Abstract
Mitotic spindle orientation in polarized cells determines whether they divide symmetrically or asymmetrically. Moreover, regulated spindle orientation may be important for embryonic development, stem cell biology, and tumor growth. Drosophila neuroblasts align their spindle along an apical/basal cortical polarity axis to self-renew an apical neuroblast and generate a basal differentiating cell. It is unknown whether the spindle alignment requires both apical and basal cues, nor have molecular motors been identified that regulate spindle movement. Using live imaging of neuroblasts within intact larval brains, we detect independent movement of both apical and basal spindle poles, suggesting that forces act on both poles. We show that reducing astral microtubules decreases the frequency of spindle movement, but not its maximum velocity, suggesting that one or few microtubules can move the spindle. Mutants in the Lis1/dynactin complex strongly decrease maximum and average spindle velocity, consistent with this motor complex mediating spindle/cortex forces. Loss of either astral microtubules or Lis1/dynactin leads to spindle/cortical polarity alignment defects at metaphase, but these are rescued by telophase. We propose that an early Lis1/dynactin-dependent pathway and a late Lis1/dynactin-independent pathway regulate neuroblast spindle orientation.
Keywords: neuroblast, Lis1, asymmetric cell division, cnn, dynein, telophase rescue, astral microtubules
Introduction
Asymmetric cell division involving unequal partitioning of cell fate determinants is a widely used mechanism for generation of cellular diversity. Successful asymmetric cell division in animal cells requires the polarized localization of cell fate determinants and alignment of the mitotic spindle along this polarity axis; spindle orientation defects during asymmetric cell division have been implicated in tissue overgrowth and tumor formation (Caussinus and Hirth, 2007; Gonzalez, 2007; Knoblich, 2008; Morrison and Kimble, 2006). A central question is how cell polarity and spindle positioning are coordinated.
We are using Drosophila larval neuroblasts as a model system to investigate spindle-to-cortex interactions during asymmetric cell division. Drosophila neuroblasts repeatedly divide in a stem cell-like fashion, each division regenerating a larger apical neuroblast and producing a smaller basal ganglion mother cell (GMC) that typically forms two neurons. Mitotic neuroblasts are highly polarized, with distinct protein complexes localized to the apical and basal cell cortex. The apical cortical domain is formed around an evolutionary conserved protein complex containing Bazooka (Baz, the Drosophila Par-3 homolog), Cdc42, Par-6, and atypical Protein Kinase C (aPKC), and the associated Inscuteable, Partner-of Inscuteable (Pins), Gαi, and Mushroom body defect (Mud) and Locomotion defects proteins. Basal proteins include the GMC cell fate determinants Miranda (Mira), Brain tumor (Brat), Prospero, and Numb (reviewed in Knoblich, 2008). The precise alignment of the mitotic spindle along the apical/basal polarity axis ensures segregation of these cell fate determinants into distinct daughter cells, which is correlated with the proper establishment of neuroblast/GMC cell fates (Caussinus and Hirth, 2007; Gonzalez, 2007). Despite the potential importance of spindle orientation in neuroblast/GMC cell fate specification, surprisingly little is known about the mechanism of spindle positioning in neuroblasts.
Studies in yeast and Caenorhabditis elegans (C. elegans) have provided strong evidence that cortex-spindle forces are generated at the astral microtubule-cortex interface by the action of cortically attached microtubule-based motors, proteins affecting microtubule length, and/or by proteins controlling the dynamics of astral microtubule-cortex interactions. Thus, spindle positioning requires dynamic attachment of astral microtubules to polarized cortical domains, and generation of forces exerted on these cortically-attached astral microtubules aligns the mitotic spindle (Cowan and Hyman, 2004; Pearson and Bloom, 2004). Consistent with this general model, reduction of either astral microtubule number or polarized cortical proteins can disrupt spindle positioning in Drosophila larval neuroblasts (Basto et al., 2006; Bowman et al., 2006; Giansanti et al., 2001; Izumi et al., 2006; Megraw et al., 2001; Nipper et al., 2007; Rebollo et al., 2007; Rusan and Peifer, 2007; Siegrist and Doe, 2005; Siller et al., 2006). Nevertheless, several important questions remain to be answered for Drosophila neuroblasts: Do both apical and basal spindle poles interact with the cortex during spindle orientation? What microtubule-binding proteins are required for productive spindle/cortex interactions? Here we address these questions using live imaging of larval Drosophila neuroblasts -- including wild type, mutants that reduce astral microtubule number, and Lis1/dynactin mutants. We find that the apical and basal spindle poles show independent dynamic movement, suggesting that forces act on both spindle poles; that virtually eliminating all astral microtubules decreases the frequency of spindle movement, without altering the maximum velocity of spindle movement, suggesting that individual microtubules are sufficient to alter spindle orientation; and that the Lis1/dynactin complex is required for spindle movement and metaphase spindle orientation, indicating that this evolutionarily-conserved protein complex is responsible for spindle/cortex forces.
Results
Neuroblast spindle alignment occurs in prophase
We first determined when spindle orientation was established during the cell cycle, so we could focus our analysis of spindle/cortex interactions at the functionally relevant stage. Previous studies showed that larval neuroblasts establish spindle orientation at prophase relative to extrinsic cellular landmarks – i.e. GMCs in contact with the neuroblast basal cortex (Nipper et al., 2007; Rebollo et al., 2007; Rusan and Peifer, 2007; Siller et al., 2006; Siller et al., 2005), and relative to the intrinsic polarity marker Bazooka-YFP (Baz-YFP) at the neuroblast apical cortex (Siller et al., 2006). We used live imaging and fixed preparations to confirm that one centrosome is fixed at the apical cortex opposite the GMC contact site and adjacent to the apical Baz-YFP cortical crescent by prophase (Figure 1; Movies 1, 2; Table 1). In rare cases, centrosomes rotated during prophase, but their final position was established before the end of prophase (mean deviation from the final division axis 9.4°±6.8°; n=14; Table 1), and spindles stayed aligned along this axis throughout mitosis. Together these data confirm that neuroblast spindle positioning relative to extrinsic and intrinsic cues is established during prophase.
Figure 1. Spindle position is determined during prophase in larval neuroblasts.
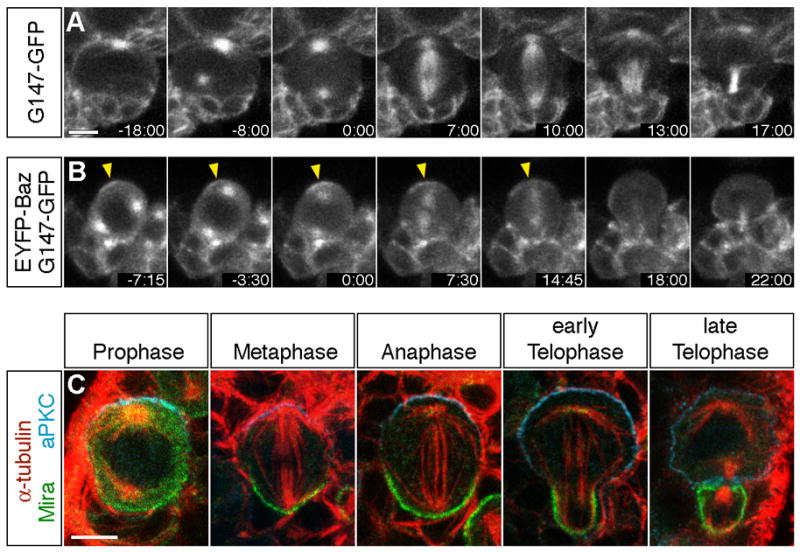
(A–B) Temporal sequence of spindle alignment in wild type larval neuroblasts visualized in neuroblasts expressing the G147-GFP microtubule-associated protein (A) or co-expressing G147-GFP and EYFP-Baz (B). Time is given in min:sec relative to onset of prometaphase (0:00), which was evident by extension of microtubules into the cell center (A+B) and detection of EYFP-Baz in the cell center (B) after nuclear envelope breakdown (NEB).
