Abstract
The DYT1 gene encodes for torsinA, a protein with widespread tissue distribution, involved in Early Onset Dystonia. Numerous studies have focused on torsinA function but no information is available on its transcriptional regulation. We cloned mouse and human 5′ upstream DYT1-DNA fragments, exhibiting high transcriptional activity, as well as tissue specificity. We identified a proximal minimal DYT1 promoter within -141bp for mouse and -191bp for human, with respect to the ATG codon. Primer extension analysis indicated multiple transcription start sites. In silico analysis, of an ∼500bp-5′ upstream DYT1 fragment, demonstrated lack of a classical TATA or CAAT box and the presence of a highly conserved direct repeat of two Ets binding cores, within -86bp to -77bp and -78bp to -69bp of the mouse and human DYT1 gene, respectively. A single or a two base nucleotide alteration within the more downstream Ets core resulted in ∼ 90% (mouse) or ∼45 to 60% (human) drop of activity. Interestingly, a 3bp distance increase between the two Ets cores, dramatically decreased transcriptional activity which was partially restored when the distance was increased up to 10bp. Ets-like dominant negatives confirmed the Ets factors as DYT1 transcriptional activators.
Keywords: Ets factors, DYT1 gene, transcriptional regulation
Introduction
The DYT1 gene encodes for torsinA (Ozelius et al. 1997), an ER-resident glycoprotein (Kustedjo et al. 2000; Liu et al. 2003) of unknown function which shares high homology with the class 2-type HSP100/CLp (Ozelius et al. 1997), a subfamily of AAA+ proteins (ATPases Associated with a variety of cellular Activities) (Ogura and Wilkinson 2001). The genetic defect linked to Early Onset Dystonia (EOD) is a GAG-deletion in the DYT1 gene, resulting in loss of a glutamic acid in the C-terminus of torsinA (ΔE-torsinA) (Ozelius et al. 1997). EOD is a neurological movement disorder, inherited in an autosomal, dominant mode (Fahn 1988), with 30-40% penetrance (Bressman et al. 2000). Despite the widespread distribution of torsinA in both peripheral and neuronal tissues (Walker et al. 2002; Augood et al. 2003; Xiao et al. 2004), effects of ΔE torsinA are limited to the nervous system (Fahn 1988). Thus, the tissue expression pattern of torsinA in itself is insufficient to account for a specific neuronal dysfunction.
TorsinA levels have been shown to be reduced in fibroblasts of EOD patients (Goodchild et al. 2005). Differential levels of mutant torsinA in ΔE-torsinA carriers could modify disease risk between individuals and account for the reduced EOD penetrance. This emphasizes the importance of elucidating the mechanisms of DYT1 gene regulation. Towards that end, we cloned the proximal 5′ upstream DYT1 region of the mouse and human gene and analyzed cis-acting elements, responsible for gene expression. In silico analysis of an ∼500bp-5′ flanking DYT1 fragment revealed lack of a canonical TATA or CAAT box (Singer et al. 1990) and the presence of highly consensus binding motifs for members of the Ets family of transcription factors.
The Ets family of transcription factors is composed of more than 30 members, covering a large spectrum of functions from hematopoiesis and extracellular matrix remodeling to neuronal differentiation and expression of synapse specific genes (Sharrocks 2001). Ets proteins bear an Activating transcription Domain (AD) and an ∼85aa highly conserved DNA binding domain (Ets-domain), located in the majority of the Ets proteins in their C-terminus (Degnan et al. 1993; Oikawa and Yamada 2003). The Ets consensus binding core is GGA(A/T) (Degnan et al. 1993; Laudet et al. 1993), while specificity at DNA binding level is provided by the flanking sequences (Sharrocks et al. 1997).
The objective of this study was to analyze the 5′ upstream region of the human and mouse DYT1 gene, delineate the proximal minimal DYT1 promoter and characterize physically as well as functionally cis-regulatory Ets-like motifs that potentially influence DYT1 expression.
Materials and methods
Cell Cultures
HEK293T, N2A cells and human primary astrocytes were maintained in Dulbecco’s modified Eagle’s medium, HepG2 in RPMI 1640 medium, and SH-SYS5 in DMEM/F-HAM, supplemented with 10% fetal bovine serum. All cell lines were from ATCC (Manassas, VA) while human primary astrocytes were kindly provided by Dr G. John (Mount Sinai School of Medicine, NY, NY10029).
Primer extension analysis
Mouse total brain or liver RNA was extracted using TRIzol Reagent. 50,000 cpm of a [32P]ATP 5′-end labeled oligodeoxynucleotide (Table I), complementary to the region from +61 to +80 relative to the translation start site of the mouse DYT1 cDNA, was hybridized to 20–50μg of total RNA, for 90 min at 65°C, in hybridization buffer (1 M NaCl, 0.1 M PIPES, pH 6.4, 25 mM EDTA, pH 8.0). The annealed primer was extended by avian myeloblastosis virus reverse transcriptase for 1 h at 42°C. The reaction products were treated with DNase-free RNase at 37 °C for 15 min to remove the RNA templates, purified by phenol-chloroform extraction followed by ethanol precipitation. Using the same primer, a DNA sequencing size marker was established on DNA constructs encompassing 1kb of the mouse 5′ upstream DYT1 region. Yeast t-RNA template was used as negative control. Products were analyzed by electrophoresis on a 6% denaturing acrylimide gel containing 8M urea.
TABLE I.
Primer used for Primer Extension Analysis
| +61′ CTCAGGCTGATGGGCTCCAC +80′ |
Generation of 5′ upstream DYT1/luc and 5′ upstream DYT1/torsinA expression constructs
Reporter constructs used in this study are as follows: a) -2416mDYT1 through -63mDYT1 containing 2416-, 1962-, 1400-, 912-, 505-, 243-, 196-, 141-, 92- and 63-bp of the mouse 5′ upstream DYT1 region, and b) -2731hDYT1 through -45hDYT1 containing 2731-, 1973-, 1446-, 931-, 445-, 301-, 191-, 121-, 80- and 45-bp of the human 5′ upstream DYT1 region. All 5′ upstream DYT1 fragments were generated by direct PCR amplification of mouse and human Bac clones (BACPAC Resources, Oakland, CA) obtained from chromosomes 2 (GenBank® accession no. AL844532) and 9 (GenBank® accession no. AL158207), respectively, and appropriate set of primers (Table II) designed based on the published mouse and human genome sequence. Restriction sites for KpnI and NheI were included at the 5′ and 3′ end of the forward and reverse PCR primers, respectively. PCR products were gel purified (QIAguick gel extraction kit, Qiagen, Valencia, CA) and ligated into the KpnI and NheI sites of the pGL3-basic vector (pGL3-BV, Promega, Madison, WI), upstream of the luciferase gene. Mutant reporter constructs -243mDYT1-S1, -S2, -S2S3, -243mDYT1+3G, +3C, +6 and +10, were generated from the parental -243mDYT1 (Fig. 4, 5A and 8A), while mutant constructs -301hDYT1-S1, -S2, -S2S3 from the -301hDYT1 (Fig. 4 and 5E), using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and primers listed in Table II. Human torsinA, previously isolated in our laboratory (Shashidharan et al. 2000) was ligated a) into the NheI and XhoI sites of the -2731hDYT1 construct, while the luciferase cDNA was excised, giving rise to the -2731hDYT1-hwtTA construct or b) into the HindIII and NotI sites of the pcDNA3.1 vector (Invitrogen, Carlsbad, CA) giving rise to the pCMV-hwtTA construct. All inserts were confirmed by DNA sequencing.
