Approximately 60% of the P. furiosus genes are predicted to be in operons (32); however, which of these encode stable protein complexes is not known. Our approach led to the initial identification of 106 potential Type 1 PCs. While this work was in progress, a similar method was reported in which aerobic purification of E. coli proteins was coupled to a sophisticated, quantitative MS-based analysis. However, in that study, only three previously known and very abundant protein complexes were identified (104). Similarly quantitative MS-based proteomics have also been applied to high molecular weight complexes in organelles separated by gradient centrifugation (105).
Analysis of Potential PCs Identified—
Analysis of all the fractions at each level of column chromatography using MS/MS allowed us to follow individual proteins through each fractionation level. As a result, 30 potential PCs identified at the c1 level were categorized as “tentative” based on the fact that the component proteins were not identified in the same fraction in any of the downstream columns (Table V and supplemental Table 5). Although some of these c1 complexes probably represent weak or false PCs, it is also possible that a number of them are true complexes that were not identified in c2–c4 fractions by MS/MS. This is supported by the fact that 35 of the 106 complexes found in downstream columns were not identified as PCs in the c1 column. Thus, although these proteins were clearly present in the fractionation, they were not detected in the MS/MS analysis of the more complex c1 fractions. The HT MS approach makes the procedure much more high throughput, but it is not as sensitive as the CV method.
The effectiveness of the MS/MS approaches used in the identification of proteins is understandably affected by the complexity of the protein mixture and the presence of abundant proteins. This is evident at the column level where new proteins were detected as the complexity decreased (supplemental Table 4) and also at the fraction level where, even for known PCs, all of the component proteins were either not identified or were not identified together at every column level (see Table II and supplemental Table 5). In addition, one known complex (PC-106) and several expected complexes were identified as PCs only in post-c1 columns (supplemental Table 5). Clearly MS/MS does not identify all of the component proteins in PCs even when they are present. This can be extrapolated to the unknown PCs that have been identified in this study. Multilevel MS/MS also enabled us to look at the extent of overlap between potential components of PCs within the individual fractions of each column at each level. Thus, potential PCs with partners that overlapped minimally (were present in one or two fractions but separate in additional fractions) were categorized as “unlikely” or “weak.” This method also allowed the identification of “abundant” proteins that are present at relatively high intracellular concentrations and often form part of the “core” proteome or are involved in central metabolism for a specific growth condition, such as ribosomes, chaperones, and major metabolic enzymes. Identification of “high abundance” partners in PCs that could arise as a result of nonspecific co-occurrence was based on the number of fractions in which they were identified.
Based on this pilot fractionation, we are currently developing HT analytical scale HPLC-based methods to reduce the amount of native biomass required and thus widen the applicability of the approach. Moreover this approach would also enable a more complete analysis of P. furiosus PCs because only 5% of the c2 fractions were fractionated onto c3 columns, and only 12% of those fractions were analyzed on c4 columns. A more comprehensive HT fractionation would include c3 columns that represent all c2 fractions, and all c3 fractions could be analyzed by c4 columns. This would maximize the number of proteins identified as the c4 fractions would be of low complexity, making it possible to identify Type 2 complexes as well as additional low abundance complexes. The use of the calculated association values appears to be an excellent criterion for assessing the validity of the potential PCs (supplemental Table 5). In addition, a major advantage of native biomass fractionation is that it provides a road map for the (partial) purification of all identified proteins, not only potential PCs, whereas low abundance PCs can be targeted for recombinant protein production by co-expression of the relevant genes.
Supplementary Material
Acknowledgments
We thank the following for assistance with the fractionation procedure: Scott Hamilton-Brehm, Stephanie Bridger, Kristina Carswell, Sonya Clarkson, Chris Coey, Bret Dillard, Brian Gerwe, Chris Hopkins, Andrew Lancaster, Gerrit Schut, Angela Snow, Kari Strand, and Rathinam Viswanathan.