(A) Note that both spindle poles were positioned along the eventual division axis by the onset of prometaphase (0:00) and that spindle orientation was stabilized along this axis from this timepoint onward (Movie 1).
(B) EYFP-Baz was concentrated in an apical cortex crescent (arrowheads) before centrosomes complete alignment along the apical-basal axis (−7:15). Separated centrosomes rotate during prophase to align along the apical-basal cell polarity axis (−3:30 – 0:00) (Movie 2).
(C) In wild type larval neuroblasts, aPKC and Mira are localized to opposite cortical domains in late prophase, metaphase, anaphase, and telophase larval neuroblasts. Apical is up and basal is down. From late prophase until late telophase/cytokinesis the centrosome pair/mitotic spindle is tightly aligned along the cortical apical-basal polarity axis. Scale bars: 5 μm.
Table 1.
Time-lapse analysis of spindle orientation in larval neuroblasts
| wild type a (n=14) | cnnhk21 (n=10) | Lis1b centr. sep. >160° (n=4) | Lis1b centr. sep. ≤160° (n=6) | |
|---|---|---|---|---|
| Angle between centrosomes at end of prophase and division axis c,d | ||||
| mean±SD | 9.4°±6.8° | 24.7°±17.8° | 41.7°±30.7° | 53.5°±38.5° |
| max. | 28° | 56° | 84° | 88° |
| % of NBs with angle >15° | 7.1% | 60.0% | 75.0% | 66.7% |
| Angle between spindle axis at anaphase onset and division axis d | ||||
| mean±SD | 7.2°±6.5° | 20.1°±16.4° | 33.1°±24.8° | 12.7°±9.5° |
| max. | 25° | 47° | 64° | 26° |
| % of NBs with angle >15° | 7.1% | 60.0% | 50.0% | 33.3% |
Genotypes: wild type (G147-GFP); cnnhk21 (cnnhk21/cnnhk21 G147-GFP); Lis1 (Lis1k13209/Lis1G10.14 G147-GFP)
Lis1 mutant neuroblasts were divided into two classes: (1) neuroblasts with complete centrosome separation (centrosome separation >160°); and (2) neuroblasts with centrosome separation defects (centrosome separation ≤160°).
in cnnhk21 mutants the angle between spindle axis at the time of first successful bipolar focused spindle formation (during prometaphase/metaphase) and the eventual division axis
division axis was defined as the axis through the center of the neuroblast and its GMC daughter at the time when central spindle microtubules had converged into a compact midbody (~5 min after anaphase onset in wild type and cnnhk21 mutant neuroblasts; ~10–15 min after anaphase onset in Lis1 mutant neuroblasts due to slower completion of cytokinesis)
NB: neuroblasts
Apical and basal spindle poles move independently
A full understanding of the mechanism of spindle orientation requires knowing which spindle pole interacts with the cortex – apical, basal, or both – and when these interactions occur. To address this question, we traced movements of apical and basal spindle poles throughout mitosis to determine their velocity, and most importantly whether they moved independently from each other. We find that during prometaphase and metaphase both spindle poles showed periods of vigorous movement, causing rocking of the spindle perpendicular to the spindle axis (Figure 2A −7:07 to −1:27; Movies 3+4). These spindle oscillations did not affect the overall spindle orientation, which was constant from the end of prophase onwards (see above). We quantified peak velocity and frequency of high velocity movements (percent of time with a velocity ≥133 nm/s, see Materials and Methods). The peak velocity reached 320 nm/s, with apical and basal metaphase spindle poles spending an average of 24%−30% moving at a velocity ≥133 nm/s (Figure 2D+E). While rapid, this is likely an underestimate of spindle pole velocities, as we could not measure movement in the z-axis of the image stack. To determine if each spindle pole could move independently, we categorized spindle pole movement into the following categories: apical pole only, basal pole only, bipolar lateral, and bipolar rotational. We observed many examples of where only one spindle pole (apical or basal) showed movement (Figure 2A). Interestingly, the basal spindle pole showed more vigorous movement than the apical spindle pole in third instar larval neuroblasts (Figure 2A,D). This is unexpected because Gα promotes spindle rocking in C. elegans (Cowan and Hyman, 2004) but is localized over the less active apical spindle pole in neuroblasts (see Discussion). We conclude that the apical and basal spindle poles can move independently; this suggests that there are force-generating proteins associated with both spindle poles.
Figure 2. Lis1 and Cnn regulate apical and basal spindle pole movements.
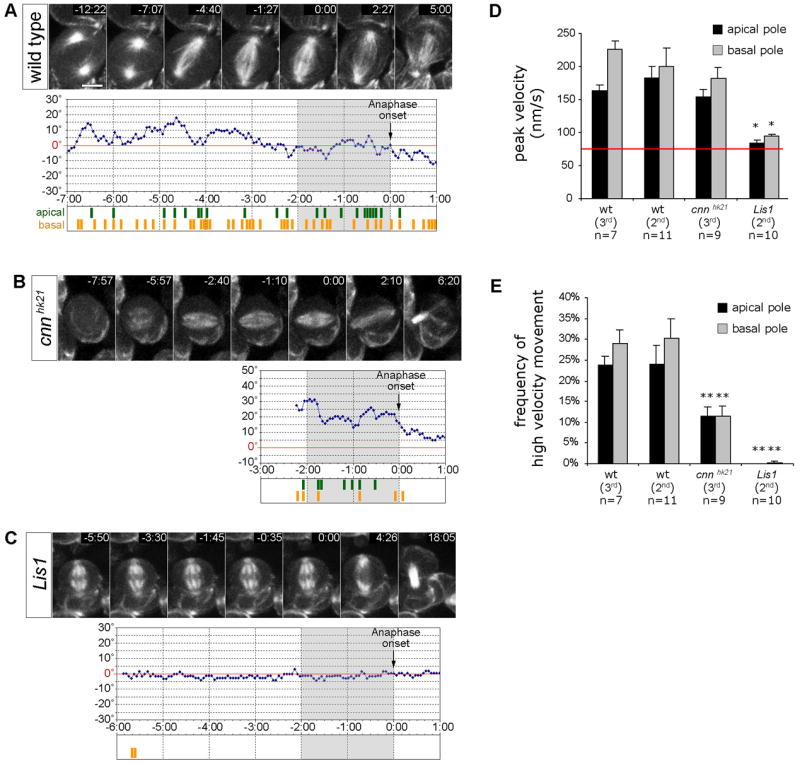
(A–C) Spindle pole movement in larval metaphase neuroblasts expressing the G147-GFP microtubule-associated protein. Time is given in min:sec relatively to the onset of anaphase (0:00). Diagrams depict the angular deviation of the spindle axis at a given time point (blue line) from the final division axis (red line = 0°, determined 5:00 min after anaphase onset). Grey shaded regions indicate the 2 min period of late prometaphase/metaphase prior to anaphase onset (0:00) used for the analysis of spindle pole movements quantified in panels D and E. High velocity movement (≥133 nm/s or ≥2 pixels/frame) of the apical spindle pole (green) and basal spindle pole (yellow) are depicted below each diagram.
(A) In 2nd instar wild type neuroblasts (48h ALH), both apical and basal spindle poles move extensively, causing oscillatory movements of the mitotic spindle around the final division axis. (Movie 3+4). No significant difference was observed between 2nd instar (48h ALH) and 3rd instar (96h ALH) larval neuroblasts (see panels D and E).
