Abstract
Current methods in the noninvasive detection and surveillance of bladder cancer via urine analysis include voided urine cytology (VUC) and some diagnostic urinary protein biomarkers; however, due to the poor sensitivity of VUC and high false-positive rates of currently available protein assays, detection of bladder cancer via urinalysis remains a challenge. In the study presented here, a rapid, high-sensitivity technique was developed to profile the N-linked glycoprotein component in naturally micturated human urine specimens. Concanavalin A (Con A) affinity chromatography coupled to nanoflow liquid chromatography was utilized to separate the complex peptide mixture prior to a linear ion trap MS analysis. Of 186 proteins identified with high confidence by multiple analyses, 40% were secreted proteins, 18% membrane proteins, and 14% extracellular proteins. In this study, the presence of several proteins appeared to be associated with the presence of bladder cancer, including α-1B-glycoprotein that was detected in all tumor-bearing patient samples but in none of the samples obtained from non-tumor-bearing individuals. The combination of Con A affinity chromatography and nano-LC/MS/MS provides an initial investigation of N-glycoproteins in complex biological samples and facilitates the identification of potential biomarkers of bladder cancer in noninvasively obtained human urine.
Keywords: glycosylation, bladder cancer, lectin
Introduction
Cancer of the urinary bladder is among the five most common malignancies worldwide.1 Transitional cell carcinomas (TCCs) are the most common urothelial tumors in Western countries and constitute approximately 95% of all cases.2 Early detection remains one of the most urgent issues in bladder cancer research. When detected early, the 5-year survival rate is approximately 94%; thus, timely intervention dramatically increases the patient survival rate. Urothelial tumors can be classified into two groups on the basis of histopathology and clinical behavior. At presentation, more than 80% of bladder tumors are non-muscle-invasive papillary tumors (pTa or pT1). The remaining 20% of tumors that show muscle invasion at the time of diagnosis have a much less favorable prognosis. While radical surgery is required for invasive bladder tumors, superficial lesions are treated more conservatively by transurethral resection. However, more than 70% of patients with Ta/T1 lesions confined to the mucosa have recurrence during the first 2 years. If left untreated, these initially superficial lesions can progress to being muscle-invasive.3 The recurrence phenomenon of superficial bladder tumors makes bladder cancer one of the most prevalent cancers worldwide. Patients with superficial tumors are under continued surveillance by routine cystoscopy examinations of the bladder for early detection of new tumor developments. Once bladder tumors are identified and removed, patients will routinely get surveillance cystoscopy every 3 months for 2 years, then every 6 months for 2 years, and then yearly thereafter. Consequently, the development of noninvasive urinalysis assays using reliable diagnostic markers would be of tremendous benefit to both patients and healthcare providers.
Voided urine cytology (VUC) remains the method of choice for the noninvasive detection of bladder cancer, with its main use being to recognize the presence of recurrence and early progression in stage and grade. VUC can be used to diagnose new malignancy, yet while it has a specificity of >93%, its sensitivity is only 25–40%, especially for low-grade and low-stage tumors.4 Furthermore, results are not available rapidly, it is prone to interobserver variation, and it is relatively expensive. Understandably, a good deal of research has focused on identifying potential urine tumor markers with higher sensitivity than urine cytology. Promising diagnostic protein markers are NMP-22, BTA, BLCA-4, and cell proliferation proteins. Unfortunately, these tests also suffer from high false-positive rates, and thus, there is no protein test available to date that can replace urine cytology.5 Thus, detecting bladder cancer using diagnostic markers still remains a challenge.
Newly developed, high-throughput techniques, i.e., time-of-flight mass spectrometry, SDS/PAGE with matrix-assisted laser desorption/ionization ion trap mass spectrometry, and liquid-phase 2-D separation techniques greatly facilitate the analysis of proteins in biological samples.6 However, only a limited number of proteomic studies utilizing the newer technologies have been conducted in the analysis of urological cancers, largely because of the lack of defined methodologies that can reduce the complexity of the sample and rapidly and accurately identify specific proteins.
In this study, we have investigated the feasibility of profiling a glycoprotein component of the naturally micturated urinary proteome and applied optimized analyses to compare the profile of a panel of urine samples obtained from patients with bladder cancer and nonmalignant bladder conditions. We utilized the ability of an immobilized α-mannose-binding lectin, concanavalin A (Con A), to enrich N-linked glycoproteins from human urine. The enriched glycoproteins were then digested with trypsin and analyzed with nano-LC/MS/MS. A total of 128 distinct N-linked glycoproteins were identified with high confidence by multiple analysis of as little as 10 mL of naturally micturated urine. The majority of identified proteins were secreted, or membranous, proteins, and a subset of proteins were identified that were commonly excreted in urine from bladder cancer patients. The combinatorial approach of Con A affinity chromatography and nano-LC/MS/MS provides high sensitivity and with relatively moderate labor demands can greatly facilitate the identification of potential biomarkers of bladder cancer from noninvasively obtained human urine.
Experimental Procedures
Urine Collection
Urine specimens were collected from the University of Florida urology clinic at Shands & UF Hospital, Jacksonville, FL. Eight samples of urine were collected from patients cystoscopically confirmed to have bladder cancer (n = 5) or to have nontumor conditions (n = 3). An additional two normal donor urine specimens (n = 2) were obtained from asymptomatic volunteers. Samples were collected from patients visiting the clinic according to University of Florida IRB approved consent protocols. Each sample consisted of 15–50 mL of midstream urine collected in a sterile cup. All specimens were processed in the same way, with no special treatment being performed to remove occasional blood prior to analysis to preserve the intrinsic components. Table 1 shows patient clinical details.
Table 1.