TABLE II.
Primers used for constructing 5′ upstream DYT1/luciferase reporter constructs
| Construct | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| -2416mDYT1 | -2416′CTCTGTATAGCCCTGGCTGTCCTGG-2392′ | -1′AACCGGACCCGCGCACCTTG-20′ |
| -1962mDYT1 | -1962′CCTGTAGCCTAGCTACCATC-1943′ | |
| -1400mDYT1 | -1400′GGCTGATCAGCCCTTTATTGAC-1379′ | |
| -912mDYT1 | -912′ACTGTAGCCCTGACTGCAC-894′ | |
| -505mDYT1 | -505′TGAGCACCACATGCACGCAG-486′ | |
| -243mDYT1 | -243′TAGGGCAGCAGTCATCATCAC-223′ | |
| -196mDYT1 | -196′CCTGTAACATCAAGGTTCAGAAGCAG-171′ | |
| -141mDYT1 | -141′GCCACTAGCGTCGGTAGACTGC-120′ | |
| -92mDYT1 | -92′CCAGCAGGAACCGGAAATAC-73′ | |
| -63mDYT1 | -63′CGGTGCGCGGGGTACGGTG-45′ | |
| -243mDYT1-S1 | -89′GCAGGAACCGTAAATACGAGTC-68′ | -68′GACTCGTATTTACGGTTCCTGC-89′ |
| -243mDYT1-S2 | -94′CGCCAGCAGGGTCCGGAAATAC-73′ | -73′GTATTTCCGGACCCTGCTGGCG-94′ |
| -243mDYT1-S1S2 | -94′CGCCAGCAGGGTCCGTAAATACGAGTCT-67′ | -67′AGACTCGTATTTACGGACCCTGCTGGCG-94′ |
| -243mDYT1+3G | -91′CAGCAGGAACCGGGGGAAATACGAG-70′ | -70′CTCGTATTTCCCCCGGTTCCTGCTG-91′ |
| -243mDYT1+3C | -91′CAGCAGGAACCCCCGGAAATACGAG-70′ | -70′CTCGTATTTCCGGGGGTTCCTGCTG-91′ |
| -243mDYT1+6 | -92′CCAGCAGGAACCGCACCCGGAAATACG-72′ | -72′CGTATTTCCGGGTGCGGTTCCTGCTGG-92′ |
| -243mDYT1+10 | -92′CCAGCAGGAACCGCACCTCCCGGAAATAC-72′ | -72′GTATTTCCGGGAGGTGCGGTTCCTGCTGG-92′ |
| -2731hDYT1 | -2674′CCAGGGCATAGCCCATAATCTGACAG-2649′ | -4′CGGACCCGCGCCACCCTGCTTG-25′ |
| -1973hDYT1 | -1976′CTGAGACTACAGGCACACGCAAC-1954′ | |
| -1446hDYT1 | -1449′CCAAGGAGGTGGAGCAAAATC-1429′ | |
| -931hDYT1 | -934′CAGGAGGCTGAGACAGGAGAATC-912′ | |
| -445hDYT1 | -448′CTCCGCCTCCTGGGTTCAAG-429′ | |
| -301hDYT1 | -304′TGACCTCAGGTGATCCGC-287′ | |
| -191hDYT1 | -193′GGGCGGAGCAGAACCGAGTTTC-173′ | |
| -121hDYT1 | -124′GTCGGGAGGAGGGCTG-109′ | |
| -80hDYT1 | -83′GAGGAGGAACCGGAAGCGTG-64′ | |
| -45hDYT1 | -48′CCGGTTCGCGGTCGGC-33′ | |
| -301hDYT1-S1 | -81′GGAGGAACCGTCAGCGTGGGTC-60′ | -60′GACCCACGCTGACGGTTCCTCC -81′ |
| -301hDYT1-S2 | -87′GCGAGAGGGGTAACCGGAAGCGTG-64′ | -64′CACGCTTCCGGTTACCCCTCTCGC-87′ |
| -301hDYT1-S1S2 | -87′GCGAGAGGGGTAACCGTCAGCGTGGGTC-60′ | -60′GACCCACGCTGACGGTTACCCCTCTCGC-87′ |
Note: numbers flanking each primer indicate binding sites on the 5′ DYT1 upstream region, relative to the ATG start codon
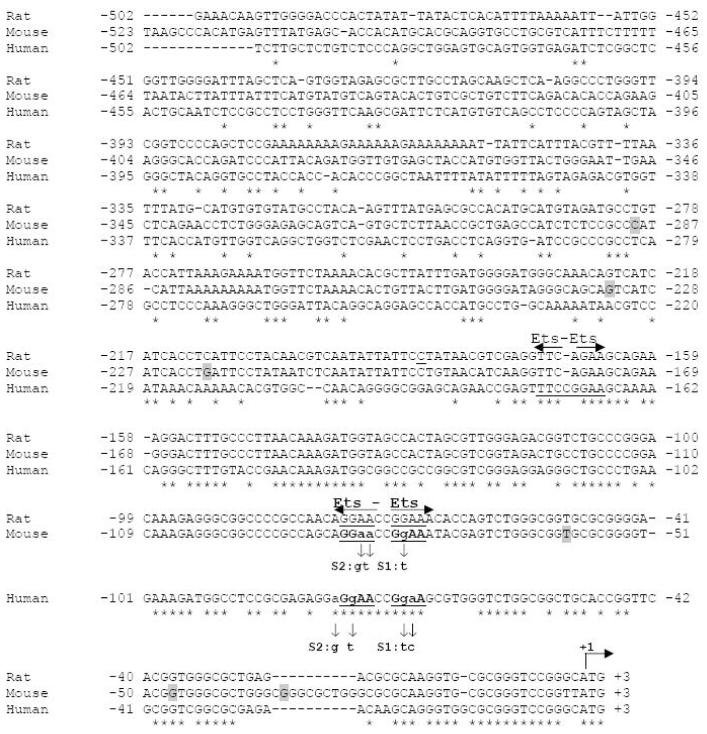
Fig. 4. Alignment of human, mouse and rat 5′ upstream DYT1 regions and Ets putative transcription binding motifs.
Potential consensus binding sites for the Ets family of transcription factors are annotated as revealed by computerized search using TRANSFAC v6.0 analysis. Underline shows the typical transcription factor recognition site and horizontal arrows point to binding orientation. Binding sites further analyzed in this study are in bold and underlined. Nucleotides are numbered from the A of the translational start site, indicated by a bent arrow as +1. asterisks (*) indicate sequence identity. Vertical arrows indicate bases altered to produce 5′ upstream DYT1 mutant versions with the substitutions annotated at the end of each arrow. Shaded nucleotides represent the transcription start sites detected by primer extension analysis.
Fig. 5. Electrophoretic mobility shift analysis of proteins interacting specifically with the mouse or human 5′ upstream DYT1 region.