(B) In 3rd instar cnnhk21 mutant neuroblasts (96h ALH), apical and basal spindle pole movements with velocities ≥133 nm/s were less frequent than in wild type counterparts. Measurements were started at the time a bipolar spindle with focused spindle poles had formed (Movie 5, see panels D and E).
(C) In 2nd instar Lis1 mutant neuroblasts (48h ALH), spindle pole movements were undetectable or significantly reduced (Movie 6; see panels D and E).
(D+E) Quantification of ‘peak velocity’, and ‘frequency of high velocity movements’ (≥133 nm/s) of apical and basal spindle poles in 2nd instar wild type (48h ALH; n=11), 3rd instar wild type (96h ALH; n=7), 3rd instar cnnhk21 (96h ALH; n=9), and 2nd instar Lis1 (48h ALH; n=10) mutant metaphase neuroblasts. For fair comparison, only the 2 min period prior to anaphase onset (−2:00 – 0:00, indicated by grey shaded diagram areas in panels A–C) was analyzed for each genotype. Depicted are means ± S.E.M. The ‘frequency of high velocity movement’ corresponds to the frequency of time periods during which spindle pole velocity was ≥133 nm/s. This velocity corresponds to spindle pole movement by ≥2 pixels between two consecutive frames; movement by 1 pixel between consecutive frames corresponds to 63–67 nm (red line) and cannot be distinguished from noise. Differences in ‘peak velocity’ and ‘frequency of high velocity movements’ between genotypes were analyzed by Student’s t-tests. Significant differences are indicated (*, p<0.001). Scale bar: 5.
Reducing astral microtubule number decreases the frequency but not maximum velocity of spindle pole movement
Spindle movements could be due to a direct interaction between astral microtubules and the neuroblast cortex, or an indirect consequence of cytoplasmic flow or movement of neighboring cells. To determine if astral microtubules were required for spindle movements, we analyzed larval neuroblasts in centrosomin (cnnhk21) mutants, which have specific defects in pericentriolar material assembly and lack most or all astral microtubules (Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999). We performed live imaging of spindle dynamics in cnnhk21 mutant neuroblasts (Figure 2B; Movie 5). We find that cnnhk21 mutant neuroblasts have a slight delay in forming a bipolar spindle and have broader spindle poles (Figure 2B −7:57 to −5:57; Movie 5), but this did not preclude analysis of spindle dynamics. We found that cnnhk21 mutant neuroblast have a significant reduction in the frequency of high velocity spindle pole movements (≥133 nm/s) for both apical and basal spindle poles (Figure 2B, E). Interestingly, both apical and basal spindle poles in cnnhk21 neuroblasts occasionally exhibited high velocity spindle pole movements with peak velocities (comparable to those seen in wild type neuroblasts; Figure 2D). This suggests that there may be one or a few microtubules still contacting the cortex, which we confirmed in fixed preparations where we occasionally detected one microtubule (or microtubule bundle) contacting the cortex (Figure 3B). We conclude that astral microtubules mediate spindle-cortex forces, and that just one or a few microtubules are capable of generating maximum velocity spindle movements.
Figure 3. Defective spindle orientation in cnn, Lis1, and dynactin (Gl) mutant neuroblasts.
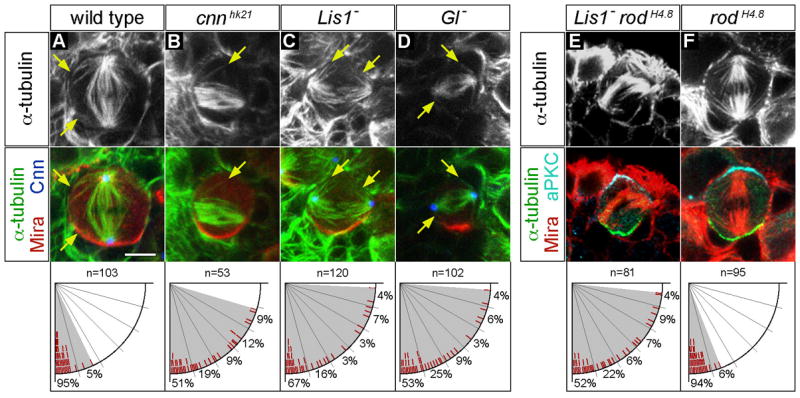
(A–D) wild type (A), cnnhk21 (B), Lis1 (C) and Gl (D) mutant neuroblasts were triple labelled for α-tubulin (top row), Cnn, and Mira (shown as merged images in bottom row).
(A) wild type metaphase neuroblasts showed spindles containing robust apical and basal asters nucleated by centrosomes (marked with Cnn). Note the proximity of astral MTs to the cortex (arrows) and the tight alignment of the spindle along the cell polarity axis (defined by basal Mira localization).
(B) cnnhk21 mutant neuroblasts lacked detectable Cnn protein, formed bipolar spindles with very few astral MTs, and often failed to align the spindle with the cell polarity axis.
(C–D) Lis1 (Lis1G10.14/Lis1k13209) and Gl (Gl1–3/Df(3L)fz-GF3b) mutant neuroblasts showed spindle alignment defects despite formation of bipolar spindles and astral microtubules (arrows) nucleated by centrosomes (marked with Cnn). The deficiency Df(3L)fz-GF3b removes the entire Gl locus.
(E–F) Lis1 rodH4.8 double and rodH4.8 single mutant neuroblasts were triple labelled for α-tubulin (top row), aPKC, and Mira (shown as merged images in bottom row).
(E) Lis1 rodH4.8 double mutant metaphase neuroblasts showed spindle alignment defects as observed in Lis1 single mutants.
(F) Metaphase spindle orientation is normal in rodH4.8 single mutant neuroblasts. Scale bar: 5 μm.
Lis1 mutants abolish spindle pole movement
Astral microtubule-dependent spindle pole movements could be due to the activity of a microtubule/cortex force generation complex, or due to growth and collapse of individual microtubules (dynamic instability). To test whether a motor complex was required for spindle movement, we investigated the role of the dynactin complex and Lis1, which associate with the dynein motor and have been shown to regulate spindle movement in several cell types (Dujardin and Vallee, 2002; Pearson and Bloom, 2004). When not attached to the cell membrane, the complex translocates to microtubule minus-ends at the centrosome; when attached to the cortex it can pull astral microtubules towards the cortex. In Drosophila, dynein is required for germ cell spindle positioning (McGrail and Hays, 1997) and dynein/dynactin/Lis1 affect spindle assembly and progression through neuroblast mitosis (Siller et al., 2005; Wojcik et al., 2001), but no member of this protein complex has been tested for a role in regulating neuroblast spindle movement. Here we use mutations in Lis1 (see methods), a known co-factor of the dynein/dynactin complex (Vallee and Tsai, 2006), to determine whether Lis1/dynactin/dynein is required for neuroblast spindle movement. We assayed a Lis1 transheterozygous mutant genotype (see methods) in which larval neuroblasts have undetectable Lis1 protein levels (Siller et al., 2005). Some of these mutant neuroblasts exhibit spindle assembly defects (Siller et al., 2005), so here we restrict our analysis to the neuroblasts that formed a normal bipolar spindle. Lis1 mutant neuroblasts have apparently normal astral microtubules that contact the cortex, but the peak velocity of spindle pole movements rarely exceeded detection threshold (Figure 2D, red line depicts detection threshold of spindle movement, see Materials and Methods), showing that Lis1 is required for spindle movement in neuroblasts. Interestingly, both apical and basal spindle poles were virtually static, with neither showing detectable rocking motion (Figure 2C, 3C; Movie 6). We conclude that the Lis1 (and by association, the dynactin complex) is required for generating apical and basal spindle/cortex force in larval neuroblasts.