Clinicopathological Data of the Study Samples
| patient sample | disease status | sex | age |
|---|---|---|---|
| Tumor-Bearing Patients | |||
| T4 | low-grade bladder cancer | female | 53 |
| T3 | high-grade bladder cancer | male | 52 |
| T9 | high-grade bladder cancer | male | 71 |
| T8 | high-grade bladder cancer | female | 75 |
| T13 | high-grade bladder cancer | male | 79 |
| Non-Tumor-Bearing Patients | |||
| N3 | hematuria, no tumor | male | 66 |
| N10 | hematuria, no tumor | male | ND |
| N2 | hematuria, no tumor | male | 53 |
| N5 | asymptomatic donor | female | 28 |
| N7 | asymptomatic donor | female | 29 |
Sample Preparation
Cells and debris were removed from urine samples by centrifugation at 5000g for 10 min at room temperature, and the supernatant was stored frozen at −80 °C until analysis. Four times the sample volumes of cold (−20 °C) acetone was added to the urine specimen for protein precipitation. The sample was left at 20 °C for 1 h followed by centrifugation at 12000g for 15 min at 4 °C. The supernatant was removed. The tube was left open at room temperature to partially remove the remaining solvent. The pellet was resuspended in binding buffer (20 mM Tris, 0.15 M NaCl, 1 mM MnCl2, and 1 mM CaCl2, pH 7.4). The sample was vortexed vigorously to completely dissolve the pelleted protein. The protein amounts in the sample concentrates were determined using the Bradford protein assay (Bio-Rad, Hercules, CA).
Con A Lectin Affinity Chromatography
An agarose-bound lectin, agarose-bound concanavalin A (Con A), was purchased from Vector Laboratories (Burlingame, CA). A 500 μL sample of agarose-bound Con A was packed into a disposable screw end cap spin column with filters at both ends. The binding and removal of nonspecific binding were done as previously described.7 The binding buffer consisted of 20 mM Tris, 1 mM MnCl2, 1 mM CaCl2, and 0.15 M NaCl, pH 7.4, and the elution buffer was 0.4 M methyl-α-D-mannopyroside in 20 mM Tris and 0.5 M NaCl, pH 7.0. The eluted fractions were pooled, and the Con A-captured samples were subjected to quantitation by Bradford assay (Bio-Rad, Hercules, CA).
Tryptic Digestion/PNGase F Treatment
The sample was concentrated using Microcon YM-10 (Millipore Corp., Bradford, MA) according to the manufacturer’s protocol. Approximately 10 μL of sample was collected from the filter device. The digestion was done as previously described.8 A 5 μL portion of 10 mM DTT (Sigma) was added and the resulting mixture incubated at 45 °C for 30 min. The digestion and reduction reaction was terminated by adding 1 μL of TFA to the digest. The tryptic digestion mixture was completely dried using a SpeedVac concentrator (Labconco Corp., Kansas City, MO) operated at 45 °C. The digest was reconstituted with 20 μL of HPLC-grade water (Fisher Scientific, Hanover Park, IL), and half of the volume of the sample (10 μL) was kept at −20 °C for later analysis by LC/MS/MS. A 50 μL portion of 100 mM ammonium bicarbonate and 1 unit of PNGase F (Sigma) were added to the rest of the digest. The deglycosylation reaction was incubated for 24 h in a water bath set at 37 °C. The reaction was stopped by incubating the digest mixture at 75 °C for 20 min. The sample was dried using a SpeedVac and kept at −20 °C for LC/MS/MS analysis.
LC/MS/MS
A Paradigm MG4 micropump (Michrom Biosciences Inc., Auburn, CA) was used for chromatographic separation of peptide mixtures. A mobile phase system of two solvents was used, where solvents A and B were composed of 0.1% formic acid (Sigma) in HPLC-grade water and ACN (Sigma), respectively. Prior to LC/MS/MS, the dried tryptic digest was reconstituted in HPLC-grade water. The final concentration of tryptic digest mixture of ~0.25–1 ng/μL was injected through a 20 μL fixed sample loop. The peptide mixture was loaded into a desalting column packed with C4 (300 μm i.d. × 50 mm) (Michrom Biosciences Inc.) and washed with 95% solvent A and 5% solvent B with a flow rate of 0.3 μL/min for 5 min. The peptides were eluted onto an analytical C18 column (100 μm i.d. × 150 mm) (Michrom Biosciences Inc.) over 45 min using a gradient elution with a flow rate of 300 nL/min. The 40 min gradient was 5–45% B in 25 min, 45–95% B in 10 min, and hold at 95% B for 5 min. The resolved peptides were analyzed on a linear ion trap mass spectrometer with a nano-LC-ESI source (LTQ, Thermo Finnigan, San Jose, CA). The capillary transfer tube was set at 200 °C, the ESI spray voltage at 2.5 kV, and the capillary voltage at 30 V. The ion activation was achieved by utilizing helium at a normalized collision energy of 35%. The data acquisition and generation of peak list files were automatically done by Xcaliber software. For each cycle of one full mass scan (range of m/z 400–2000), the three most intense ions in the spectrum were selected for tandem MS analysis, unless they appeared in the dynamic or mass exclusion lists. Precursor selection was based upon a normalized threshold of 30 counts/s. Multiple analyses (up to 10 LC/MS/MS runs) were performed for each sample. Dynamic exclusion was employed in the LC/MS/MS analysis to increase the number of identified proteins. In a few cases, precursor ion masses of highly abundant proteins such as albumin or uromodulin were listed for mass exclusion when their mass hindered the detection of relatively lowly abundant proteins.
Database Searching and Manual Validation
All MS/MS spectra were analyzed by TurboSequest of Bioworks software, version 3.1 SR1 (Thermo Finnigan). Peptide fragment lists were generated and submitted to Swiss-Prot database searching. The search parameters were as follows: (1) database species, Homo sapiens; (2) allowing two missed cleavages; (3) possible modifications, oxidation of M; (4) peptide ion mass tolerance 1.50 Da; (5) fragment ion mass tolerance 0.0 Da; (6) peptide charges +1, +2, and +3. The filter function in Bioworks browser was used to set a single threshold to consider fully tryptic peptides assigned with Xcorr9 values as follows: ≥ 1.5 for singly charged ions, ≥ 2.5 for doubly charged ions, and ≥ 3.5 for triply charged ions, while no ions at higher charged states were considered. After that, the search results that passed the first criteria were subjected to manual inspection, in which ΔCn ≥ 0.1 was considered. Data reduction was then manually performed using stringent criteria as follows: (1) preliminary score (Sp) >500, for >6 residual peptides; 200 < Sp < 500, for <6 residual peptides; (2) ranked preliminary score (Rsp) <5; (3) ions >70%; (4) less than two potential modification sites were considered; (5) if the same spectrum matched different proteins, the higher Rsp, the higher ΔCn, and/or ions >70% would be selected. In other words, peptide ion scans must correspond to a single accepted protein. All SEQUEST search parameters and data filtering were the same in all digest fractions, except the modification of asparagine was allowed by +1 Da in the tryptic digest/PNGase F fractions. The +1 Da was allowed owing to hydrolysis of the amide of the asparagine side chain to release the asparagine-linked oligosaccharides from glycopeptides.