Footnotes
Published, MCP Papers in Press, November 28, 2008, DOI 10.1074/mcp.M800246-MCP200
The abbreviations used are: PC, protein complex; ACS, acyl-CoA synthetase; CV, conventional; Fd, ferredoxin; FNOR, ferredoxin-NADPH oxidoreductase; HT, high throughput; POR, pyruvate ferredoxin oxidoreductase; SH1, soluble hydrogenase 1; VOR, 2-ketoisovalerate ferredoxin oxidoreductase; KOR, 2-keto acid oxidoreductase; CRISPR, clustered regularly interspaced short palindromic repeats; Cas, CRISPR-associated; LTQ, linear trap quadropole.
This work was carried out as part of the MAGGIE project under the direction of John A. Tainer and supported by United States Department of Energy Grant DE-FG0207ER64326.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Martin, A. C., and Drubin, D. G. ( 2003) Impact of genome-wide functional analyses on cell biology research. Curr. Opin. Cell Biol. 15, 6–13 [DOI] [PubMed] [Google Scholar]
- 2.Alberts, B. ( 1998) The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92, 291–294 [DOI] [PubMed] [Google Scholar]
- 3.Bamford, D. H., Gilbert, R. J., Grimes, J. M., and Stuart, D. I. ( 2001) Macromolecular assemblies: greater than their parts. Curr. Opin. Struct. Biol. 11, 107–113 [DOI] [PubMed] [Google Scholar]
- 4.Wilson, D. N., and Nierhaus, K. H. ( 2005) Ribosomal proteins in the spotlight. Crit. Rev. Biochem. Mol. Biol. 40, 243–267 [DOI] [PubMed] [Google Scholar]
- 5.Chiu, W., Baker, M. L., and Almo, S. C. ( 2006) Structural biology of cellular machines. Trends Cell Biol. 16, 144–150 [DOI] [PubMed] [Google Scholar]
- 6.Gilbert, H. J. ( 2007) Cellulosomes: microbial nanomachines that display plasticity in quaternary structure. Mol. Microbiol. 63, 1568–1576 [DOI] [PubMed] [Google Scholar]
- 7.Srere, P. A. ( 1987) Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 56, 89–124 [DOI] [PubMed] [Google Scholar]
- 8.Ovadi, J., and Srere, P. A. ( 2000) Macromolecular compartmentation and channeling. Int. Rev. Cytol. 192, 255–280 [DOI] [PubMed] [Google Scholar]
- 9.Ovadi, J., and Saks, V. ( 2004) On the origin of intracellular compartmentation and organized metabolic systems. Mol. Cell. Biochem. 256–257, 5–12 [DOI] [PubMed] [Google Scholar]
- 10.Berggard, T., Linse, S., and James, P. ( 2007) Methods for the detection and analysis of protein-protein interactions. Proteomics 7, 2833–2842 [DOI] [PubMed] [Google Scholar]
- 11.Piehler, J. ( 2005) New methodologies for measuring protein interactions in vivo and in vitro. Curr. Opin. Struct. Biol. 15, 4–14 [DOI] [PubMed] [Google Scholar]
- 12.Azarkan, M., Huet, J., Baeyens-Volant, D., Looze, Y., and Vandenbussche, G. ( 2007) Affinity chromatography: a useful tool in proteomics studies. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 849, 81–90 [DOI] [PubMed] [Google Scholar]
- 13.Fields, S., and Song, O. ( 1989) A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 [DOI] [PubMed] [Google Scholar]
- 14.Uetz, P., and Hughes, R. E. ( 2000) Systematic and large-scale two-hybrid screens. Curr. Opin. Microbiol. 3, 303–308 [DOI] [PubMed] [Google Scholar]
- 15.Koegl, M., and Uetz, P. ( 2007) Improving yeast two-hybrid screening systems. Brief. Funct. Genomics Proteomics 6, 302–312 [DOI] [PubMed] [Google Scholar]
- 16.Deane, C. M., Salwinski, L., Xenarios, I., and Eisenberg, D. ( 2002) Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell. Proteomics 1, 349–356 [DOI] [PubMed] [Google Scholar]
- 17.Bauer, A., and Kuster, B. ( 2003) Affinity purification-mass spectrometry. Powerful tools for the characterization of protein complexes. Eur. J. Biochem. 270, 570–578 [DOI] [PubMed] [Google Scholar]