Lis1/dynactin and astral microtubules are required to align the mitotic spindle with cortical polarity during prophase/metaphase
We next determined whether astral microtubules or Lis1/dynactin were required to align the mitotic spindle with cortical polarity. We analyzed cnn, Lis1, and dynactin mutant neuroblasts from prometaphase/metaphase to telophase, as these stages always showed spindle alignment with cortical polarity in wild type neuroblasts (95.1% of all wild type metaphase neuroblasts showed ≤15° angle between spindle and cell polarity axis; Figure 3A; Table 2). In contrast, cnnhk21 mutant metaphase neuroblasts showed spindle orientation defects at metaphase (Figure 3B; Table 2), consistent with previous studies (Megraw et al., 2001). Similarly, Lis1 and dynactin (Glued; Gl) mutant neuroblasts showed spindle positioning defects at metaphase without apparent defects in cortical polarity as assayed by normal apical Baz/aPKC and basal Mira localization (Figure 3C, D; Table 2). Although some Lis1 and Gl mutants had neuroblasts with spindle morphology defects (Siller et al., 2005), the observed spindle orientation defects were present even in mutant neuroblasts with normal bipolar spindle morphology, tightly focused spindle poles, and well-developed microtubule asters (Figure 3C, D). We confirmed these results by time lapse analysis of spindle orientation in wild type, cnnhk21, and Lis1 mutant neuroblasts. Wild type neuroblasts maintain spindle orientation from the end of prophase to telophase (Figure 1A, 2A, 4A; Movie 7; Table 2). In contrast, cnnhk21 mutant neuroblasts showed either no spindle movement or a slow drifting spindle that was unrelated to the cortical polarity axis (Figure 4B, D; Movie 8; Table 1) – consistent with reduced astral microtubule-cortex interactions described above. We conducted a similar analysis in Lis1 mutant neuroblasts, scoring only neuroblasts that formed a clear bipolar spindle with well-focused spindle poles. We found that in Lis1 mutant neuroblasts, centrosome position at the end of prophase deviated from the eventual division axis by 42°±31° (Figure 4C; Movie 9; Table 1); that is, spindle rotation occurred during metaphase-telophase, which was never observed in wild type neuroblasts. We conclude that the Lis1/dynactin complex and astral microtubules are required to align the mitotic spindle with the cortical polarity axis in prophase neuroblasts.
Table 2.
Spindle orientation relative to cell polarity axis in larval neuroblasts
| wild type a | cnnhk21 | Lis1 | Lis1 rod | rod | Gl1–3 | Gl1–3/Df | |
|---|---|---|---|---|---|---|---|
| Metaphase b | n=103 | n=53 | n=120 | n=81 | n=94 | n=101 | n=102 |
| 0°–15° | 95.1% | 50.9% | 66.7% | 54.3% | 93.6% | 62.4% | 55.9% |
| 16°–30° | 4.9% | 20.8% | 15.8% | 19.8% | 6.4% | 26.7% | 23.5% |
| 31°–45° | 0.0% | 11.3% | 3.3% | 7.4% | 0.0% | 6.9% | 7.8% |
| 46°–60° | 0.0% | 7.6% | 3.3% | 6.2% | 0.0% | 2.0% | 3.9% |
| 61°–75° | 0.0% | 9.4% | 6.7% | 8.6% | 0.0% | 1.0% | 4.9% |
| 76°–90° | 0.0% | 0.0% | 4.2% | 3.7% | 0.0% | 1.0% | 3.9% |
| Telophase | n=95 | n=86 | n=45 | n=79 | n=81 | n=19 | na |
| Mira asymmetrically segregated | 100.0% | 100.0% | 100.0% | 97.5% | 100.0% | 100.0% | Na |
| Mira symmetrically partitioned | 0.0% | 0% | 0.0% | 2.5% | 0.0% | 0.0% | na |
Genotypes: wild type (Oregon R); cnn (cnnhk21/cnnhk21); Lis1 (Lis1k13209/Lis1G10.14); Lis1 rod (Lis1k13209/Lis1G10.14 rodH4.8/rodH4.8); rod (rodH4.8/rodH4.8); Gl1–3 (Gl1–3/Gl1–3); Gl1–3/Df (Gl1–3/Df(3R)fz-Gf3b)
deviation of spindle axis from center of Mira crescent in metaphase neuroblasts (compare to Figure 5) na: not assayed
Figure 4. Live imaging reveals aberrant spindle movement during metaphase/anaphase in Lis1 and cnn mutant neuroblasts.

(A–C) Centrosome positioning and spindle orientation in larval neuroblasts expressing the G147-GFP microtubule-associated protein. Time is given in min:sec relative to onset of prometaphase (0:00).
(A) In wild type neuroblasts, centrosomes were already aligned with the final division axis by NEB (0:00) and spindle orientation remained fixed from prometaphase to telophase (0:00 –13:50; Movie 7).
(B) In cnnhk21 mutant neuroblasts, bipolar spindle formation was delayed (0:00 – 4:30). Eventually bipolar spindles formed, yet spindles could rotate during metaphase, anaphase, and telophase (4:30 – 11:10; Movie 8).
(C) In Lis1 mutant neuroblasts, centrosome position was abnormal at the onset of prometaphase (0:00) leading to formation of metaphase spindles with aberrant orientation (e.g. 38:40). Misoriented spindles could rotate during metaphase (14:40 – 38:20) and anaphase (63:00 – 66:30; Movie 9). Note the prolonged prometaphase/metaphase interval due to delayed bipolar spindle assembly and defects in mitotic checkpoint silencing.
(D) Representation of centrosome/spindle position in live larval neuroblasts. Depicted is the angular position of the centrosome pair/spindle over time relative to the centrosome pair position at the end of mitosis. Scale bar: 5 μm.
“Telophase rescue” of spindle/cortical polarity alignment in Lis1/dynactin mutants is not due to a delay in cell cycle progression
Lis1 and dynactin mutants had defective spindle alignment relative to cortical polarity at metaphase (see above), but surprisingly showed normal spindle/cortical polarity alignment by telophase (Figure 5; Table 2). One possible mechanism is that a delay in anaphase onset, characteristic of Lis1 and dynactin mutants (Siller et al., 2005), allows enough time for proper spindle/cortical polarity alignment. To address this issue, we tested whether spindle positioning defects still occurred in Lis1 rodH4.8 double mutants, where cell cycle delay is suppressed (Siller et al., 2005). Rod (Rough deal) is a checkpoint protein required to maintain metaphase arrest until kinetochore microtubules have attached to kinetochores, which is delayed in Lis1 mutants (Siller et al., 2005). We found that Lis1 rodH4.8 double mutant metaphase neuroblasts showed metaphase spindle orientation defects as well as “telophase rescue” of spindle/cortical polarity alignment (compare Figures 3, 5; Table 2). Thus, the defects in spindle/cortical polarity alignment observed in Lis1/dynactin mutants are not due to a delay in cell cycle progression.
Figure 5. cnn, Lis1, and dynactin (Gl) mutant metaphase neuroblasts show defective spindle alignment in metaphase but not telophase neuroblasts.
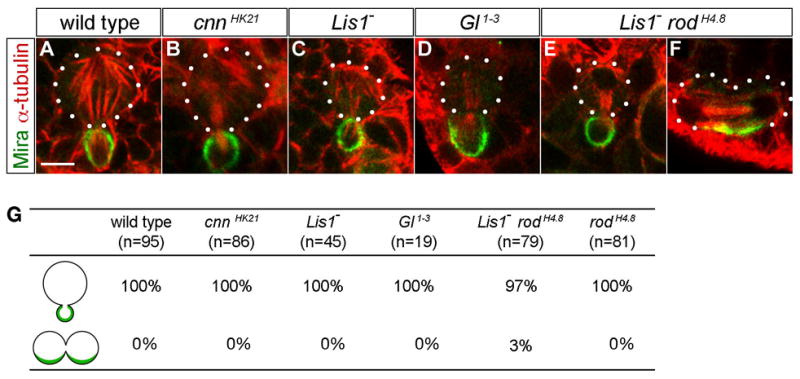
(A–F) Spindle orientation relative to cortical polarity axis in telophase.