Protein IDs were accepted if, and only if, the ID was positively identified in at least two MS/MS analyses. If the protein was identified by a single peptide matching, the spectrum was manually validated. The matched ions covering at least 70% of the peptide sequence must have a high signal-to-noise ratio (S/N). Two high S/N spectra of a single peptide matching protein were accepted for positive identification.
Results
Urinary Protein Content
In the present study, we investigated and compared the glycoprotein profile of urine obtained from patients with cystoscopically confirmed bladder cancer or no evidence of bladder cancer (Table 1). The protein concentration of normal urine is estimated to be less than 100 mg/L.10 Our measurements of urinary protein concentrations from non-tumor-bearing patients were consistent with the reported value (average of 31 mg/L) except for one patient who showed a sign of proteinuria (defined as >150 mg/day of protein) most likely due to a high level of hematuria. The average protein concentration in urine from tumor-bearing patients was 127 mg/L. The greater amount of protein recovered from bladder cancer patients’ urine meant that 7–15 mL of urine was typically sufficient for our analysis; however, a larger volume of asymptomatic patient urine was required, i.e., 15–30 mL. Agarose-bound Con A was chosen to isolate glycoproteins in human urine owing to its broad specificity and preferential binding to oligomannosidic, hybrid, and biantennary N-glycans.11 Con A-capture experiments were performed in duplicate with starting material of lower than 400 μg. The measured protein content of eluted fractions from urine samples indicated that ~4% of the total input protein bound to the lectin column.
Mass Spectrometric Analysis
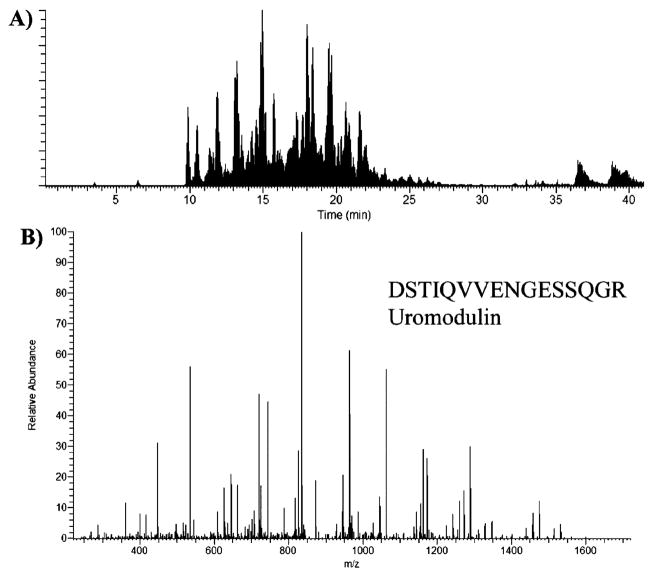
Con A-bound proteins were subjected to mass spectrometric analysis. A semishotgun approach was used where the bound fractions were enzymatically digested and analyzed by nano-LC/MS/MS. For each scan, the three most abundant peptides were sequenced. Figure 1A is a representative nano-LC/MS/MS base peak chromatogram, showing the detection of the more abundant ions across a 40 min gradient separation. The peak capacity of our nano-LC separation was estimated to be ~60–120 on the basis of examining the full width at half-maximum (fwhm). This implies that we can sequence >60 peptides within a single run; however, it is important to note that multiple peptides were detected within a single resolved peak from all experimentally based peak chromatograms, which is commonly observed from LC/MS/MS analysis of highly complex samples.
Figure 1.
(A) A representative nano-LC/MS/MS base peak chromatogram, showing the detection of the peptide ions across the 40 min gradient separation. (B) MS/MS sequencing data of a peptide from uromodulin, identified in the eluted fraction of a bladder-tumor-bearing patient urine sample.
Figure 1B shows a representative MS/MS spectrum of a peptide sequence from uromodulin, one of the most abundant proteins in urine, identified in the eluted fraction of a cancer-bearing patient sample. Eight other ion peaks were also detected and assigned to this protein. Uromodulin is an N-linked glycoprotein as annotated by Swiss-Prot and was detected in 7 of 10 urine samples. The reproducibility of the Con A-capture experiment coupled to nano-LC/MS/MS was evaluated by running samples in triplicate in independently packed Con-A affinity columns. The data showed that the Con A-capture experiments were very reproducible. Approximately 75% of the identified proteins were N-linked glycoproteins, and these were observed in all three parallel, lectin-captured samples (Table 2). To evaluate the reproducibility of nano-LC/MS/MS analysis alone, the retention time of several peptides detected in a normal captured fraction was determined.
Table 2.
Reproducibility Study of Con A Lectin-Captured Urinary Proteinsa
| Con A-captured sample | no. of protein IDs | no. of N-linked glycoproteinsb |
|---|---|---|
| 1 | 35 | 25 |
| 2 | 31 | 25 |
| 3 | 35 | 27 |
| common proteinsc | 25 | 20 |
| found in two samplesd | 6 | 4 |
| found in only one samplee | 14 | 9 |
Proteins from a tumor-bearing patient urine sample were loaded equally onto three replicate lectin affinity columns. The number of proteins identified in each eluted fraction is summarized.
N-linked glycoproteins as annotated in the Swiss-Prot database and as predicted by the software NetNGlyc 1.0 server.
The number of proteins found in all three Con A-captured samples.
The number of proteins found in two samples.
The number of proteins found in only one sample.
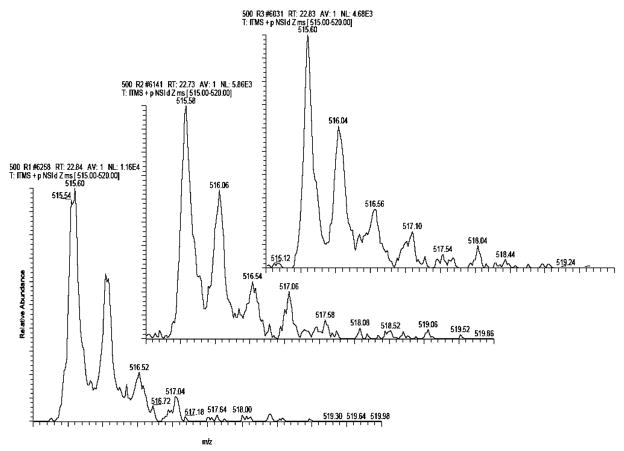
Figure 2 shows representative MS spectra of the +2 precursor mass of YFIDFVAR from the protein kinnogen-1 in three independent runs. The retention times of this peptide in these runs were not significantly different. On the basis of the results, the standard deviation of the elution time of a peptide was estimated to be less than 10 s in our experimental system employing a capillary C18 column.