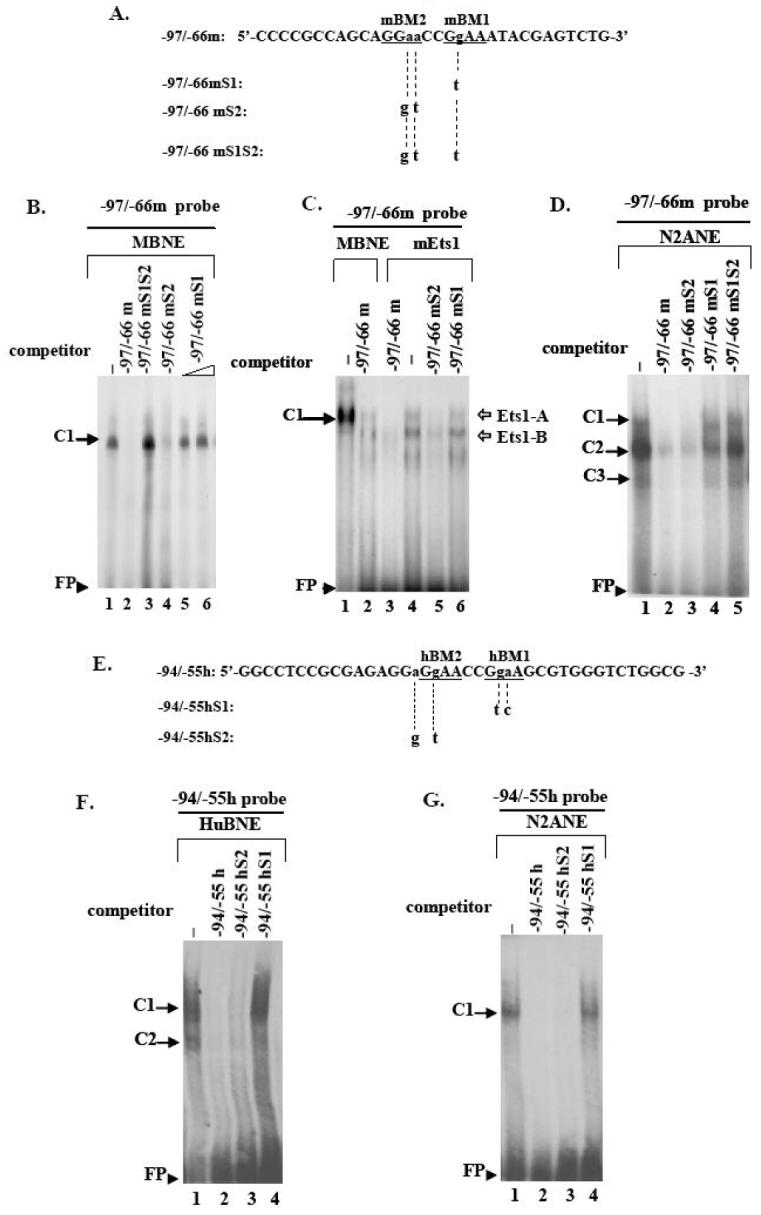
(A, E) Coding strand nucleotide sequence of mouse -97/-66 (-97/-66m, A) and human -94/-55 wildtype probes (-94/-55h, E) used in EMSAs. Underlined bases indicate examined Ets binding sites; the -86bp to -83bp and -80bp to -77bp Ets mouse cores have been designated as mEts2 and mEts1, respectively (A) while the -78bp to -75bp and -72bp to -69bp human Ets cores have been designated as hEts2 and hEts1, respectively (E). Small letters indicate bases substituted to produce the mouse mutant versions -97/-66mS1, -97/-66mS2, -97/-66mS1S2 (A) or the human mutant versions -94/-55hS1, -94/-55hS2 (E), for which only the altered bases are shown. Substitutions are also shown in Fig. 4. (A2). A retarded DNA-protein complex C1 was formed after incubation of mouse brain nuclear extract (MBNE) with the -97/-66m probe in the absence of competitor (lane 1), which was abrogated in the presence of 100-fold excess of homologous -97/-66m (lane 2) or heterologous -97/-66mS2 (lane 4) competitor. C1 was retained in the presence of 100-fold excess of -97/-66mS1S2 (lane3) or 100 to 200-fold excess of -97/-66mS1 (lanes 5, 6) competitor. (A3). Incubation of in vitro translated mouse Ets1 protein (mEts1) with the -97/-66m probe led to two retarded DNA-mEts1 complexes (Ets1-A and Ets1-B) (lane 4). Ets1-A complex exhibits the same retardation velocity as the C1 complex resulting from interaction of the -97/-66m probe with mouse nuclear extract (lane 1). Competition with 100-fold excess of either homologous (lane 3) or heterologous -97/-66mS2 (lane 5) competitor abolishes both complexes, while competition with -97/-66mS1 probe does not (lane 6). (B2). Human brain nuclear extract (HuBNE) incubated with the -94/-55h probe in absence of competitor gave rise to two DNA-protein complexes C1, C2 (lane 2), both of which were abolished in the presence of 100-fold excess of homologous -94/-55h (lane 1), or heterologous -94/-55hS2 (lane 3) competitor, while they were retained intact when competed with 100 fold excess of -94/-55hS1 probe (lane 4). DNA-nuclear extract protein retarded complexes are indicated by black arrows while DNA-mEts1 protein complexes by white arrows. Free probe is indicated by arrowheads. Negative controls in the absence of nuclear extract or mEts1 protein did not produce any specific binding (data not shown).
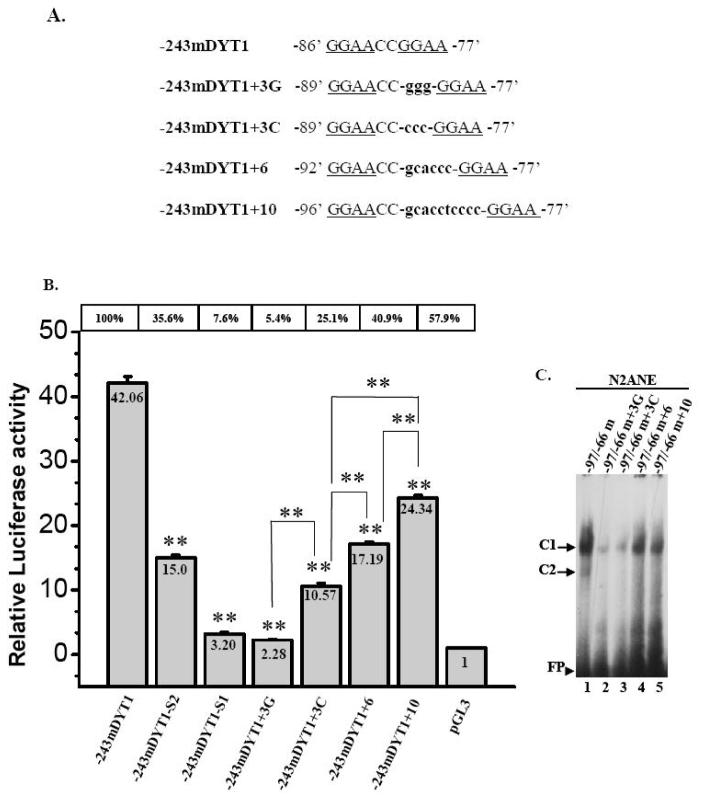
Fig. 8. Effects of increasing the distance between the -86/-83mEts2 and -80/-77mEts1 cores present within the mouse 5′ upstream DYT1 region.
(A). Three, six or ten nucleotides were inserted at -81 position, between the -86/-83 and -80/-77 Ets elements of the -243mDYT1 DNA/luciferase construct giving rise to mutant constructs -243mDYT1+3C or +3G, -243mDYT1+6 and -243mDYT1+10 (B). HEK293 and N2A cells were transiently transfected with 400ng of either -243mDYT1, -243mDYT1-S1, -243mDYT1-S2, -243mDYT1+3C, -243mDYT1-3G, -243mDYT1+6, -243mDYT1+10 or pGL3 vector and 250ng pCMV-β gal. Each experiment was done in four repeats; a representative experiment out of three is shown. Bars show means ±S.E.M. of normalized luciferase activity (based on pCMV-β-gal co-transfection) over pGL3 vector. The per cent activity of each construct over the parental construct is shown above each case. Statistically significant inter-individual differences are also indicated. **= p<0.005.