- 18.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., Yang, L., Wolting, C., Donaldson, I., Schandorff, S., Shewnarane, J., Vo, M., Taggart, J., Goudreault, M., Muskat, B., Alfarano, C., Dewar, D., Lin, Z., Michalickova, K., Willems, A. R., Sassi, H., Nielsen, P. A., Rasmussen, K. J., Andersen, J. R., Johansen, L. E., Hansen, L. H., Jespersen, H., Podtelejnikov, A., Nielsen, E., Crawford, J., Poulsen, V., Sorensen, B. D., Matthiesen, J., Hendrickson, R. C., Gleeson, F., Pawson, T., Moran, M. F., Durocher, D., Mann, M., Hogue, C. W., Figeys, D., and Tyers, M. ( 2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 19.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. ( 1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 20.Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Seraphin, B. ( 2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 21.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., Remor, M., Hofert, C., Schelder, M., Brajenovic, M., Ruffner, H., Merino, A., Klein, K., Hudak, M., Dickson, D., Rudi, T., Gnau, V., Bauch, A., Bastuck, S., Huhse, B., Leutwein, C., Heurtier, M. A., Copley, R. R., Edelmann, A., Querfurth, E., Rybin, V., Drewes, G., Raida, M., Bouwmeester, T., Bork, P., Seraphin, B., Kuster, B., Neubauer, G., and Superti-Furga, G. ( 2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 22.Masters, S. C. ( 2004) Co-immunoprecipitation from transfected cells. Methods Mol. Biol. 261, 337–350 [DOI] [PubMed] [Google Scholar]
- 23.Farhoud, M. H., Wessels, H. J., Steenbakkers, P. J., Mattijssen, S., Wevers, R. A., van Engelen, B. G., Jetten, M. S., Smeitink, J. A., van den Heuvel, L. P., and Keltjens, J. T. ( 2005) Protein complexes in the archaeon Methanothermobacter thermautotrophicus analyzed by blue native/SDS-PAGE and mass spectrometry. Mol. Cell. Proteomics 4, 1653–1663 [DOI] [PubMed] [Google Scholar]
- 24.Krause, F. ( 2006) Detection and analysis of protein-protein interactions in organellar and prokaryotic proteomes by native gel electrophoresis: (membrane) protein complexes and supercomplexes. Electrophoresis 27, 2759–2781 [DOI] [PubMed] [Google Scholar]
- 25.Lasserre, J. P., Beyne, E., Pyndiah, S., Lapaillerie, D., Claverol, S., and Bonneu, M. ( 2006) A complexomic study of Escherichia coli using two-dimensional blue native/SDS polyacrylamide gel electrophoresis. Electrophoresis 27, 3306–3321 [DOI] [PubMed] [Google Scholar]
- 26.Pyndiah, S., Lasserre, J. P., Menard, A., Claverol, S., Prouzet-Mauleon, V., Megraud, F., Zerbib, F., and Bonneu, M. ( 2007) Two-dimensional blue native/SDS gel electrophoresis of multiprotein complexes from Helicobacter pylori. Mol. Cell. Proteomics 6, 193–206 [DOI] [PubMed] [Google Scholar]
- 27.Gygi, S. P., Corthals, G. L., Zhang, Y., Rochon, Y., and Aebersold, R. ( 2000) Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc. Natl. Acad. Sci. U. S. A. 97, 9390–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiala, G., and Stetter, K. O. ( 1986) Pyrococcus furiosus sp. nov., represents a novel genus of marine heterotrophic Archaebacteria growing optimally at 100°C. Arch. Microbiol. 145, 56–61 [Google Scholar]
- 29.Robb, F. T., Maeder, D. L., Brown, J. R., DiRuggiero, J., Stump, M. D., Yeh, R. K., Weiss, R. B., and Dunn, D. M. ( 2001) Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 330, 134–157 [DOI] [PubMed] [Google Scholar]