(A) In wild type telophase neuroblasts the mitotic spindle is aligned such that Mira gets exclusively segregated into the smaller GMC.
(B–F) Telophase spindle alignment was normal in the vast majority of cnn (B), Lis1 (C), Gl1–3 (D), Lis1 rodH4.8 (E), and rodH4.8 (data not shown) mutant neuroblasts. Only in Lis1 rodH4.8 mutants we observed a few telophase neuroblasts with oblique spindle orientation causing equal partitioning of Mira into both daughter cells (F).
(G) Quantification of spindle alignment in telophase neuroblasts for the indicated genotypes; see Table 2 for complete quantification.
Scale bar: 5μm.
Discussion
Here we show that spindle/cortical polarity alignment is established at prophase in Drosophila larval neuroblasts, and that both apical and basal spindle poles move independently, as if spindle/cortex forces are applied to both poles. We show that reducing astral microtubule number reduces the frequency of spindle pole movements, but that maximum spindle pole velocity is unaffected, suggesting that maximum velocity may occur when only one or a few microtubules are simultaneously contacting the cortex. We show that the Lis1/dynactin complex is required for spindle pole movement; reducing Lis1/dynactin complex activity reduces the maximum and average spindle velocity, even though astral microtubules still contact the cortex. This suggests that Lis1/dynactin is required to translate microtubule-cortex contact into spindle movement. Finally, we show that Lis1/dynactin is required for spindle orientation at metaphase but not at telophase. These findings are summarized in Figure 6.
Figure 6. Summary of spindle positioning mechanism in larval neuroblasts.
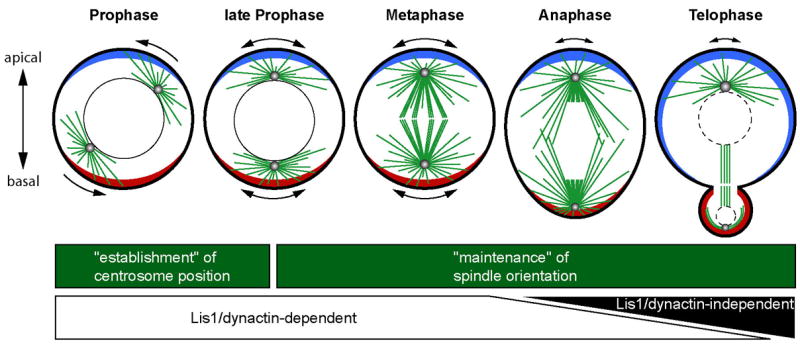
In wild type neuroblasts, apical-basal positioning of centrosomes is established during prophase and maintained from prometaphase onward. Apical-basal centrosome/spindle positioning during the period from prophase to metaphase depends on Lis1/dynactin function, presumably due to their role in regulation of cortex-spindle forces. The short-lived imbalance of theses forces is manifest in the rocking movements of both apical and basal spindle poles (indicated by double headed arrows outside the neuroblasts). Spindle rocking ceases during anaphase and telophase –only the apical pole shows sporadic lateral movements – possibly as a consequence of reduced or more balanced force generation/transduction. Spindle positioning in anaphase and telophase neuroblasts is largely Lis1/dynactin-independent, suggesting the existence of a novel yet unidentified spindle positioning pathway that can re-establish correct spindle alignment before the end of telophase in the absence of Lis1/dynactin (see Discussion for details).
Apical and basal spindle poles move independently
We observed Lis1-dependent dynamic microtubule-cortex interactions at both apical and basal spindle poles, as well as asynchronous movements of apical and basal spindle poles. What are the candidate apical or basal cortical proteins that might regulate spindle pole movement? Insight into the role of cortical proteins in regulating spindle movement has been made in both C. elegans and mammals, and can be used to model spindle dynamics in Drosophila. Apical proteins in neuroblasts known to regulate spindle force in C. elegans include Gαi, Pins and Mud. During the first division of the C. elegans zygote, enrichment of the Gα-binding and activating Pins-related GPR1/2 proteins at the posterior cortex leads to increased Gα activity, resulting in higher cortex-spindle force generation, spindle pole rocking, and posterior spindle displacement (Colombo et al., 2003; Couwenbergs et al., 2007; Gotta et al., 2003; Grill et al., 2003; Nguyen-Ngoc et al., 2007; Srinivasan et al., 2003). This suggests that Gαi/Pins/Mud may promote movement of the apical spindle pole in Drosophila neuroblasts, which is supported by our recent finding that reducing Gαi can decrease spindle rocking (Bowman et al., 2006; Izumi et al., 2006; Nipper et al., 2007; Parmentier et al., 2000; Schaefer et al., 2001; Schaefer et al., 2000; Siller et al., 2006; Yu et al., 2000).
In C. elegans, Lin-5 mediates the physical interaction of Lis1/dynein/dynactin with the cortical Gα and the Pins-related GPR1/2 proteins (Couwenbergs et al., 2007; Nguyen-Ngoc et al., 2007), and reduction of dynein or Lis1 function also reduces spindle pole rocking and posterior spindle displacement (Couwenbergs et al., 2007; Nguyen-Ngoc et al., 2007; Schmidt et al., 2005). Furthermore, in mammalian tissue culture cells Gαi overexpression can induce robust spindle rocking that requires LGN (a Pins/GPR-related protein) and NuMA (a Mud/Lin-5-related protein that binds dynein/dynactin (Du and Macara, 2004; Merdes et al., 1996). An attractive model is that Gαi/LGN activates NuMA, which interacts with dynein/dynactin/Lis1-loaded astral microtubules. In Drosophila neuroblasts, Gαi, Pins (LGN-related) and Mud (NuMA-related Pins-binding protein) are all enriched at the apical cortex and required for proper metaphase spindle orientation (Bowman et al., 2006; Izumi et al., 2006; Nipper et al., 2007; Parmentier et al., 2000; Schaefer et al., 2001; Schaefer et al., 2000; Siller et al., 2006; Yu et al., 2000). Thus, it is tempting to propose that apical Gαi/Pins/Mud interacts with dynein/dynactin/Lis1-loaded astral microtubules to center the apical spindle pole with the apical cortical domain. Identifying a physical link between Mud and dynein/dynactin/Lis1, and determining its functional importance in spindle orientation, would be a good test of this model.
Surprisingly, we find that third instar larval neuroblasts have more vigorous basal spindle pole rocking than apical spindle pole rocking, revealing a Gαi-independent spindle force generation mechanism at the basal cortex. Basal cortical proteins include Armadillo, DE-cadherin, β-catenin, APC2, and Mud (Akong et al., 2002; Bowman et al., 2006; Izumi et al., 2006; McCartney et al., 1999; Siller et al., 2006). Components of the APC2/DE-cadherin/α-catenin/β-catenin complex physically interact with the dynein complex in mammalian cells, and are required for spindle positioning in the Drosophila pre-cellular embryo, epithelial cells, and germline stem cells (Akong et al., 2002; Bowman et al., 2006; Izumi et al., 2006; Ligon et al., 2001; Lu et al., 2001; McCartney et al., 1999; McCartney et al., 2001; McCartney et al., 2006; Yamashita et al., 2003). Previous studies indicated no spindle positioning defects in neuroblasts after reduction of APC2 function (Akong et al., 2002), however these findings do not rule out a role for APC2 in spindle orientation because it may function redundantly with an apical cue, such as the Gαi/Pins/Mud pathway (Akong et al., 2002; Bowman et al., 2006; Izumi et al., 2006; Nipper et al., 2007; Siller et al., 2006).