Figure 2.
Reproducibility of nano-LC/MS/MS of Con A-captured fractions from a human urine sample. The retention times of a +2 charged precursor mass of YFIDFVAR from the protein kinnogen-1 from three independent runs were measured to be 22.84, 22.73, and 22.83 min.
Glycoprotein Identification
Both tryptic digest fractions and tryptic digest/PNGase F fractions were analyzed, and their results were combined to increase the confidence in protein identification. Removing glycans from digested peptides with PNGase F is reported to provide a stronger signal for the peptide ions compared to the glycan remaining intact.12 On the basis of our results, we did indeed find that glycopeptides were poorly identified when only the tryptic digest fractions were analyzed, but after tryptic digest/PNGase F treatment, a number of glycopeptides were positively identified from the same fraction. In our experiments, all SEQUEST search parameters and data filtering were the same, except that 1 Da was allowed for modification of Asn for deglycosylated digests. Table 3 lists the peptides that were not detected in the fraction with the tryptic digestion alone, where “#” denotes the glycosylated site of the peptide. These peptides have consensus sequence NXS/T, where X is not proline, and these sites have been annotated using the Swiss-Prot database. Several potential novel glycopeptides were identified using this approach; however, improved mass accuracy MS/MS instrumentation or other types of experiments are required to confirm these sites. Locating glycosylation sites was not the primary goal of our experiments, but our data support the claims of others that improved signals for glycopeptides are revealed when their glycans have been enzymatically released.12 The analysis of the deglycosylated fraction as well as the tryptic digest fraction facilitated detection of unique peptide sequences and increased the total number of positive protein IDs. Accurate identification of components in a complex biological material, such as urinary proteomes, using the LC/tandem mass spectrometry technique, requires strict attention to data interpretation and handling. Multiple analyses of samples and manual data validation with stringent criteria are particularly important for correct identification. In this study, the Xcorr for each ion charge and ΔCn were set at the accepted values for a high confidence of protein identification.9 In addition, Rsp, Sp, and the percentage ion match were also considered in order to increase the confidence level. Visual assessment of the spectra was conducted if a single peptide resulted in a positive protein ID and was detected in at least two LC/MS/MS runs of the same sample. The protein was considered present in the sample if its corresponding peptides passed all the stringent criteria (described in the Experimental Procedures) and the protein was observed in at least two MS/MS analyses.
Table 3.
A List of Glycopeptides That Were Successfully Identified in Deglycosylated Fractions but Went Undetected in Tryptic Digests
| protein name | potential glycosylated site | peptide mass, MH+ | peptide sequence | glycosylation sitea | no. of samples detected |
|---|---|---|---|---|---|
| uromodulin | K.QDFN#ITDISLLEHR.L | 1701.85 | 319–332 | 322 N-linked (GlcNAc…) | 2 |
| prostaglandin-H2 D-isomerase | R.WFSAGLASN#SSWLR.E | 1582.77 | 45–56 | 51 N-linked (GlcNAc…) | 5 |
| K.SVVAPATDGGLN#LTSTFLR.K | 1920.01 | 67–85 | 78 N-linked (GlcNAc…) | 10 | |
| lysosomal α-glucosidase | K.LEN#LSSSEMGYTATLTR.T | 1873.90 | 138–154 | 140 N-linked (GlcNAc…) | 5 |
| R.GVFITN#ETGQPLIGK.V | 1574.85 | 465–479 | 470 N-linked (GlcNAc…) | 5 | |
| R.N#NTIVNELVR.V | 1172.63 | 882–890 | 882 N-linked (GlcNAc…) | 7 | |
| CD44 antigen | K.AFN#STLPTMAQMEK.A | 1569.74 | 55–68 | 57 N-linked (GlcNAc…) | 8 |
| kininogen-1 | K.YNSQN#QSNNQFVLYR.I | 1875.87 | 44–58 | 48 N-linked (GlcNAc…) | 2 |
| K.LNAENN#ATFYFK.I | 1432.68 | 289–300 | 294 N-linked (GlcNAc…) | 7 | |
| α-1-antitrypsin | K.YLGN#ATAIFFLPDEGK.L | 1756.89 | 268–283 | 271 N-linked (GlcNAc…) | 4 |
| clusterin | R.LAN#LTQGEDQYYLR.V | 1725.94 | 372–385 | 374 N-linked (GlcNAc…) | 4 |
| polymeric immunoglobulin receptor | K.VPGN#VTAVLGETLK.V | 1398.79 | 466–479 | 469 N-linked (GlcNAc…) | 6 |
| galectin-3-binding protein | R.ALGFEN#ATQALGR.A | 1348.69 | 64–76 | 69 N-linked (GlcNAc…) | 3 |
| cathepsin D | K.GSLSYLN#VTR.K | 1110.59 | 257–266 | 263 N-linked (GlcNAc…) | 2 |
| lysosomal acid phosphatase | R.YEQLQN#ETR.Q | 1181.55 | 162–170 | 167 N-linked (GlcNAc…) | 3 |
| Ig α-2 chain C region | K.TPLTAN#ITK.S | 959.55 | 200–208 | 205 N-linked (GlcNAc…) | 3 |
| β-glucuronidase | K.VVAN#GTGTQGQLK.V | 1273.69 | 269–281 | 272 N-linked (GlcNAc…) | 4 |
| haptoglobin | K.VVLHPN#YSQVDIGLIK.L | 1796.01 | 236–251 | 241 N-linked (GlcNAc…) | 4 |
| α-1-antichymotrypsin | K.YTGN#ASALFILPDQDK.M | 1753.88 | 268–283 | 271 N-linked (GlcNAc…) | 2 |
| lysosome-associated membrane glycoprotein 2 | K.IAVQFGPGFSWIAN#FTK.A | 1883.98 | 88–104 | 101 N-linked (GlcNAc…) | 3 |
| fibrillin-1 | K.TAIFAFN#ISHVSNK.V | 1549.81 | 2761–2774 | 2767 N-linked (GlcNAc…) | 2 |