- 30.Poole, F. L., II, Gerwe, B. A., Hopkins, R. C., Schut, G. J., Weinberg, M. V., Jenney, F. E., Jr., and Adams, M. W. ( 2005) Defining genes in the genome of the hyperthermophilic archaeon Pyrococcus furiosus: implications for all microbial genomes. J. Bacteriol. 187, 7325–7332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergman, N. H., Passalacqua, K. D., Hanna, P. C., and Qin, Z. S. ( 2007) Operon prediction for sequenced bacterial genomes without experimental information. Appl. Environ. Microbiol. 73, 846–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran, T. T., Dam, P., Su, Z. C., Poole, F. L., Adams, M. W. W., Zhou, G. T., and Xu, Y. ( 2007) Operon prediction in Pyrococcus furiosus. Nucleic Acids Res. 35, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schut, G. J., Brehm, S. D., Datta, S., and Adams, M. W. ( 2003) Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J. Bacteriol. 185, 3935–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinberg, M. V., Schut, G. J., Brehm, S., Datta, S., and Adams, M. W. ( 2005) Cold shock of a hyperthermophilic archaeon: Pyrococcus furiosus exhibits multiple responses to a suboptimal growth temperature with a key role for membrane-bound glycoproteins. J. Bacteriol. 187, 336–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verhagen, M. F., Menon, A. L., Schut, G. J., and Adams, M. W. ( 2001) Pyrococcus furiosus: large-scale cultivation and enzyme purification. Methods Enzymol. 330, 25–30 [DOI] [PubMed] [Google Scholar]
- 36.Ma, K., and Adams, M. W. W. ( 1994) Sulfide dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus—a new multifunctional enzyme involved in the reduction of elemental sulfur. J. Bacteriol. 176, 6509–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinberg, M. V., Jenney, F. E., Cui, X. Y., and Adams, M. W. W. ( 2004) Rubrerythrin from the hyperthermophilic archaeon Pyrococcus furiosus is a rubredoxin-dependent, iron-containing peroxidase. J. Bacteriol. 186, 7888–7895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schut, G. J., Menon, A. L., and Adams, M. W. W. ( 2001) 2-Ketoacid oxidoreductases from Pyrococcus furiosus and Thermococcus litoralis. Methods Enzymol. 331, 144–158 [DOI] [PubMed] [Google Scholar]
- 39.Mukund, S., and Adams, M. W. W. ( 1991) The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an aldehyde ferredoxin oxidoreductase. Evidence for its participation in a unique glycolytic pathway. J. Biol. Chem. 266, 14208–14216 [PubMed] [Google Scholar]
- 40.Ma, K., and Adams, M. W. W. ( 1999) A hyperactive NAD(P)H:rubredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 181, 5530–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bryant, F. O., and Adams, M. W. ( 1989) Characterization of hydrogenase from the hyperthermophilic archaebacterium, Pyrococcus furiosus. J. Biol. Chem. 264, 5070–5079 [PubMed] [Google Scholar]
- 42.Blake, P. R., Park, J. B., Bryant, F. O., Aono, S., Magnuson, J. K., Eccleston, E., Howard, J. B., Summers, M. F., and Adams, M. W. W. ( 1991) Determinants of protein hyperthermostability—purification and amino-acid-sequence of rubredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus and secondary structure of the zinc adduct by NMR. Biochemistry 30, 10885–10895 [DOI] [PubMed] [Google Scholar]
- 43.Aono, S., Bryant, F. O., and Adams, M. W. ( 1989) A novel and remarkably thermostable ferredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus. J. Bacteriol. 171, 3433–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bradford, M. M. ( 1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 45.Pruitt, K. D., Tatusova, T., and Maglott, D. R. ( 2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., and Lopez, R. ( 2005) InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holden, J. F., Tollaksen, S. L., Giometti, C. S., Lim, H., Yates, J. R., and Adams, M. W. W. ( 2001) Identification of membrane proteins in the hyperthermophilic archaeon Pyrococcus furiosus using proteomics and prediction programs. Comp. Funct. Genomics 2, 275–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blamey, J. M., and Adams, M. W. W. ( 1993) Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim. Biophys. Acta 1161, 19–27 [DOI] [PubMed] [Google Scholar]