Lis1/dynactin regulation of spindle positioning forces
We have demonstrated that both apical and basal spindle pole movements are greatly diminished in Lis1 mutant larval neuroblasts (even in those with well-formed bipolar spindles and asters), providing first evidence that Lis1/dynactin is a critical component in the regulation of both apical and basal cortex-spindle forces. How does Lis1 regulate cortex-spindle forces? One possibility is that translocation of cortically associated motor proteins towards microtubule(−) ends results in movement of the microtubule towards the cortex. Consistent with this hypothesis, Lis1 colocalizes with and binds the microtubule minus-end motor dynein/dynactin complex (Dujardin and Vallee, 2002; Vallee et al., 2001; Xiang, 2003). Specifically, the budding yeast Lis1 homologue (Pac1) targets dynein to astral microtubule plus-ends (Lee et al., 2003; Sheeman et al., 2003) where it promotes movement of astral microtubules towards the cortex, resulting in translocation of the spindle apparatus through the bud neck (Adames and Cooper, 2000). By analogy, Lis1 may regulate spindle pole movement in neuroblasts by promoting dynein-dependent movement of astral microtubules towards the cortex. Alternatively, Lis1 may modulate the polymerization/depolymerization cycle (dynamic instability) of cortically-attached astral microtubules or the duration of astral microtubule-cortex interactions. In support of this latter hypothesis, loss of Lis1 or dynein function in Aspergillus nidulans or budding yeast results in reduced microtubule catastrophe and/or decreased shrinkage rates, thereby promoting assembly of overly long microtubules (Carvalho et al., 2003). Currently, we are not able to visualize astral microtubule plus-ends with sufficient spatial and temporal resolution to distinguish between these models for Lis1 function.
Both models for Lis1 function described above would require association of Lis1 protein with astral microtubules and/or the neuroblast cortex. Indeed, Lis1/dynactin complex proteins have been detected on astral microtubule plus-ends or at the cortex in mammalian, nematode, and yeast cells (Wynshaw-Boris, 2007). We previously analyzed the localization of HA-tagged Lis1, GFP-tagged Lis1, endogenous Lis1, and endogenous dynactin protein distribution using various fixation and live imaging methods in embryonic and larval neuroblasts, but were unable to detect enrichment of Lis1/dynactin at the cortex or at astral microtubule plus-ends (Siller et al., 2005). The most likely explanation is that Lis1/dynactin at these sites is masked by the high level of cytoplasmic protein present in neuroblasts.
Two spindle orientation pathways
The Lis1/dynactin complex is required for reliable spindle orientation with the apical/basal polarity axis in metaphase neuroblasts. These spindle orientation defects may be due in part to failure in anchoring one centrosome at the apical cortex during interphase, as recently reported for wild type neuroblasts (Rebollo et al., 2007; Rusan and Peifer, 2007); we have observed mis-positioned interphase centrosomes in Lis1 mutants, but have not analyzed this phenotype in detail. We were surprised to find that spindle orientation was essentially normal at telophase in Lis1 and dynactin (Gl) mutant neuroblasts, despite severe defects at metaphase. This indicates that there are two pathways for regulating spindle orientation: an early Lis1/dynactin-dependent pathway (prophase/metaphase), and a late Lis1/dynactin-independent pathway (anaphase/telophase). There are several models consistent with these findings: (1) Lis1 and dynactin mutants have a delay in anaphase onset (Siller et al., 2005) which allows sufficient time for “telophase rescue” to occur. (2) A spindle orientation checkpoint -- analogous to the yeast spindle orientation checkpoint (Lew and Burke, 2003) -- may delay cytokinesis until proper spindle orientation has occurred. These first two hypotheses are disproven by our finding that Lis1 rod double mutants have normal metaphase progression but still show metaphase defects and “telophase rescue” of spindle orientation. (3) The cleavage furrow may be positioned by cortical polarity cues, such that cell elongation at early anaphase may mechanically re-orient the spindle along the long axis of the neuroblast. This model is unlikely because it is commonly accepted that the position of the cleavage furrow is determined by the position of the mitotic spindle and not by cortical cues (Burgess and Chang, 2005). (4) Additional microtubule-cortex regulators unrelated to Lis1/dynactin promote telophase spindle orientation.
The fourth model is the most likely, except that microtubule-cortex regulators unrelated to Lis1/dynactin have not yet been identified in Drosophila neuroblasts. Help may come from analysis of budding yeast spindle orientation pathways, where Lis1/dynactin-dependent and -independent pathways have been identified (Huisman and Segal, 2005). Several components of the yeast Lis1/dynactin-independent pathway are evolutionarily conserved, including the microtubule plus-end binding protein Bim1p, called EB1 in Drosophila. It is tempting to speculate that these proteins may regulate the Lis1/dynactin-independent pathway in Drosophila neuroblasts.
Experimental Procedures
Fly Genetics
The Oregon R strain was used as wild type control in all experiments. To analyze the Lis1 mutant phenotype we used Lis1G10.14/Lis1k13209 transheterozygotes (Liu et al., 1999). Other strains used include: cnnhk21/CyO (Megraw et al., 1999); Gl1–3/TM6B (Harte and Kankel, 1982; Siller et al., 2005); Df(3R)fz-GF3b/TM6B (Bloomington stock center); rodH4.8/TM6B (Karess and Glover, 1989); the gene trap line P{PTT-GA}JupiterG00147, expressing a GFP-tagged microtubule-associated protein CG17238 (Morin et al., 2001). EYFP-Baz was expressed in larval neuroblasts by crossing pUAST-3xEYFP-Baz transgenic flies (Siller et al., 2006) to a worniu-Gal4 driver line (Albertson and Doe, 2003). Mutant alleles were rebalanced over Cyo actin-GFP or TM3 actin-GFP Ser. Newly hatched mutant larvae (identified based on absence of GFP expression in the gut) were aged to appropriate stage at 25ºC.
Time-lapse analysis of larval neuroblast divisions
Larval brains of wild type, cnnHk21, or Lis1 mutants were dissected and neuroblasts were imaged using a BioRad Radiance 2000 laser scanning confocal microscope equipped with a 60× 1.4NA oil immersion objective as previously described (Siller et al., 2005). Time-lapse sequences were processed using MetaMorph (Universal Imaging Corp) and ImageJ and were converted into movies using QuickTime Pro.
Analysis of spindle pole movements in live neuroblasts
Stacks of 3 focal planes, 1.5 μm apart, were imaged in 3.3 to 3.5s intervals with a resolution of 0.222 μm/pixel (Figure 2, Movies 3–5). The ‘frequency of high velocity movement’ of spindle poles corresponds to the frequency of time periods with a velocity ≥133 nm/s (corresponding to spindle pole movement by ≥2 pixels between two consecutive frames). Thus, movement by 1 pixel between consecutive frames corresponds to 63–67 nm, which corresponds to the “detection threshold” (red line in Figure 2D).