In silico analysis
Computerized search of an ∼500bp-5′-upstream DYT1 region of mouse, rat and human gene was performed using TRANSFAC v6.0 library of transcription factor binding sites (URL: http://www.gene-regulation.com). To detect evolutionarily conserved elements, we aligned the proximal 5′ upstream DYT1 region of human (DYT1; GenBank® accession no. NC_000009: 131626000..131630000, complement), mouse (dyt1; GenBank® accession no. NC_000068: 30823300..30827300, complement) and rat (dyt1; GenBank® accession no. NC_005102: 10033000..10037125, complement) genes, using ClustalW (URL: http://www.ebi.ac.uk/Tools/clustalw/).
Electrophoretic mobility-shift assays (EMSAs)
Nuclear extracts were prepared from cortex human brain (Brain and tissue bank, University of Maryland, Baltimore, MD), whole mouse brain, N2A or HEK293 cell lines, using a nuclear extraction kit (PIERCE, Rockford, IL). Protein concentrations were determined by a BCA protein Assay kit (PIERCE, Rockford, IL). Mouse Ets1 protein (mEts1) was produced in vitro, using a pCDNA3-mEts1 expression vector (Mattot et al. 2000) and a Quick coupled transcription/translation kit (Promega, Madison, WI). Complementary single-stranded deoxyoligonucleotides (Integrated DNA Technologies, Coralville, IA), covering the 5′ upstream DYT1 regions of interest (Fig. 5A and E), were annealed in 10mM Tris HCl, 100mM NaCl, 1mM EDTA buffer, by heating at 80 °C for 10 min and cooling down to RT, to produce double-strand probes. Double-strand probes were then [γ-32P]-5′end labeled (PerkinElmer Life Sciences, Waltham, MA) using T4 polynucleotide kinase (Invitrogen, Carlsbad, CA). The unincorporated isotope was removed using a Quick-spin G25 column. Nuclear extracts (6 to 10 μg of protein) or in vitro translated Ets1 protein were incubated in presence of 20-50 fmol of [γ-32P]-5′-labeled double-strand probes (∼300.000 cpms), in a 20 μl binding buffer (10mM HEPES, pH 7.9, 150mM NaCl, 1mM EDTA, 0.5 mM dithiothreitol, 10% glycerol and protease inhibitor mixture) plus 2μg poly(dI:dC) (Roche Diagnostics, Indianapolis, IN), for 20min at RT. Competition assays were performed by addition of 100-fold excess (unless otherwise specified) of unlabelled wildtype (homologous) or mutant (heterologous) probes. Negative controls were included for which no nuclear extract was added in the binding reaction. Gels were then dried and visualized by autoradiography film.
Transfections and dual luciferase assays
Transient transfections were performed using lipofectamine, according to manufacturer’s instruction (Invitrogen, Carlsbad, CA) in 1.5 to 2 × 106 cells plated onto 12-well tissue culture plates. For each construct used in transfections, at least two independent plasmid Midipreps (Qiagen, Valencia, CA) were prepared and tested. Each transfection comprised of 0.4μg of DNA/luciferase reporter construct and 0.25 μg of pCMV-βgal vector (Stratagene, La Jolla, CA) bearing the β-galactosidase gene under the cytomegalovirus promoter, as an internal control of transfection efficiency. Promoter-less pGL3-BV served as negative control. The mEts1-DB, a mouse Ets1 dominant-negative lacking its transactivation domain (Mattot et al. 2000), was used in co-transfections with DNA/luciferase constructs at 1:2 ratio. Total amount of transfected DNA was maintained the same in all cases. After incubation for 3hrs (HEK293, HepG2 cells) or 6hrs (N2A, SH-SY5Y cells and primary astrocytes), transfection medium was replaced with culture medium. Luciferase activity was analyzed 40-48hrs post transfection, using a luciferase based reporter activity kit (Promega, Madison, WI) and an LKB-Wallac 1251 luminometer (Turku, Finland). Results were expressed as fold increase of luciferase activity over pGL3-BV normalized with β-galactosidase values, unless otherwise stated. Each experiment was repeated at least four times with each construct transfected four to six repeats. Statistical significance was determined by Student’s t-test.
Immunocytochemistry
HEK293, HepG2, N2A, SH-SY5Y cell lines and cultured human primary astrocytes were plated on coverslips pre-treated with poly-L-lysine. The 5′ upstream DYT1/torsinA expression construct -2731hDYT1-hwtTA (Fig. 2A), the pCMV-hwtTA or the -2731mDYT1 lacking torsinA cDNA (Fig. 1A) were transfected using lipofectamine as described above. Cells were fixed with 4% paraformaldehyde in PBS 40hrs post-transfection. Immuno-fluorescence was performed using a polyclonal antibody against human torsinA (1:10.000), kindly provided by Dr P. Gonzalez-Alegre (University of Iowa) and visualized by Alexa598 (Molecular probes, Eugene, Oregon). In primary cultured astrocytes, co-labelling was performed using the same polyclonal torsinA antibody, visualized by Alexa598 and a monoclonal antibody for GFAP (1:500, Millipore,), visualized by Alexa488. Stained cells were observed using fluorescence microscopy. Nuclei were stained with bis-benzimide (Sigma Aldrich, St Louis, MO).
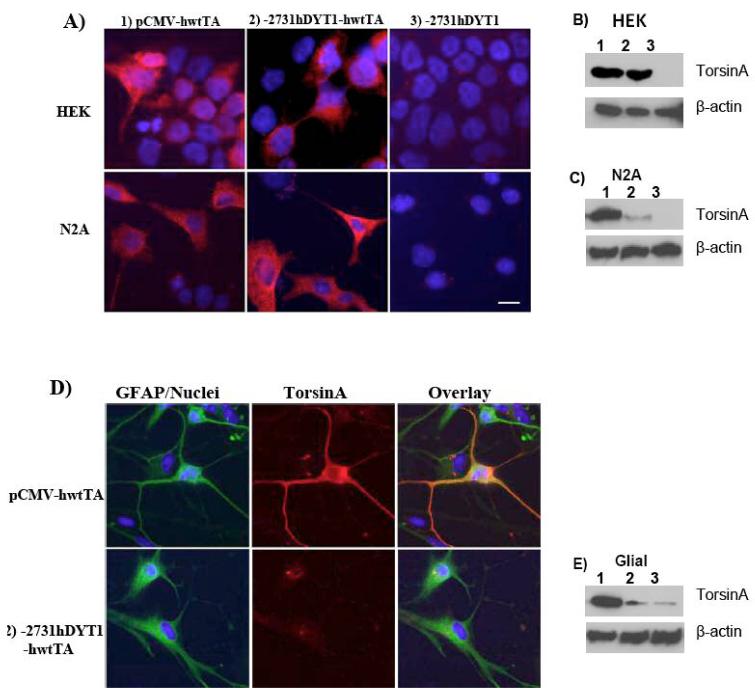
Fig. 2. The human -2731bp - 5′ upstream DYT1 fragment directs expression of torsinA in a neuronal but not in a non-neuronal environment.