| lysosome-associated membrane glycoprotein 1 | K.N#MTFDLPSDATVVLN#R.S | 1794.87 | 61–76 | 61, 75 N-linked (GlcNAc…) | 3 |
| proactivator polypeptide | R.TN#STFVQALVEHVK.E | 1573.83 | 214–227 | 215 N-linked (GlcNAc…) | 2 |
Glycosylated sites as annotated in Swiss-Prot database
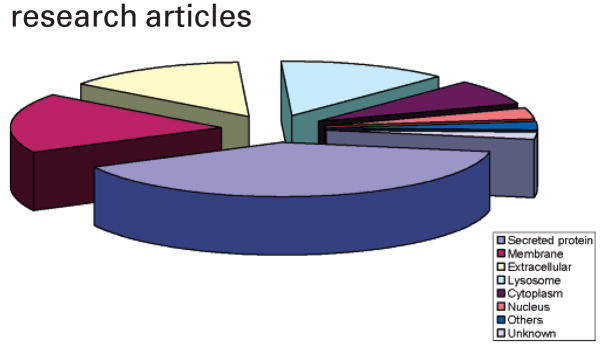
A total of 186 urinary proteins were positively identified in our study, with ~40–65 proteins being detected in each urine specimen. A table (Table S1) summarizing all of the protein IDs obtained from analysis of all of the urine specimens is available in supplemental data (http://genomics.biotech.ufl.edu/people/goodison/table_s1.pdf) and Supporting Information. The majority of the glycoproteins had molecular masses within the range 30000–80000 kDa, but glycoproteins as large as 300 000 kDa were detected. Of the 186 identified proteins, 128 (69%) were glycoproteins as annotated by the Swiss-Prot database. Other proteins could be accounted for through nonspecific lectin column binding, or unconventional glycan sites. Not surprisingly, Figure 3 shows that the majority of identified proteins were secreted (40%), membrane (18%), and extracellular (14%) proteins. Zinc α-2-glycoprotein and α-1-microglobulin (AMBP protein) were the most frequent positive protein IDs and were identified in all urine samples (Table S1 in Supporting Information). These two proteins are expected to be abundant in urine; however, we observed that their sequenced peptides were not always in the top five protein identifications per sample. Table 4 ranks the identified proteins that were most discriminatory between samples from tumor-and non-tumor-bearing patients by comparing the occurrence of a specific protein in urine samples obtained from tumor-bearing and non-tumor-bearing individuals. Urinary serotransferrin and haptoglobin were associated with the presence of bladder cancer, but most discriminatory was α-1B-glycoprotein (A1BG-human). This protein was detected in all Con A-captured fractions of bladder cancer patients’ samples but, more importantly, was never found to occur in non-tumor-bearing patients’ urine.
Figure 3.
Subcellular location of proteins identified in naturally micturated urine samples using a Con A lectin column and MS/MS analysis. A total of 186 urinary proteins were identified.
Table 4.
Urinary Proteins Detected in >4 out of 5 Samples Tested in Either a Bladder-Tumor-Bearing Donor Group (n = 5) or a Non-Tumor-Bearing Donor Group (n = 5)a
| Swiss-Prot ID | protein name | subcellular location | glycoprotein | protein class | bladder cancer (n = 5) | normal (n = 5) |
|---|---|---|---|---|---|---|
| KLK1_HUMAN | kallikrein-1 | secreted protein | O,N-linked | protease | 1 | 4 |
| ATRN_HUMAN | attractin | secreted protein | N-linked | immune system | 1 | 4 |
| ARSA_HUMAN | arylsulfatase A | lysosome | N-linked | enzyme | 3 | 4 |
| LAMP2_HUMAN | LAMP-2 | membrane | O,N-linked | adhesion | 3 | 4 |
| LYAG_HUMAN | lysosomal α-glucosidase | lysosome | N-linked | glycosidase | 3 | 4 |
| UROM_HUMAN | uromodulin | secreted protein | N-linked | unknown | 3 | 4 |
| ITIH4_HUMAN | inter-α-inhibitor heavy chain 4 | extracellular | O,N-linked | protease inhibitor | 3 | 4 |
| CD44_HUMAN | CD44 antigen | membrane | N-linked | adhesion | 4 | 4 |
| KNG1_HUMAN | kininogen-1 | secreted protein | O,N-linked | protease inhibitor | 4 | 4 |
| KV2A_HUMAN | Ig κ chain V-II region Cum | extracellular | no sites | immune system | 4 | 4 |
| PIGR_HUMAN | polymeric immunoglobulin receptor | secreted protein | N-linked | carrier/transport protein | 4 | 4 |
| CD59_HUMAN | CD59 glycoprotein | membrane | O,N-linked | inhibitor | 4 | 5 |
| KAC_HUMAN | Ig κ chain C region | extracellular | no sites | immune system | 4 | 5 |
| LAC_HUMAN | Ig λ chain C regions | extracellular | no sites | immune system | 4 | 5 |
| PTGDS_HUMAN | prostaglandin-H2 D-isomerase | secreted protein | N-linked | enzyme | 4 | 5 |
| ZA2G_HUMAN | zinc α-2-glycoprotein | secreted protein | N-linked | unknown | 4 | 5 |
| ALBU_HUMAN | serum albumin | secreted protein | N-linked | carrier/transport protein | 4 | 5 |
| IGHA1_HUMAN | Ig α-1 chain C region | secreted protein | O,N-linked | immune system | 5 | 4 |
| AMBP_HUMAN | α-1-microglobulin | secreted protein | O,N-linked | protease inhibitor | 5 | 5 |
| KV3A_HUMAN | Ig κ chain V-III region B6 | extracellular | no sites | immune system | 4 | 3 |
| KV3B_HUMAN | Ig κ chain V-III region SIE | extracellular | no sites | immune system | 4 | 3 |
| CADH1_HUMAN | epithelial cadherin | membrane | N-linked | adhesion | 4 | 3 |
| APOA_HUMAN | apolipoprotein(a) | extracellular | O,N-linked | protease | 4 | 3 |
| A1AT_HUMAN | α-1-antitrypsin | secreted protein | N-linked | protease inhibitor | 4 | 2 |
| TRFE_HUMAN | serotransferrin | secreted protein | O,N-linked | carrier/transport protein | 4 | 2 |
| HPT_HUMAN | haptoglobin | secreted protein | N-linked | carrier/transport protein | 5 | 1 |
| A1BG_HUMAN | α-1B-glycoprotein | secreted protein | N-linked | unknown | 5 | not detected |
The proteins were ranked on the basis of their relative abundances in each group.