Antibodies and immunofluorescent staining
Wild type (48h ALH), cnnhk21 (96–120h ALH), Lis1G10.14/Lis1k13209 (48h ALH), Lis1G10.14/Lis1k13209 rodH4.8 (48h ALH), rodH4.8 (48h ALH), Gl1–3 (96–120h ALH), and Gl1–3/Df(3R)fz-GF3b (96–120h ALH) larval brains were dissected, fixed and immunofluorescently labelled as previously described (Siller et al., 2005). Primary antibodies were: rat anti-Miranda (Irion et al., 2004); rabbit anti-nPKCλ (C20, Santa Cruz Biotechnology Inc; 1:1000); rabbit anti-Baz (gift from Andreas Wodarz; 1:500); mouse anti α-tubulin (DM1A, Sigma; 1:2000); rabbit anti-Cnn (Heuer et al., 1995). Fluorescently conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories and Molecular Probes. Confocal images were acquired on a BioRad Radiance 2000 or Leica TCS SP2 laser scanning confocal microscope equipped with a 60× or 63× 1.4 NA oil immersion oil respectively. Final figures were arranged using ImageJ and Adobe Photoshop.
Supplementary Material
Movie 1. In wild type neuroblasts, apical-basal spindle positioning is established during prophase.
Centrosome and spindle movements were analyzed in 2nd instar wild type neuroblasts (48h ALH) expressing the G147-GFP microtubule-associated protein. Separated centrosomes were positioned along the apical-basal division axis by the end of prophase. Stable centrosome/spindle orientation was maintained from the end of prophase onward (Figure 1). Similar observations were made on 3rd instar larval neuroblasts (data not shown).
Movie 2. Alignment of the mitotic spindle along the apical-basal cortical polarity axis in wild type neuroblasts
Coordination of cortical polarity and spindle positioning was analyzed in 2nd instar wild type neuroblasts (48h ALH) co-expressing EYFP-Baz and the G147-GFP microtubule-associated protein. An apical EYFP-Baz crescent was established before the centrosomes reach their final position (Figure 1).
Movie 3+4. Dynamics of spindle pole movement in wild type larval neuroblast.
Dynamic movements of spindle poles in 2nd instar (48h ALH) wild type larval neuroblasts expressing the G147-GFP microtubule-associated protein. Note that periods of rapid apical and basal spindle pole movement could occur independently from each other. No significant difference was observed between 2nd instar (48h ALH) and 3rd instar (96h ALH) larval neuroblasts (see Figure 2D).
Movie 5. Reduced frequency of rapid spindle pole movement in cnn mutant larval neuroblast
Dynamic movements of spindle poles were analyzed in 3rd instar (96h ALH) cnnhk21 larval neuroblasts expressing the G147-GFP microtubule-associated protein. Due to compromised centrosome function bipolar spindle formation was delayed, yet bipolar spindles form eventually. Note the reduction of astral microtubule number and decrease in frequency of high velocity spindle pole movements compared to wild type counterparts (Figure 2).
Movie 6. Reduced spindle pole movement in Lis1 mutant larval neuroblasts.
Dynamic movements of apical and basal spindle poles were analyzed in 2nd instar (48h ALH) Lis1 larval neuroblasts expressing the G147-GFP microtubule-associated protein. Despite robust bipolar spindle formation and presence of astral microtubules, peak velocity and frequency of high velocity spindle pole movements were greatly reduced compared to wild type counterparts (Figure 2).
Movie 7: In wild type neuroblasts, spindle orientation is stable during the propmetaphase-telophase interval.
Centrosome and spindle movements were analyzed in 2nd instar wild type neuroblasts (48h ALH) expressing the G147-GFP microtubule-associated protein. Separated centrosomes were positioned along the eventual division (apical-basal) axis by the end of prophase. Stable centrosome/spindle orientation was maintained from the end of prophase onward (Figure 4).
Movie 8: Spindle positioning defects in cnn mutant larval neuroblast.
Spindle position was analyzed in cnnhk21 larval neuroblasts (96h ALH) expressing the G147-GFP-tagged microtubule-associated protein. Orientation of the metaphase spindle was not fixed and final spindle orientation was achieved through rotation during of anaphase/early telophase spindle (Figure 4).
Movie 9. Centrosome/spindle positioning defects in Lis1 mutant larval neuroblast.
Centrosome and spindle position was analyzed in Lis1 larval neuroblasts expressing the G147-GFP-tagged microtubule-associated protein. At NEB, centrosomes were not positioned along the eventual division axis. Orientation of the metaphase spindle was not fixed and final spindle orientation was achieved through rotation during of anaphase/early telophase spindle (Figure 4).
Acknowledgments
We are grateful to Drs. Doug Kankel, Andreas Wodarz, and Thom Kaufman for generously sharing reagents. We also like to thank Clemens Cabernard and Sarah Siegrist for stimulating discussions and helpful comments on the manuscript. This study was supported by the American Heart Association (predoctoral fellowship to K.H.S.) and the Howard Hughes Medical Institute, where C.Q.D. is an Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–74. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akong K, et al. Drosophila APC2 and APC1 have overlapping roles in the larval brain despite their distinct intracellular localizations. Dev Biol. 2002;250:71–90. doi: 10.1006/dbio.2002.0777. [DOI] [PubMed] [Google Scholar]
- Albertson R, Doe CQ. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat Cell Biol. 2003;5:166–70. doi: 10.1038/ncb922. [DOI] [PubMed] [Google Scholar]
- Basto R, et al. Flies without centrioles. Cell. 2006;125:1375–86. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Bowman SK, et al. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–42. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Burgess DR, Chang F. Site selection for the cleavage furrow at cytokinesis. Trends Cell Biol. 2005;15:156–62. doi: 10.1016/j.tcb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Carvalho P, et al. Surfing on microtubule ends. Trends Cell Biol. 2003;13:229–37. doi: 10.1016/s0962-8924(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Caussinus E, Hirth F. Asymmetric stem cell division in development and cancer. Prog Mol Subcell Biol. 2007;45:205–25. doi: 10.1007/978-3-540-69161-7_9. [DOI] [PubMed] [Google Scholar]
- Colombo K, et al. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 2003;300:1957–61. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- Couwenbergs C, et al. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J Cell Biol. 2007;179:15–22. doi: 10.1083/jcb.200707085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu Rev Cell Dev Biol. 2004;20:427–53. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–16. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Dujardin DL, Vallee RB. Dynein at the cortex. Curr Opin Cell Biol. 2002;14:44–9. doi: 10.1016/s0955-0674(01)00292-7. [DOI] [PubMed] [Google Scholar]
- Giansanti MG, et al. The role of centrosomes and astral microtubules during asymmetric division of Drosophila neuroblasts. Development. 2001;128:1137–45. doi: 10.1242/dev.128.7.1137. [DOI] [PubMed] [Google Scholar]
- Gonzalez C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat Rev Genet. 2007;8:462–72. doi: 10.1038/nrg2103. [DOI] [PubMed] [Google Scholar]
- Gotta M, et al. Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr Biol. 2003;13:1029–37. doi: 10.1016/s0960-9822(03)00371-3. [DOI] [PubMed] [Google Scholar]
- Grill SW, et al. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–21. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- Harte PJ, Kankel DR. Genetic analysis of mutations at the Glued locus and interacting loci in Drosophila melanogaster. Genetics. 1982;101:477–501. doi: 10.1093/genetics/101.3-4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer JG, et al. The Drosophila homeotic target gene centrosomin (cnn) encodes a novel centrosomal protein with leucine zippers and maps to a genomic region required for midgut morphogenesis. Development. 1995;121:3861–76. doi: 10.1242/dev.121.11.3861. [DOI] [PubMed] [Google Scholar]
- Huisman SM, Segal M. Cortical capture of microtubules and spindle polarity in budding yeast - where’s the catch? J Cell Sci. 2005;118:463–71. doi: 10.1242/jcs.01650. [DOI] [PubMed] [Google Scholar]
- Irion U, et al. Abstrakt, a DEAD box protein, regulates Insc levels and asymmetric division of neural and mesodermal progenitors. Curr Biol. 2004;14:138–44. doi: 10.1016/j.cub.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Izumi Y, et al. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006;8:586–93. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- Karess RE, Glover DM. rough deal: a gene required for proper mitotic segregation in Drosophila. J Cell Biol. 1989;109:2951–61. doi: 10.1083/jcb.109.6.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–97. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Lee WL, et al. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–64. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Burke DJ. The spindle assembly and spindle position checkpoints. Annu Rev Genet. 2003;37:251–82. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- Ligon LA, et al. Dynein binds to beta-catenin and may tether microtubules at adherens junctions. Nat Cell Biol. 2001;3:913–7. doi: 10.1038/ncb1001-913. [DOI] [PubMed] [Google Scholar]