- 49.Uemori, T., Sato, Y., Kato, I., Doi, H., and Ishino, Y. ( 1997) A novel DNA polymerase in the hyperthermophilic archaeon, Pyrococcus furiosus: gene cloning, expression, and characterization. Genes Cells 02, 499–512 [DOI] [PubMed] [Google Scholar]
- 50.Cann, I. K. O., Ishino, S., Yuasa, M., Daiyasu, H., Toh, H., and Ishino, Y. ( 2001) Biochemical analysis of replication factor C from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183, 2614–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedone, E., Ren, B., Ladenstein, R., Rossi, M., and Bartolucci, S. ( 2004) Functional properties of the protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus: a member of a novel protein family related to protein disulfide-isomerase. Eur. J. Biochem. 271, 3437–3448 [DOI] [PubMed] [Google Scholar]
- 52.Pisa, K. Y., Huber, H., Thomm, M., and Muller, V. ( 2007) A sodium ion-dependent A1AO ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus. FEBS J. 274, 3928–3938 [DOI] [PubMed] [Google Scholar]
- 53.Mai, X., and Adams, M. W. ( 1994) Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J. Biol. Chem. 269, 16726–16732 [PubMed] [Google Scholar]
- 54.Massant, J., and Glansdorff, N. ( 2005) New experimental approaches for investigating interactions between Pyrococcus furiosus carbamate kinase and carbamoyltransferases, enzymes involved in the channeling of thermolabile carbamoyl phosphate. Archaea 1, 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monaghan, P. J., Leys, D., and Scrutton, N. S. ( 2007) Mechanistic aspects and redox properties of hyperthermophilic l-proline dehydrogenase from Pyrococcus furiosus related to dimethylglycine dehydrogenase/oxidase. FEBS J. 274, 2070–2087 [DOI] [PubMed] [Google Scholar]
- 56.Ma, K., Weiss, R., and Adams, M. W. ( 2000) Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 182, 1864–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sapra, R., Verhagen, M. F. J. M., and Adams, M. W. W. ( 2000) Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 182, 3423–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goede, B., Naji, S., von Kampen, O., Ilg, K., and Thomm, M. ( 2006) Protein-protein interactions in the archaeal transcriptional machinery. Binding studies of isolated RNA polymerase subunits and transcription factors. J. Biol. Chem. 281, 30581–30592 [DOI] [PubMed] [Google Scholar]
- 59.Bocs, C., Buhler, C., Forterre, P., and Bergerat, A. ( 2001) DNA topoisomerases VI from hyperthermophilic archaea. Methods Enzymol. 334, 172–179 [DOI] [PubMed] [Google Scholar]
- 60.Komori, K., and Ishino, Y. ( 2001) Replication protein A in Pyrococcus furiosus is involved in homologous DNA recombination. J. Biol. Chem. 276, 25654–25660 [DOI] [PubMed] [Google Scholar]
- 61.Kletzin, A., and Adams, M. W. W. ( 1996) Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J. Bacteriol. 178, 248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guagliardi, A., de Pascale, D., Cannio, R., Nobile, V., Bartolucci, S., and Rossi, M. ( 1995) The purification, cloning, and high level expression of a glutaredoxin-like protein from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 270, 5748–5755 [DOI] [PubMed] [Google Scholar]