Discussion
Recent improvements in technologies to detect, identify, and characterize proteins, particularly two-dimensional electrophoresis and mass spectrometry, enable the detailed and systematic isolation, identification, and characterization of proteins in a given sample. Proteomics is regarded as a sister technology to genomics; however, although the pattern of gene activity may be abnormal in a tissue with a pathological lesion, there can be a poor correlation between the level of the transcription of different genes and the relative abundance within the tissue of the corresponding proteins. Consequently, the information about a pathological process that can be derived at the level of gene transcription is incomplete. It is the high-throughput approach that defines and characterizes modern proteomics.
Despite the high complexity of components in urine, the urinary proteome is highly amenable to clinical research due to the wide availability of the samples, the noninvasive nature of collection, and the possibility of repeat sampling. The urinary protein profile may reflect not only renal disease,13 but also other diseases of the urinary tract, including bladder cancer. Thus, analysis of proteins in patient urine with a defined disease may provide detailed knowledge of the pathological process associated with the disease. Furthermore, the characterization of differences between asymptomatic and disease-associated urinary proteomes should provide markers for diagnosis and prognosis and as potential targets for drug development.
Two-dimensional electrophoresis (2-DE) of proteins has been the conventional method for biomarker assessment in urological proteomics,14,15 and investigators have performed a systematic evaluation of sample preparation methods for gel-based human urinary proteomes.16 A near-standard 2-D map17 and high-resolution 2-D gels18 of urological tissues have contributed to the construction of a 2-D database.11,19 The database also contains a listing of 339 proteins detectable in urine, 124 of which have been identified. Using the urological 2-DE databases, pathologic subtypes of solid bladder tumor tissue specimens could be distinguished on the basis of protein fingerprints.12,20,21 Although a number of dysregulated proteins have been identified in a variety of tissue-based studies, it is disappointing that no reliable markers have been identified for transitional cell carcinoma, the most common type of bladder cancer. Through the proteomic study of a set of only six urine samples, Kageyama et al.22 were able to identify a potential tumor marker, calreticulin, which is found in the urine of patients with bladder carcinoma. However, as is common in the search for cancer biomarkers, the authors used a differential display method and mass spectrometry to identify proteins that are increased in solid tumor tissues first and then subsequently monitored the presence of a reduced set of candidate biomarkers in urine samples. Unfortunately, in a larger cohort of patients, the diagnostic accuracy of calreticulin in urine was vulnerable to urinary tract infections.22 Because none of the markers described to date have sufficient sensitivity and specificity for diagnostic use, the quest for identifying additional bladder cancer biomarkers continues. Investigators have also applied gel-free methodologies to urine analysis. Pang et al. employed three different strategies to identify biomarkers related to acute inflammation,23 and Cutillas et al. used a 2-D LC/MS/MS approach to generate a peptide profile of urine from patients with Dent’s disease.24 Most recently, Ru et al. used multidimensional protein identification technology (MudPIT) to identify 87 proteins in human urine.25 It is interesting to note that no more than 30 mL of urine was needed in our experiments while hundreds of milliliters of urine is typically used for gel-based analysis.16–19,23 The pooling of samples from different individuals is typically observed in the published gel analysis of urines to increase the amount of proteins.17,19 By pooling samples, one is likely to lose the individual information of the intrinsic components present in urine from a single patient at a given time. Thus, a reliable and accurate profiling technique employing a small amount of urine is essential for detecting urinary proteomes and for marker identification. Low sample consumption also minimizes sample handling steps, which reduces potential sample losses as well as analysis time.
In this study, we focused on the analysis of one specific fraction of the urinary proteome, that of glycosylated proteins. Selection and concentration of one component of the sample proteome reduced the complexity of the sample, which in turn enabled the rapid and accurate identification of multiple unique proteins in sample volumes that are routinely, and noninvasively, obtained in the clinic. Protein glycosylation has long been recognized as one of the most prevalent post-translational modifications,26 playing a fundamental role in many biological processes such as immune response and cellular regulation.27 Accordingly, the alteration in protein glycosylation which occurs through varying the heterogeneity of glycosylation sites or changing the glycan structure of proteins on the cell surface and in body fluids has been shown to correlate with the development of numerous disease states, including cancer.28 Indeed, many clinical biomarkers and therapeutic targets in cancer are glycoproteins29 such as CA125 in ovarian cancer, Her2/neu in breast cancer, and prostate-specific antigen (PSA) in prostate cancer. PSA is one of the best characterized examples of a secreted glycoprotein used in cancer diagnostics, and its glycoforms have been described.30 Such carbohydrate differences allow a distinction to be made between proteins from normal and tumor origins and suggest a valuable biochemical tool for diagnosis.31
The use of Con A affinity chromatography combined with a powerful nano-LC/MS/MS platform provided an efficient capture of N-glycoproteins from a relatively complex sample and enabled the identification of 186 unique proteins. This combinatorial approach provides high sensitivity and with relatively moderate labor demands should facilitate the identification of potential biomarkers of disease from body fluids. Our pilot comparative study of a panel of clinical samples revealed that differential urinary glycoprotein profiles exist that can distinguish bladder-cancer-bearing patients from individuals with noncancer conditions. It is important to note that only relatively high abundance peptides/proteins will be positively identified by LC/MS/MS analysis. In particular, ion suppression can occur when many peptides elute from the column simultaneously. Increasing the number of sequenced peptides per scan or using mass exclusion may increase the number of identified proteins, but these improvements are still restricted by the MS performance. Obtaining a large sequence coverage as well as sequencing large peptides is also difficult with the shotgun approach. In this study, repeated analysis of the sample enabled us to detect low-level peptides/proteins and obtain confident IDs of N-linked glycoproteins in a small amount of patient urine. In addition to the use of high Xcorr criteria for peptide/protein assignment, restricted SEQUEST scores were determined and manual analysis was performed to ensure the accuracy of our assignments. Using this concerted approach, we were able to distinguish the marked differences between a group of healthy individuals and cancer-bearing patients and propose potential diagnostic biomarkers.