(A) Immuno-fluorescence using a polyclonal antibody against human torsinA (red) in HEK293 or N2A cell lines, transiently transfected with pCMV-hwtTA (I), -2731hDYT1-hwtTA (II) or -2731hDYT1 construct (III). (B) Immuno-fluorescence using a polyclonal antibody against human torsinA (red) and a monoclonal against GFAP (green) in cultured primary human astrocytes transfected with pCMV-hwtTA (I), -2731hDYT1-hwtTA (II) or -2731hDYT1 construct (III). Overlay shows that torsinA was not expressed in glial cells (GFAP positive) when driven by the -2731- 5′ upstream DYT1 fragment (II) while its expression was achieved under a cytomegalovirus promoter (I). Nuclei were stained with bis-benzimide. Bar indicates 20μm.
Fig. 1. A -2731bp- and -2416bp-5′ upstream fragment of the human and mouse DYT1 gene, respectively, drive expression of a reporter gene in a cell type specific manner.
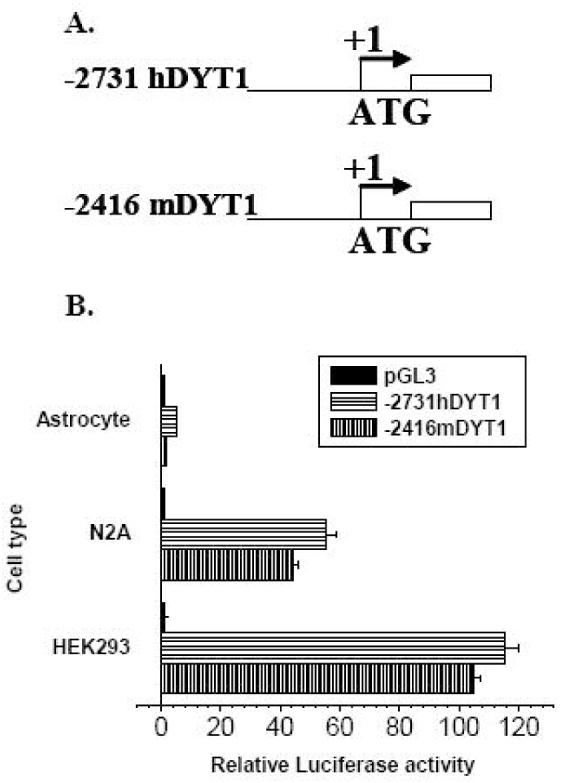
(A) Schematic representation of the 5′ upstream DYT1/luciferase constructs for human (-2731hDYT1) and mouse (-2416mDYT1) DYT1 gene. The constructs were named based on the length between the 5′ and the 3′ boundary which corresponds to the first (mouse) or the fourth (human) nucleotide upstream of the ATG start codon, designated as +1 (bent arrow). The white rectangle represents the luciferase gene. (B) Both mouse and human 5′ upstream sequences exhibit over 100- or 40-fold induction of luciferase activity in HEK293 and N2A cell lines, respectively. The experiment was repeated at least three times. Results shown are means of luciferase activity over promoterless pGL3-BV, normalized to pCMV-βgal ± S.E.M. of a representative experiment, where each transfection was done in triplicates. mDYT1=mouse DYT1 gene, hDYT1=human DYT1 gene.
Western Blotting
HEK293, HepG2, N2A, SH-SY5Y cell lines and primary human astrocytes were transfected in 6-well plates, with 1μg of pCMV-hwtTA, -2731hDYT1-hwtTA or -2731hDYT1 construct. Cell homogenates containing 60-80 μg of protein were resolved on SDS—PAGE and electroblotted onto a PVDF membrane. The blot was incubated with polyclonal antibody to torsinA or a monoclonal to β-actin (1: 1000, sc-1616) (Santa Cruz, California) for 1 h. Detection was performed using an ECL western blot chemiluminescence kit (Amersham).
RESULTS
Primer extension analysis
Primer extension analysis demonstrated more than one extension products, corresponding to -35, -48, -61, -220, -233 and -289 in regards to the ATG translation start codon (Fig. 4 and Suppl. Fig. 1). The same pattern was observed using either liver or brain mouse total RNA.
Activity of cloned 5′ upstream regions of mouse and human DYT1 gene
The -2416mDYT1 and -2731hDYT1 reporter constructs, bearing a -2416bp- and a -2731bp-5′ upstream fragment of the mouse and human DYT1 gene, respectively (Fig.1A), were tested for their ability to direct luciferase expression in transient transfection assays in HEK293, HepG2, N2A, SH-SY5Y cell lines and cultured human primary astrocytes. Both constructs exhibited over 100- (HEK293) or 40-fold (N2A) induction of luciferase activity, over the pGL3-BV which only gave rise to basal levels of activity (Fig. 1B). Similar results were obtained using HepG2 or SH-SY5Y cell lines (data not shown). Luciferase induction in human primary astrocytes was very low (Fig. 1B) while β-galactosidase values were similar as in N2A cell lines (data not shown), indicating successful transfection of primary astrocytes.
Wildtype human torsinA was homogenously expressed in the cytoplasm of HEK293 or N2A cells when driven by either the -2731bp-5′ upstream human DYT1 fragment (Fig. 2A, II) or by the CMV promoter (positive control) (Fig. 2A, I). No signal for torsinA was detected when transfected with the -2731hDYT1 construct which lacks torsinA cDNA (negative control) (Fig. 2A, III). Western Blot confirmed expression of torsinA under the -2731bp DYT1 promoter in both cell lines (Fig. 2B and 2C, lane 2) but in the case of neuronal N2A, it was less potent than the pCMV promoter (Fig. 2C, lane 2). No signal was detected by western blot in HEK293 or N2A cells transfected with the -2731hDYT1 negative control (Fig. 2B, and 2C, lane 3). In the case of human primary astrocytes (GFAP positive cells, green), torsinA (red) was only expressed under the CMV promoter (Fig. 2D, upper lane) but not under the -2731bp-5′ upstream fragment (Fig. 2D middle lane). Western Blot analysis in transfected astrocytes confirmed successful expression of torsinA under the CMV promoter (Fig. 2E, lane 1) while considerably lower signal was observed from cells transfected with either the -2731hDYT1-hwtTA or the -2731hDYT1 lacking torsinA cDNA (Fig. 2E, lane 2 and 3 respectively). Results obtained with HepG2 cells were similar to HEK293 cells, while for the SH-SY5Y cells similar to N2A cells (data not shown).
Identification of the proximal minimal promoter of mouse and human DYT1 gene
To identify the proximal 5′ upstream DYT1 fragment necessary for minimal promoter activity, we took advantage of 5′ upstream DYT1 unidirectional truncated reporter constructs. From -2416mDYT1 to -141mDYT1 constructs, containing 2416-, 1962-, 1400-, 912-, 505-, 243-, 196- and 141-bp of the mouse 5′ upstream DYT1 region, an augmentation in luciferase activity was observed which was more intense in HEK293 than in N2A cells (Fig. 3A). An abrupt drop in activity was detected from -141mDYT1 to -92mDYT1, bearing a -92bp-5′ upstream DYT1 fragment. Luciferase activity reached background levels using the -63mDYT1 construct (Fig. 3A), bearing just 63bp of the 5′ upstream DYT1 region. Very low activity was conferred by any of the reporter constructs in cultured astrocytes with the exception of the -243mDYT1 and -196mDYT1 exhibiting ∼3.3- and 2.5-fold induction, respectively (Fig. 3A, insert). Human DYT1 reporter constructs -2731hDYT1 through -931hDYT1, bearing 2731-, 1973-, 1446- and 931-bp fragments of the human 5′ upstream DYT1 region, gave rise to gradually increased luciferase activity until there was a slight drop in the -445hDYT1 construct, followed again by an increase in the -301hDYT1 or -191hDYT1 constructs (Fig. 3B). When the human 5′ upstream DYT1 region was truncated from 191bp to 121bp (-121hDYT1 construct) there was a dramatic decrease in luciferase activity, which reached background levels in the -45hDYT1 construct, bearing just 45bp of the 5′ upstream human DYT1 region (Fig. 3B). Very low activity was observed by most of the truncated constructs in astrocytes, with the exception of -301hDYT1 and -191hDYT1 constructs, showing an ∼8- and ∼6.5- fold luciferase induction, respectively (Fig. 3B, insert).