- 63.Shiraki, K., Tsuji, M., Hashimoto, Y., Fujimoto, K., Fujiwara, S., Takagi, M., and Imanaka, T. ( 2003) Genetic, enzymatic, and structural analyses of phenylalanyl-tRNA synthetase from Thermococcus kodakaraensis KOD1. J. Biochem. 134, 567–574 [DOI] [PubMed] [Google Scholar]
- 64.Oshikane, H., Sheppard, K., Fukai, S., Nakamura, Y., Ishitani, R., Numata, T., Sherrer, R. L., Feng, L., Schmitt, E., Panvert, M., Blanquet, S., Mechulam, Y., Soll, D., and Nureki, O. ( 2006) Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science 312, 1950–1954 [DOI] [PubMed] [Google Scholar]
- 65.Schmitt, E., Panvert, M., Blanquet, S., and Mechulam, Y. ( 2005) Structural basis for tRNA-dependent amidotransferase function. Structure ( Lond.) 13, 1421–1433 [DOI] [PubMed] [Google Scholar]
- 66.Ramos, C. R., Oliveira, C. L., Torriani, I. L., and Oliveira, C. C. ( 2006) The Pyrococcus exosome complex. Structural and functional characterization. J. Biol. Chem. 281, 6751–6759 [DOI] [PubMed] [Google Scholar]
- 67.Mai, X., and Adams, M. W. ( 1996) Characterization of a fourth type of 2-keto acid-oxidizing enzyme from a hyperthermophilic archaeon: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J. Bacteriol. 178, 5890–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawakami, R., Sakuraba, H., Tsuge, H., Goda, S., Katunuma, N., and Ohshima, T. ( 2005) A second novel dye-linked l-proline dehydrogenase complex is present in the hyperthermophilic archaeon Pyrococcus horikoshii OT-3. FEBS J. 272, 4044–4054 [DOI] [PubMed] [Google Scholar]
- 69.Kerrien, S., Alam-Faruque, Y., Aranda, B., Bancarz, I., Bridge, A., Derow, C., Dimmer, E., Feuermann, M., Friedrichsen, A., Huntley, R., Kohler, C., Khadake, J., Leroy, C., Liban, A., Lieftink, C., Montecchi-Palazzi, L., Orchard, S., Risse, J., Robbe, K., Roechert, B., Thorneycroft, D., Zhang, Y., Apweiler, R., and Hermjakob, H. ( 2007) IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 35, D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deluca, T. F., Wu, I. H., Pu, J., Monaghan, T., Peshkin, L., Singh, S., and Wall, D. P. ( 2006) Roundup: a multi-genome repository of orthologs and evolutionary distances. Bioinformatics 22, 2044–2046 [DOI] [PubMed] [Google Scholar]
- 71.Steen, I. H., Madern, D., Karlstrom, M., Lien, T., Ladenstein, R., and Birkeland, N. K. ( 2001) Comparison of isocitrate dehydrogenase from three hyperthermophiles reveals differences in thermostability, cofactor specificity, oligomeric state, and phylogenetic affiliation. J. Biol. Chem. 276, 43924–43931 [DOI] [PubMed] [Google Scholar]
- 72.Muir, J. M., Russell, R. J., Hough, D. W., and Danson, M. J. ( 1995) Citrate synthase from the hyperthermophilic Archaeon, Pyrococcus furiosus. Protein Eng. 8, 583–592 [DOI] [PubMed] [Google Scholar]
- 73.Mai, X., and Adams, M. W. ( 1996) Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 178, 5897–5903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laksanalamai, P., Pavlov, A. R., Slesarev, A. I., and Robb, F. T. ( 2006) Stabilization of Taq DNA polymerase at high temperature by protein folding pathways from a hyperthermophilic archaeon, Pyrococcus furiosus. Biotechnol. Bioeng. 93, 1–5 [DOI] [PubMed] [Google Scholar]
- 75.Riera, J., Robb, F. T., Weiss, R., and Fontecave, M. ( 1997) Ribonucleotide reductase in the archaeon Pyrococcus furiosus: a critical enzyme in the evolution of DNA genomes? Proc. Natl. Acad. Sci. U. S. A. 94, 475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsunasawa, S., Izu, Y., Miyagi, M., and Kato, I. ( 1997) Methionine aminopeptidase from the hyperthermophilic archaeon Pyrococcus furiosus: molecular cloning and overexpression in Escherichia coli of the gene, and characteristics of the enzyme. J. Biochem. 122, 843–850 [DOI] [PubMed] [Google Scholar]