Of the 186 proteins identified in this study, 146 have been detected in urine using other techniques;10,19 thus, we have added a significant list to the urinary proteome database. We did identify several potential novel glycosylated peptides in this study, but these would require confirmational studies focused upon the detailed analysis of specific proteins. Even though all patients visiting the clinic presented with hematuria, the presence of several glycoproteins was associated with samples obtained from patients with transitional cell carcinoma of the bladder. The most discriminatory protein associated with bladder cancer in this pilot study was A1BG. Although the human A1BG protein was purified and sequenced in 1986, its physiological role is unknown. Ishioka et al. determined the complete amino acid sequence of A1BG and showed it to consist of a single polypeptide chain N-linked to four glucosamine oligosaccharides. Analysis of the amino acid sequence revealed significant homology to variable regions of certain immunoglobulin light and heavy chains and to other members of the immunoglobulin supergene family.32 No specific function has been ascribed to A1BG to date, but an indication of a function comes from the characterization of the opossum A1BG homologue, oprin.33 Oprin is a metalloproteinase inhibitor, which in some properties, but not in sequence, resembles the tissue inhibitor of metalloproteinases (TIMPs), a family of proteins with complex roles in tumor progression and angiogenesis.34 An association of A1BG expression with cancer has been described in one previous study. In a study that utilized expressed sequence tag (EST) profiling of cDNA libraries, Yoon et al. identified A1BG as 1 of 14 genes confirmed to be highly expressed in human hepatocellular carcinoma (HCC) cell lines and liver tumor tissue specimens.35 While many more cases are required to be screened, the reported correlation of A1BG expression with liver cancer supports the notion that this protein is worthy of further study in the context of a potential biomarker for bladder cancer. Haptoglobin was also associated with bladder cancer in this study, and this glycoprotein has been reported to be diagnostically useful in both ovarian and liver cancer.34,35 Further studies are needed in large cohorts of bladder cancer patients with a broad range of disease stage and grade and with noncancer conditions to evaluate the true potential of the glycoproteins implicated in this study in bladder disease management. Beyond the investigation of individual proteins identified here, the described technical approach will facilitate a more focused analysis of clinically relevant samples, such as urine and other bodily fluids, which in turn will lead to the identification of reliable biomarkers for improved detection, surveillance, and screening regimens for a range of diseases.
Supplementary Material
Acknowledgments
This study was supported in part by a John and Esther King Young Investigator Award from the Florida State Department of Health (C.R., S.G.) and NIH Grant RO1 CA108597 (S.G.).
Footnotes
Supporting Information Available: Table summarizing all of the protein IDs obtained from analysis of all of the urine specimens. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pisani P, Parkin DM, Bray F, Ferlay J. Estimates of the worldwide mortality from 25 cancers in 1990. Int J Cancer. 1999;83(1):18–29. doi: 10.1002/(sici)1097-0215(19990924)83:1<18::aid-ijc5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 2.Aben KK, Kiemeney LA. Epidemiology of bladder cancer. Eur Urol. 1999;36(6):660–72. doi: 10.1159/000020069. [DOI] [PubMed] [Google Scholar]
- 3.Millan-Rodriguez F, Chechile-Toniolo G, Salvador-Bayarri J, Palou J, Algaba F, Vicente-Rodriguez J. Primary superficial bladder cancer risk groups according to progression, mortality and recurrence. J Urol. 2000;164(3 Part 1):680–4. doi: 10.1016/s0022-5347(05)67280-1. [DOI] [PubMed] [Google Scholar]
- 4.Cajulis RS, Haines GK, 3rd, Frias-Hidvegi D, McVary K, Bacus JW. Cytology, flow cytometry, image analysis, and interphase cytogenetics by fluorescence in situ hybridization in the diagnosis of transitional cell carcinoma in bladder washes: a comparative study. Diagn Cytopathol. 1995;13(3):214–23. doi: 10.1002/dc.2840130307. discussion 224. [DOI] [PubMed] [Google Scholar]
- 5.Hautmann S, Toma M, Lorenzo Gomez MF, Friedrich MG, Jaekel T, Michl U, Schroeder GL, Huland H, Juenemann KP, Lokeshwar VB. Immunocyt and the HA-HAase urine tests for the detection of bladder cancer: a side-by-side comparison. Eur Urol. 2004;46(4):466–71. doi: 10.1016/j.eururo.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Kreunin P, Urquidi V, Lubman DM, Goodison S. Identification of metastasis-associated proteins in a human tumor metastasis model using the mass-mapping technique. Proteomics. 2004;4(9):2754–65. doi: 10.1002/pmic.200300767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, Simeone DM, Heidt D, Anderson MA, Lubman DM. Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: application to pancreatic cancer serum. J Proteome Res. 2006;5(7):1792–802. doi: 10.1021/pr060034r. [DOI] [PubMed] [Google Scholar]
- 8.Kreunin P, Yoo C, Urquidi V, Lubman DM, Goodison S. Proteomic profiling identifies breast tumor metastasis-associated factors in an isogenic model. Proteomics. 2007;7(2):299–312. doi: 10.1002/pmic.200600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen EI, Hewel J, Felding-Habermann B, Yates JR., 3rd Large scale protein profiling by combination of protein fractionation and multidimensional protein identification technology (MudPIT) Mol Cell Proteomics. 2006;5(1):53–6. doi: 10.1074/mcp.T500013-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7(9):R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wuhrer M, Catalina MI, Deelder AM, Hokke CH. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J Chromatogr, B: Anal Technol Biomed Life Sci. doi: 10.1016/j.jchromb.2006.09.041. in press. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Wu SL, Hancock WS. Approaches to the study of N-linked glycoproteins in human plasma using lectin affinity chromatography and nano-HPLC coupled to electrospray linear ion trap-Fourier transform mass spectrometry. Glycobiology. 2006;16(6):514–23. doi: 10.1093/glycob/cwj091. [DOI] [PubMed] [Google Scholar]