Fig. 3. Promoter activity of 5′ upstream serial deletion/luciferase constructs for the mouse (A) and human (B) DYT1 gene.
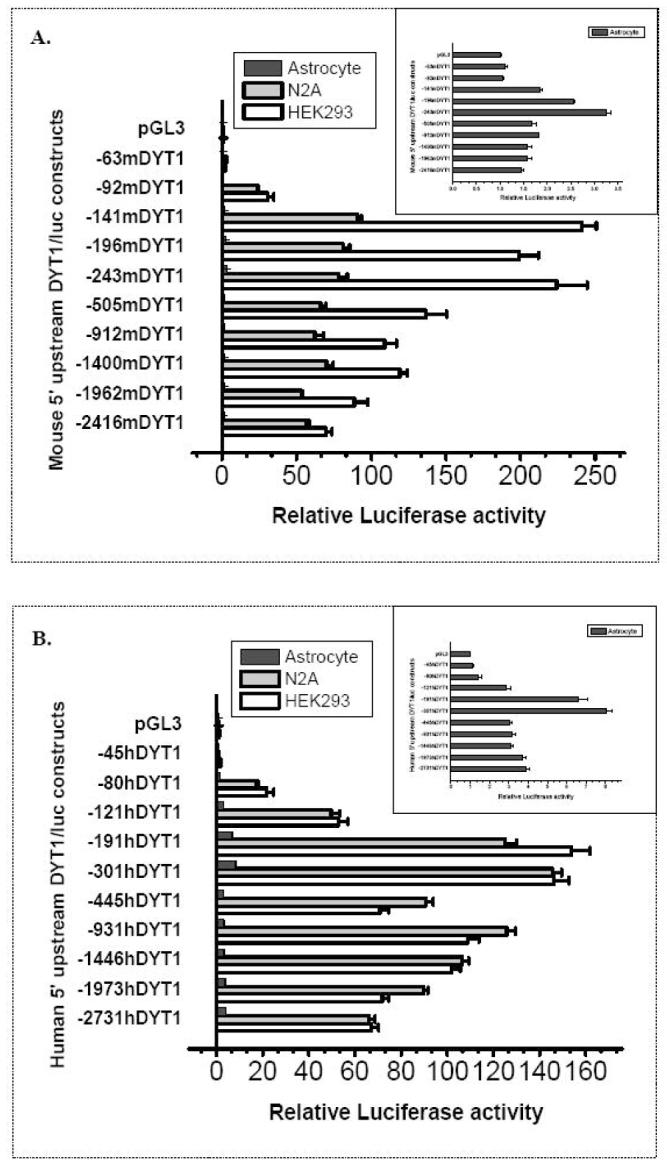
5′-upstream DYT1/luciferase serial deletion constructs for the mouse (A) or human (B) gene, were transiently transfected in HEK293, N2A cell lines and in primary human astrocytes. Constructs were named based on the length between their 5′ boundary and 3′ boundary which corresponds to the first (mouse) or the fourth (human) nucleotide upstream of the ATG start codon. Each experiment was done in triplicates and repeated at least 4 times; a representative experiment is shown. Bars show means ±S.E.M. of normalized luciferase activity over pGL3-BV and normalized with β-galactosidase values. The 5′ upstream DYT1 region necessary for minimal promoter activity is laying within -141bp or -191bp with regards to the ATG start codon of the mouse and human DYT1 gene, respectively. Note that in astrocytes, luciferase activity of reporter constructs is closer to background levels as more clearly exhibited by insert graphs. mDYT1=mouse DYT1 gene, hDYT1=human DYT1 gene.
In silico analysis
TRANSFAC v6.0 library of binding sites revealed absence of a canonical TATA or CAAT box, corresponding to the consensus sequence TATA(A/T)A(A/T) or GG(T/C)CAATCT (Singer et al. 1990) within an ∼500-bp 5′ upstream fragment of the mouse or human DYT1 gene (Fig. 4). TRANSFACT analysis combined with alignment of the 5′ upstream DYT1 regions for mouse, rat and human, demonstrated evolutionarily conserved Ets-like motifs for all three species within their -220bp 5′ upstream region (Fig. 4). In particular, we identified a consensus direct repeat of two Ets cores, interrupted by a di-nucleotide (5′-GGAACCGGAA-3′), spanning -78bp to -69bp of human, -86bp to -77bp of mouse and -76bp to -67bp of rat DYT1 gene. An additional palindrome of two Ets cores (5′-TTCCGGAA -3′) was identified between -175bp to -168bp of the human DYT1 gene.
Determining the binding pattern of proteins to the putative 5′ upstream DYT1 Ets-like motifs
EMSA studies determined the binding pattern of proteins within the -97bp to -66bp 5′upstream mouse DYT1 region (-97/-66m probe, Fig. 5A). This region contains the -86bp to -83bp and -80bp to -77bp Ets Binding Motifs of the direct repeat (-86′GGAA’-83′CC-80′GGAA-77′, Fig. 4 and 5) designated as mBM2-and mBM1, respectively (Fig. 5A). Interaction of the -97/-66m probe with mouse brain nuclear extract gave rise to a single retarded DNA-protein complex, designated as C1 (Fig. 5B, lane 1). The C1 complex was abolished in the presence of excess homologous competitor (Fig. 5B, lane 2) but not the heterologous -97/-66mS1S2 (Fig. 5B, lane 3) having both mBM2 and mBM1 cores mutated (Fig. 4 and 5A). Competition with excess of -97/-66mS2, bearing mutations in the mBM2 core (Fig. 4 and 5A), was also sufficient to abolish the C1 complex (Fig. 5B, lane 4). The -97/-66mS1 probe, bearing just a single mutation at -79bp of the mBM1 core (Fig. 4 and 5A), did not abrogate the C1 complex when used as a competitor even at 200-fold excess (Fig. 5B, lanes 5, 6). Similar results were observed using N2A nuclear cell extracts (Fig. 5D) but in this case the -97/-66mS1 probe partially abrogated the main complex (C2) (Fig. 5D, lane 4). Nuclear cell extracts from HEK cells gave similar results (data not shown).