- 77.Cann, I. K., Ishino, S., Hayashi, I., Komori, K., Toh, H., Morikawa, K., and Ishino, Y. ( 1999) Functional interactions of a homolog of proliferating cell nuclear antigen with DNA polymerases in Archaea. J. Bacteriol. 181, 6591–6599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mayer, K. L., Qu, Y., Bansal, S., LeBlond, P. D., Jenney, F. E., Jr., Brereton, P. S., Adams, M. W., Xu, Y., and Prestegard, J. H. ( 2006) Structure determination of a new protein from backbone-centered NMR data and NMR-assisted structure prediction. Proteins 65, 480–489 [DOI] [PubMed] [Google Scholar]
- 79.Maeshima, H., Okuno, E., Aimi, T., Morinaga, T., and Itoh, T. ( 2001) An archaeal protein homologous to mammalian SRP54 and bacterial Ffh recognizes a highly conserved region of SRP RNA. FEBS Lett. 507, 336–340 [DOI] [PubMed] [Google Scholar]
- 80.Komori, K., Miyata, T., DiRuggiero, J., Holley-Shanks, R., Hayashi, I., Cann, I. K., Mayanagi, K., Shinagawa, H., and Ishino, Y. ( 2000) Both RadA and RadB are involved in homologous recombination in Pyrococcus furiosus. J. Biol. Chem. 275, 33782–33790 [DOI] [PubMed] [Google Scholar]
- 81.Chen, H. Y., Chu, Z. M., Ma, Y. H., Zhang, Y., and Yang, S. L. ( 2007) Expression and characterization of the chaperonin molecular machine from the hyperthermophilic archaeon Pyrococcus furiosus. J. Basic Microbiol. 47, 132–137 [DOI] [PubMed] [Google Scholar]
- 82.Hutchins, A. M., Holden, J. F., and Adams, M. W. W. ( 2001) Phosphoenolpyruvate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183, 709–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Borges, K. M., Bergerat, A., Bogert, A. M., DiRuggiero, J., Forterre, P., and Robb, F. T. ( 1997) Characterization of the reverse gyrase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 179, 1721–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roy, R., Mukund, S., Schut, G. J., Dunn, D. M., Weiss, R., and Adams, M. W. ( 1999) Purification and molecular characterization of the tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus: the third of a putative five-member tungstoenzyme family. J. Bacteriol. 181, 1171–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mukund, S., and Adams, M. W. ( 1995) Glyceraldehyde-3-phosphate ferredoxin oxidoreductase, a novel tungsten-containing enzyme with a potential glycolytic role in the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 270, 8389–8392 [DOI] [PubMed] [Google Scholar]
- 86.Siebers, B., Brinkmann, H., Dorr, C., Tjaden, B., Lilie, H., van der Oost, J., and Verhees, C. H. ( 2001) Archaeal fructose-1,6-bisphosphate aldolases constitute a new family of archaeal type class I aldolase. J. Biol. Chem. 276, 28710–28718 [DOI] [PubMed] [Google Scholar]
- 87.Adams, M. W. W. ( 1999) The biochemical diversity of life near and above 100°C in marine environments. J. Appl. Microbiol. 85, 108S–117S [DOI] [PubMed] [Google Scholar]
- 88.Heider, J., Mai, X., and Adams, M. W. ( 1996) Characterization of 2-ketoisovalerate ferredoxin oxidoreductase, a new and reversible coenzyme A-dependent enzyme involved in peptide fermentation by hyperthermophilic archaea. J. Bacteriol. 178, 780–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chou, C. J., Shockley, K. R., Conners, S. B., Lewis, D. L., Comfort, D. A., Adams, M. W., and Kelly, R. M. ( 2007) Impact of substrate glycoside linkage and elemental sulfur on bioenergetics of and hydrogen production by the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 73, 6842–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lombo, T., Takaya, N., Miyazaki, J., Gotoh, K., Nishiyama, M., Kosuge, T., Nakamura, A., and Hoshino, T. ( 2004) Functional analysis of the small subunit of the putative homoaconitase from Pyrococcus horikoshii in the Thermus lysine biosynthetic pathway. FEMS Microbiol. Lett. 233, 315–324 [DOI] [PubMed] [Google Scholar]