- Lu B, et al. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature. 2001;409:522–5. doi: 10.1038/35054077. [DOI] [PubMed] [Google Scholar]
- McCartney BM, et al. Drosophila APC2 is a cytoskeletally-associated protein that regulates wingless signaling in the embryonic epidermis. J Cell Biol. 1999;146:1303–18. doi: 10.1083/jcb.146.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney BM, et al. Drosophila APC2 and Armadillo participate in tethering mitotic spindles to cortical actin. Nat Cell Biol. 2001;3:933–8. doi: 10.1038/ncb1001-933. [DOI] [PubMed] [Google Scholar]
- McCartney BM, et al. Testing hypotheses for the functions of APC family proteins using null and truncation alleles in Drosophila. Development. 2006;133:2407–18. doi: 10.1242/dev.02398. [DOI] [PubMed] [Google Scholar]
- McGrail M, Hays TS. The microtubule motor cytoplasmic dynein is required for spindle orientation during germline cell divisions and oocyte differentiation in Drosophila. Development. 1997;124:2409–19. doi: 10.1242/dev.124.12.2409. [DOI] [PubMed] [Google Scholar]
- Megraw TL, et al. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–20. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- Megraw TL, et al. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 1999;126:2829–39. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- Merdes A, et al. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–58. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Morin X, et al. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15050–5. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–74. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Nguyen-Ngoc T, et al. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 2007;9:1294–302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- Nipper RW, et al. Galphai generates multiple Pins activation states to link cortical polarity and spindle orientation in Drosophila neuroblasts. Proc Natl Acad Sci U S A. 2007;104:14306–11. doi: 10.1073/pnas.0701812104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier ML, et al. Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J Neurosci. 2000;20:RC84. doi: 10.1523/JNEUROSCI.20-14-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Bloom K. Dynamic microtubules lead the way for spindle positioning. Nat Rev Mol Cell Biol. 2004;5:481–92. doi: 10.1038/nrm1402. [DOI] [PubMed] [Google Scholar]
- Rebollo E, et al. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–74. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, et al. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–94. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- Schaefer M, et al. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr Biol. 2000;10:353–62. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- Schmidt DJ, et al. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol Biol Cell. 2005;16:1200–12. doi: 10.1091/mbc.E04-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B, et al. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–72. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–35. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Siller KH, et al. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- Siller KH, et al. Live Imaging of Drosophila Brain Neuroblasts Reveals a Role for Lis1/Dynactin in Spindle Assembly and Mitotic Checkpoint Control. Mol Biol Cell. 2005 doi: 10.1091/mbc.E05-04-0338. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan DG, et al. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev. 2003;17:1225–39. doi: 10.1101/gad.1081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaizel-Ohayon D, Schejter ED. Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr Biol. 1999;9:889–98. doi: 10.1016/s0960-9822(99)80393-5. [DOI] [PubMed] [Google Scholar]
- Vallee RB, et al. LIS1: cellular function of a disease-causing gene. Trends Cell Biol. 2001;11:155–60. doi: 10.1016/s0962-8924(01)01956-0. [DOI] [PubMed] [Google Scholar]
- Vallee RB, Tsai JW. The cellular roles of the lissencephaly gene LIS1, and what they tell us about brain development. Genes Dev. 2006;20:1384–93. doi: 10.1101/gad.1417206. [DOI] [PubMed] [Google Scholar]
- Wojcik E, et al. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat Cell Biol. 2001;3:1001–7. doi: 10.1038/ncb1101-1001. [DOI] [PubMed] [Google Scholar]
- Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- Xiang X. LIS1 at the microtubule plus end and its role in dynein-mediated nuclear migration. J Cell Biol. 2003;160:289–90. doi: 10.1083/jcb.200212168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, et al. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–50. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- Yu F, et al. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. In wild type neuroblasts, apical-basal spindle positioning is established during prophase.
Centrosome and spindle movements were analyzed in 2nd instar wild type neuroblasts (48h ALH) expressing the G147-GFP microtubule-associated protein. Separated centrosomes were positioned along the apical-basal division axis by the end of prophase. Stable centrosome/spindle orientation was maintained from the end of prophase onward (Figure 1). Similar observations were made on 3rd instar larval neuroblasts (data not shown).
Movie 2. Alignment of the mitotic spindle along the apical-basal cortical polarity axis in wild type neuroblasts
Coordination of cortical polarity and spindle positioning was analyzed in 2nd instar wild type neuroblasts (48h ALH) co-expressing EYFP-Baz and the G147-GFP microtubule-associated protein. An apical EYFP-Baz crescent was established before the centrosomes reach their final position (Figure 1).
Movie 3+4. Dynamics of spindle pole movement in wild type larval neuroblast.
Dynamic movements of spindle poles in 2nd instar (48h ALH) wild type larval neuroblasts expressing the G147-GFP microtubule-associated protein. Note that periods of rapid apical and basal spindle pole movement could occur independently from each other. No significant difference was observed between 2nd instar (48h ALH) and 3rd instar (96h ALH) larval neuroblasts (see Figure 2D).
Movie 5. Reduced frequency of rapid spindle pole movement in cnn mutant larval neuroblast
Dynamic movements of spindle poles were analyzed in 3rd instar (96h ALH) cnnhk21 larval neuroblasts expressing the G147-GFP microtubule-associated protein. Due to compromised centrosome function bipolar spindle formation was delayed, yet bipolar spindles form eventually. Note the reduction of astral microtubule number and decrease in frequency of high velocity spindle pole movements compared to wild type counterparts (Figure 2).
Movie 6. Reduced spindle pole movement in Lis1 mutant larval neuroblasts.
Dynamic movements of apical and basal spindle poles were analyzed in 2nd instar (48h ALH) Lis1 larval neuroblasts expressing the G147-GFP microtubule-associated protein. Despite robust bipolar spindle formation and presence of astral microtubules, peak velocity and frequency of high velocity spindle pole movements were greatly reduced compared to wild type counterparts (Figure 2).
Movie 7: In wild type neuroblasts, spindle orientation is stable during the propmetaphase-telophase interval.
Centrosome and spindle movements were analyzed in 2nd instar wild type neuroblasts (48h ALH) expressing the G147-GFP microtubule-associated protein. Separated centrosomes were positioned along the eventual division (apical-basal) axis by the end of prophase. Stable centrosome/spindle orientation was maintained from the end of prophase onward (Figure 4).
Movie 8: Spindle positioning defects in cnn mutant larval neuroblast.
Spindle position was analyzed in cnnhk21 larval neuroblasts (96h ALH) expressing the G147-GFP-tagged microtubule-associated protein. Orientation of the metaphase spindle was not fixed and final spindle orientation was achieved through rotation during of anaphase/early telophase spindle (Figure 4).
Movie 9. Centrosome/spindle positioning defects in Lis1 mutant larval neuroblast.
Centrosome and spindle position was analyzed in Lis1 larval neuroblasts expressing the G147-GFP-tagged microtubule-associated protein. At NEB, centrosomes were not positioned along the eventual division axis. Orientation of the metaphase spindle was not fixed and final spindle orientation was achieved through rotation during of anaphase/early telophase spindle (Figure 4).