We further tested binding of an in vitro transcribed/translated mouse Ets1 protein to the -97/-66m probe (Fig. 5A) which resulted in two retarded DNA-Ets1 protein complexes, Ets1-A and Ets1-B (Fig. 5C, lane 4). The Ets1-A exhibited slower mobility than Ets1-B, but similar to the C1 complex resulting from mouse nuclear extracts interacting with the -97/-66m probe (Fig. 5C, lane 1). Both Ets1-A and -B complexes were abolished in the presence of excess homologous -97/-66m (Fig. 5C, lane 3) or heterologous competitor -97/-66mS2 (Fig. 5C, lane 5) bearing mutations in mBM2 core (Fig. 4 and 5A). However, both Ets1-A and -B complexes were retained in the presence of excess -97/-66mS1 (Fig. 5C, lane 6) bearing a single mutation at -79 position of the mBM1 core (Fig. 4 and 5A). The above observations strongly suggest that the -80/-77mBM1 site (Fig. 4 and 5A) is the one mainly involved in DNA-protein interactions.
We next incubated human brain nuclear extract with the -94/-55h probe, bearing within the -78bp to -75bp and -72bp to -69bp Ets cores of the direct repeat (-78‘GGAA’-75′CC-72′GGAA’-69, Fig. 4) designated as hBM2 and hBM1, respectively (Fig. 5E). Autoradiogram revealed two retarded complexes C1, C2 (Fig. 5F, lane 1), both of which were abolished in the presence of excess homologous -94/-55h competitor (Fig. 5F, lane 2) or heterologous -94/-55hS2 competitor (Fig. 5F, lane 3) bearing mutations only in the hBM2 core (Fig. 4 and 5E). However, the C1 complex was retained when competed with excess of -94/-55hS1 (Fig. 5F, lane 4) bearing mutations within the hBM1 core (Fig. 4 and 5E). When N2A nuclear extract was used (Fig. 5G), a single complex was formed (C1) which was completely abolished by competition with excess of -94/-55h (Fig. 5G, lane 2) or heterologous -94/-55hS2 competitor (Fig. 5G, lane 3) while it was retained when competed with -94/-55hS1 competitor (Fig. 5G, lane 4). Similar results were obtained using HEK nuclear extract (data not shown). These results indicate that the -72/-69hBM1 core (Fig. 5E), - which has complete homology with the -80/-77mBM1 core (Fig. 4) -, is the most critical one for DNA/protein interactions.
Biological importance of the Ets-elements for proximal 5′ upstream DYT1 activity
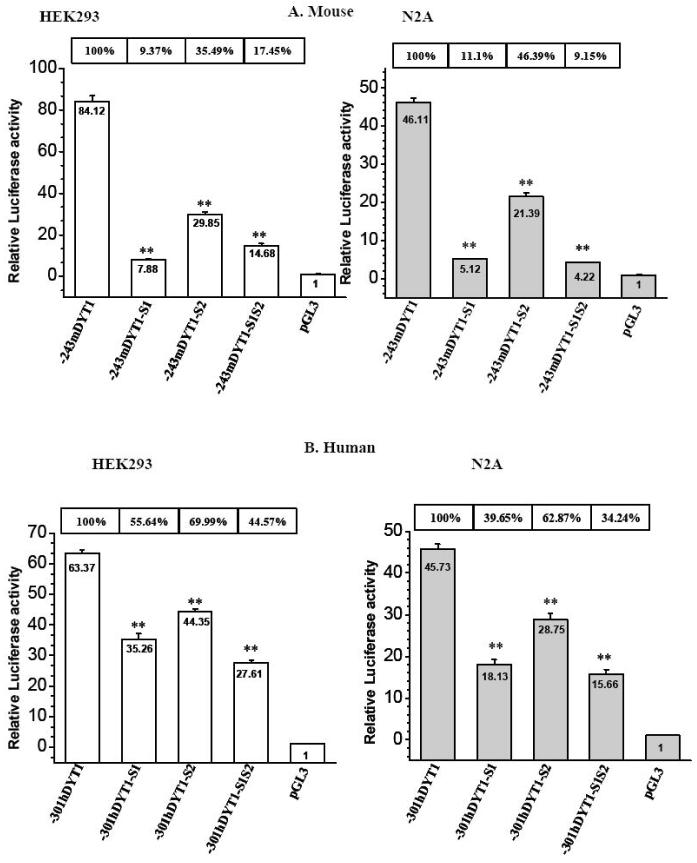
In order to determine if the mBM and hBM cores are critical elements for DYT1 transcriptional activity, we mutated these sites within the -243mDYT1 and -301hDYT1 constructs and assayed their activity in HEK293 and N2A cell lines. Introduced mutations are annotated in Fig. 4 (vertical arrows) and are identical to those introduced in mutant probes used for EMSAs (Fig. 5A and E). Mutant construct -243mDYT1-S1, bearing a single mutation at -79bp of the -80/-77mBM1 core (Fig. 4 and 5A), exhibited markedly reduced activity of 9.37% in HEK293 and 11.1% in N2A cells, compared to the parental -243mDYT1 construct (Fig. 6A, p< 0.005). Mutant construct -243mDYT1-S2, bearing mutations at -83bp and -84bp of the mBM2 core (Fig. 4 and 5A) also showed a lower degree reduced luciferase activity, of 35.49% in HEK293 and 46.39% in N2A cells (Fig. 6A, p< 0.005). The -243mDYT1-S1S2 construct, containing mutations in both mBM1 and mBM2 cores (Fig. 4 and 5A) also showed reduction in luciferase activity (17.45% in HEK293 and 9.15% in N2A cells) as the -243mDYT1-S1 (Fig. 6A, p< 0.005). The biological relevance of the hBM1 and hBM2 cores was subsequently examined with the -301hDYT1-S1 construct, bearing mutations at -70bp and -71bp of the -72/-69hBM1 core (Fig. 4 and 5E) exhibiting reduced luciferase expression of 55.64% in HEK293 and 39.65% in N2A transiently transfected cells, compared to the parental -301hDYT1 construct (Fig. 6B, p< 0.005). The -301hDYT1-S2 construct, having mutations at -77bp and -79bp of the -78/-75 hBM2, also exhibited reduced activity of 69.99% in HEK293 and 62.87% in N2A cells (Fig. 6B, p< 0.01 and p<0.005) while the -301hDYT1-S1S2 construct, having eliminated both hBM1 and hBM2 cores, of 44.57% in HEK293 and 34.24% in N2A cells (Fig. 6B, p < 0.005).
Fig. 6. Effects of mutating putative Ets1 and 2 binding sites on 5′ upstream DYT1 driven gene luciferase expression.
(A) HEK293 and N2A cells were transiently transfected with 400ng of either -243mDYT1, -243mDYT1-S1, -243mDYT1-S2, -243mDYT1-S1S2 or pGL3-BV and 250ng pCMV-β gal. (B) HEK293 and N2A cells were transiently transfected with 400ng of either -301hDYT1, -301hDYT1-S1, -301hDYT1-S2, -301hDYT1-S1S2 or pGL3-BV and 250ng pCMV-β gal. Each experiment was done in four repeats and repeated at least 3 times; a representative experiment is shown. Bars show means ±S.E.M. of relative luciferase activity over pGL3-BV, normalized with β-galactosidase values. The percent activity of each construct compared to the parental ones (-241mDYT1 or -301hDYT1) is shown above each bar. ** = p<0.005.