- 91.Durbecq, V., Legrain, C., Roovers, M., Pierard, A., and Glansdorff, N. ( 1997) The carbamate kinase-like carbamoyl phosphate synthetase of the hyperthermophilic archaeon Pyrococcus furiosus, a missing link in the evolution of carbamoyl phosphate biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 94, 12803–12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang, S., Mura, C., Sawaya, M. R., Cascio, D., and Eisenberg, D. ( 2002) Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets. Acta Crystallogr. Sect. D Biol. Crystallogr. 58, 571–578 [DOI] [PubMed] [Google Scholar]
- 93.Fontecave, M., Choudens, S. O., Py, B., and Barras, F. ( 2005) Mechanisms of iron-sulfur cluster assembly: the SUF machinery. J. Biol. Inorg. Chem. 10, 713–721 [DOI] [PubMed] [Google Scholar]
- 94.Jenney, F. E., and Adams, M. W. W. ( 2008) The impact of extremophiles on structural genomics (and vice versa). Extremophiles 12, 39–50 [DOI] [PubMed] [Google Scholar]
- 95.Blokesch, M., and Bock, A. ( 2002) Maturation of [NiFe]-hydrogenases in Escherichia coli: the HypC cycle. J. Mol. Biol. 324, 287–296 [DOI] [PubMed] [Google Scholar]
- 96.Blokesch, M., Albracht, S. P., Matzanke, B. F., Drapal, N. M., Jacobi, A., and Bock, A. ( 2004) The complex between hydrogenase-maturation proteins HypC and HypD is an intermediate in the supply of cyanide to the active site iron of [NiFe]-hydrogenases. J. Mol. Biol. 344, 155–167 [DOI] [PubMed] [Google Scholar]
- 97.Forterre, P. ( 2002) A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 18, 236–237 [DOI] [PubMed] [Google Scholar]
- 98.Tadesse, S., and Graumann, P. L. ( 2006) Differential and dynamic localization of topoisomerases in Bacillus subtilis. J. Bacteriol. 188, 3002–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Godde, J. S., and Bickerton, A. ( 2006) The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 62, 718–729 [DOI] [PubMed] [Google Scholar]
- 100.Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., Romero, D. A., and Horvath, P. ( 2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 [DOI] [PubMed] [Google Scholar]
- 101.Makarova, K. S., Grishin, N. V., Shabalina, S. A., Wolf, Y. I., and Koonin, E. V. ( 2006) A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Overbeek, R., Fonstein, M., D'Souza, M., Pusch, G. D., and Maltsev, N. ( 1999) The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. U. S. A. 96, 2896–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ogata, H., Fujibuchi, W., Goto, S., and Kanehisa, M. ( 2000) A heuristic graph comparison algorithm and its application to detect functionally related enzyme clusters. Nucleic Acids Res. 28, 4021–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dong, M., Yang, L. L., Williams, K., Fisher, S. J., Hall, S. C., Biggin, M. D., Jin, J., and Witkowska, H. E. ( 2008) A “tagless” strategy for identification of stable protein complexes genome-wide by multidimensional orthogonal chromatographic separation and iTRAQ reagent tracking. J. Proteome Res. 7, 1836–1849 [DOI] [PubMed] [Google Scholar]
- 105.Hartman, N. T., Sicilia, F., Lilley, K. S., and Dupree, P. ( 2007) Proteomic complex detection using sedimentation. Anal. Chem. 79, 2078–2083 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.