- 13.O’Riordan E, Goligorsky MS. Emerging studies of the urinary proteome: the end of the beginning? Curr Opin Nephrol Hypertens. 2005;14(6):579–85. doi: 10.1097/01.mnh.0000168425.60729.36. [DOI] [PubMed] [Google Scholar]
- 14.Tantipaiboonwong P, Sinchaikul S, Sriyam S, Phutrakul S, Chen ST. Different techniques for urinary protein analysis of normal and lung cancer patients. Proteomics. 2005;5(4):1140–9. doi: 10.1002/pmic.200401143. [DOI] [PubMed] [Google Scholar]
- 15.Nabi G, N’Dow J, Hasan TS, Booth IR, Cash P. Proteomic analysis of urine in patients with intestinal segments transposed into the urinary tract. Proteomics. 2005;5(6):1729–33. doi: 10.1002/pmic.200401125. [DOI] [PubMed] [Google Scholar]
- 16.Thongboonkerd V, Chutipongtanate S, Kanlaya R. Systematic evaluation of sample preparation methods for gel-based human urinary proteomics: quantity, quality, and variability. J Proteome Res. 2006;5(1):183–91. doi: 10.1021/pr0502525. [DOI] [PubMed] [Google Scholar]
- 17.Oh J, Pyo JH, Jo EH, Hwang SI, Kang SC, Jung JH, Park EK, Kim SY, Choi JY, Lim J. Establishment of a near-standard two-dimensional human urine proteomic map. Proteomics. 2004;4(11):3485–97. doi: 10.1002/pmic.200401018. [DOI] [PubMed] [Google Scholar]
- 18.Pieper R, Gatlin CL, McGrath AM, Makusky AJ, Mondal M, Seonarain M, Field E, Schatz CR, Estock MA, Ahmed N, Anderson NG, Steiner S. Characterization of the human urinary proteome: a method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics. 2004;4(4):1159–74. doi: 10.1002/pmic.200300661. [DOI] [PubMed] [Google Scholar]
- 19.Zerefos PG, Vougas K, Dimitraki P, Kossida S, Petrolekas A, Stravodimos K, Giannopoulos A, Fountoulakis M, Vlahou A. Characterization of the human urine proteome by preparative electrophoresis in combination with 2-DE. Proteomics. 2006;6(15):4346–55. doi: 10.1002/pmic.200500671. [DOI] [PubMed] [Google Scholar]
- 20.Liu T, Qian WJ, Gritsenko MA, Camp DG, 2nd, Monroe ME, Moore RJ, Smith RD. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4(6):2070–80. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bunkenborg J, Pilch BJ, Podtelejnikov AV, Wisniewski JR. Screening for N-glycosylated proteins by liquid chromatography mass spectrometry. Proteomics. 2004;4(2):454–65. doi: 10.1002/pmic.200300556. [DOI] [PubMed] [Google Scholar]
- 22.Kageyama S, Isono T, Iwaki H, Wakabayashi Y, Okada Y, Kontani K, Yoshimura K, Terai A, Arai Y, Yoshiki T. Identification by proteomic analysis of calreticulin as a marker for bladder cancer and evaluation of the diagnostic accuracy of its detection in urine. Clin Chem. 2004;50(5):857–66. doi: 10.1373/clinchem.2003.027425. [DOI] [PubMed] [Google Scholar]
- 23.Pang JX, Ginanni N, Dongre AR, Hefta SA, Opitek GJ. Biomarker discovery in urine by proteomics. J Proteome Res. 2002;1(2):161–9. doi: 10.1021/pr015518w. [DOI] [PubMed] [Google Scholar]
- 24.Cutillas PR, Norden AG, Cramer R, Burlingame AL, Unwin RJ. Detection and analysis of urinary peptides by on-line liquid chromatography and mass spectrometry: application to patients with renal Fanconi syndrome. Clin Sci (London) 2003;104(5):483–90. doi: 10.1042/CS20020342. [DOI] [PubMed] [Google Scholar]
- 25.Ru QC, Katenhusen RA, Zhu LA, Silberman J, Yang S, Orchard TJ, Brzeski H, Liebman M, Ellsworth DL. Proteomic profiling of human urine using multi-dimensional protein identification technology. J Chromatogr, A. 2006;1111(2):166–74. doi: 10.1016/j.chroma.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 26.Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- 27.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291(5512):2370–6. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 28.Block TM, Comunale MA, Lowman M, Steel LF, Romano PR, Fimmel C, Tennant BC, London WT, Evans AA, Blumberg BS, Dwek RA, Mattu TS, Mehta AS. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proc Natl Acad Sci USA. 2005;102(3):779–84. doi: 10.1073/pnas.0408928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dube DH, Bertozzi CR. Glycans in cancer and inflammation-potential for therapeutics and diagnostics. Nat Rev Drug Discovery. 2005;4(6):477–88. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 30.Prakash S, Robbins PW. Glycotyping of prostate specific antigen. Glycobiology. 2000;10(2):173–6. doi: 10.1093/glycob/10.2.173. [DOI] [PubMed] [Google Scholar]
- 31.Peracaula R, Tabares G, Royle L, Harvey DJ, Dwek RA, Rudd PM, de Llorens R. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology. 2003;13(6):457–70. doi: 10.1093/glycob/cwg041. [DOI] [PubMed] [Google Scholar]
- 32.Ishioka N, Takahashi N, Putnam FW. Amino acid sequence of human plasma alpha 1B-glycoprotein: homology to the immunoglobulin supergene family. Proc Natl Acad Sci USA. 1986;83(8):2363–7. doi: 10.1073/pnas.83.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Catanese JJ, Kress LF. Isolation from opossum serum of a metalloproteinase inhibitor homologous to human alpha 1B-glycoprotein. Biochemistry. 1992;31(2):410–8. doi: 10.1021/bi00117a015. [DOI] [PubMed] [Google Scholar]
- 34.Chirco R, Liu XW, Jung KK, Kim HR. Novel functions of TIMPs in cell signaling. Cancer Metastasis Rev. 2006;25(1):99–113. doi: 10.1007/s10555-006-7893-x. [DOI] [PubMed] [Google Scholar]
- 35.Yoon SY, Kim JM, Oh JH, Jeon YJ, Lee DS, Kim JH, Choi JY, Ahn BM, Kim S, Yoo HS, Kim YS, Kim NS. Gene expression profiling of human HBV- and/or HCV-associated hepatocellular carcinoma cells using expressed sequence tags. Int J Oncol. 2006;29(2):315–27. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.