We finally examined the response of the -243mDYT1 construct to an ectopically expressed mouse Ets1 dominant-negative (mEts1-DB) lacking its trans-activation domain. Co-expression of -243mDYT1 and mEts1-DB constructs resulted in 54% and 50% reduction of luciferase activity in HEK293 and N2A cells, respectively (Fig. 7A, p<0.005). The mEts1-DB also evoked a significant drop of luciferase expression exhibited by the -243mDYT1-S2 construct, bearing mutations in the mBM2 core (34% in HEK293 and 33% in N2A, Fig. 7A, p<0.005). A slighter Ets-DB-dependent decrease was also observed on the -243mDYT1-S1 construct (12% in HEK293 and 14% in N2A, Fig. 7A, p<0.005 and 0.05, respectively) in which the mBM1 has been mutated, while no Ets-DB effect was observed on the -243mDYT1-S1S2 construct bearing mutations on both mBM1 and mBM2 cores (Fig. 4 and 5A). Likewise, the mEts1-DB construct evoked 49% (HEK293) and a 54% (N2A) reduction of luciferase expression exhibited by the -301hDYT1 construct, as well as a 29% (HEK293) and 38% (N2A) reduction of the -301hDYT1-S2 activity (Fig. 7B, p < 0.005). A slighter effect of the Ets1-DB was observed on the -301hDYT1 (11% in HEK293 and 17% in N2A cells), while it had no effect on the -301hDYT1-S1S2 construct (Fig. 7B, p < 0.005).
Fig. 7. Response of mouse and human 5′ upstream DYT1 regions to an ectopically expressed dominant negative for the Ets-family of transcription factors.
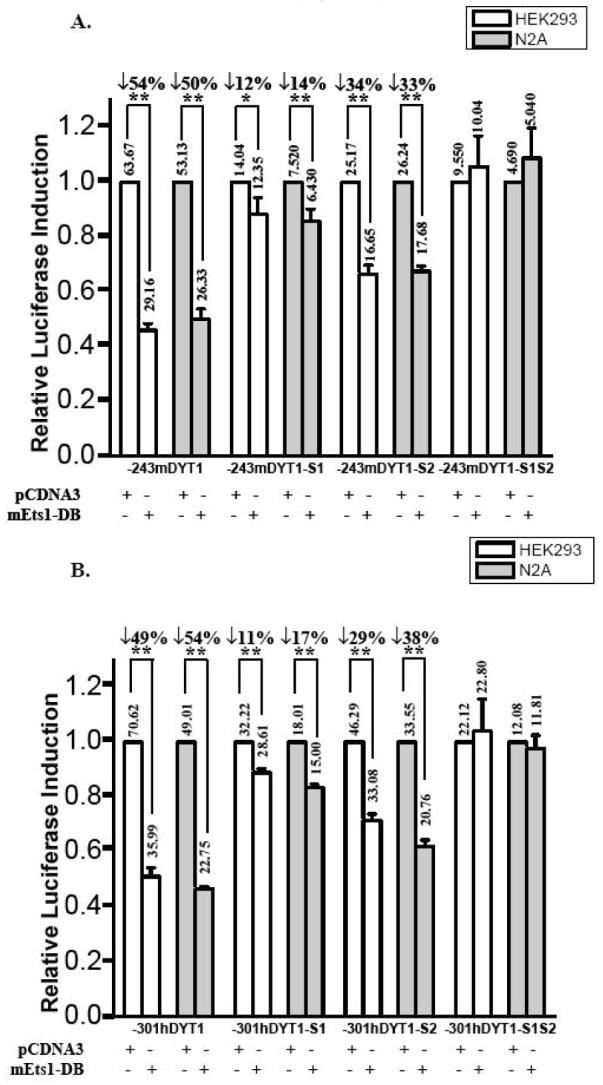
HEK293 and N2A cell lines were transiently co-transfected with 350ng mEts1-DB and (A) 350ng of either -243mDYT1, -243mDYT1-S1, -243mDYT1-S2 or -243mDYT1-S1S2 or (B) 350ng of either -301mDYT1, -301hDYT1-S1, -301hDYT1-S2 or -301hDYT1-S1S2. To maintain the same amount of transfected DNA and avoid squelching artifacts in control wells the absence of mEts1-DB was complemented by using 350ng of plain pCDNA3.1 vector. Transfection efficiency was normalized using pCMV-β-gal. Results are presented as histograms indicating the induction by mEts1-DB (in fold) with respect to the activity of each 5′ upstream DYT1/luciferase reporter construct in the absence of mEts1-DB, which was assigned a value of 1. Means ± S.E.M. are shown. Numbers above each bar are actual values of normalized luciferase activity. Vertical arrows indicate reduction in activity and numbers next to them express the percent difference between pcDNA3.1 and mEts1-DB co-transfections. A representative experiment of four independent transfections with at least two different DNA preparations is shown. ** = p<0.005, * = p<0.05, compared to parental -241mDYT1 or -301hDYT1 construct.
Effects of increasing the distance between the mBM1 and mBM2 cores of the -86/-77Ets direct repeat
To examine the effects of increasing the distance between the two Ets cores of the direct repeat, we introduced up to ten bases between the -86/-83mBM2 and -80/-77mBM1 cores, at position -81 of the -243mDYT1 construct (Fig. 8A). Luciferase assays following transfection in N2A cells, showed that increasing the distance between the mBM1 and mBM2 sites by 3C’s or 3G’s (Fig. 8A) decreased transcriptional activity to 25.1% and 5.4%, respectively compared to the parental -243mDYT1 (Fig. 8B). Interestingly, adding six extra nucleotides between the mBM1 and 2 cores resulted in 40.9% activity which is still lower than the parental construct but higher than the activity exhibited by the 243mDYT1+3C construct. Finally, introducing 10 nucleotides, restored even further promoter activity, reaching 57.9% compared to the parental construct (Fig. 8B, p < 0.005). We also examined the effects of increasing the mBM1-mBM2 cores on their binding properties. We compared binding of N2A nuclear extracts on the -97/-66m wildtype primer or on primers bearing plus 3G’s (-97/-66m+3G), 3C’s (-97/-66m+3C), 6 (-97/-66m+6) or 10 (-97/-66m+10) bases between the mBM1 and mBM2 cores. In all cases, wildtype primer formed a main complex (C1) of higher intensity compared to the other probes (Fig. 8C). Probes bearing three additional bases (Fig. 8C, lanes 2, 3) exhibited weaker binding than the ones bearing six additional bases (Fig. 8C, lanes 4, 5). A second C2 complex, present in wildtype primer (lane 1) was not present in any of the other primers (lanes 2 - 5).
DISCUSSION
We identified functional proximal 5′ upstream DYT1 DNA fragments and demonstrated the Ets family of proteins as critical DYT1 transcriptional regulators. A GGAA core located at -80/-77 and -72/-69 of the mouse and human DYT1 gene, respectively, was proven critical for DNA-protein interactions; a point mutation at -79bp of the -80/-77mBM1 core or a double mutation at -71bp and -72bp of the -72/-69 hBM1 core almost abolished protein binding, accompanied by decreased 5′ upstream DYT1 driven reporter gene expression. An ectopically expressed Ets dominant negative had an Ets-box dependent inhibitory effect on the proximal 5′ upstream DYT1 fragment.
Primer extension analysis demonstrated multiple transcription start sites which is more commonly observed in TATA-less promoters (Swick et al. 1989) such as the DYT1 gene (see In silico analysis). Search of the NCBI EST database revealed multiply potential transcription start sites for the mouse DYT1 gene, with the majority of them at -47bp and -48bp (http://dbtss_old.hgc.jp), which is actually the more intense band observed in our primer extension analysis (Suppl., Fig. 1). Thus, the Ets elements examined in this study are probably located upstream of the 5′ untranslated region.
In neuronal (N2A, SH-SY5Y) and peripheral (HEK293, HepG2) cell lines we achieved very strong induction of reporter gene by a -2416 (mouse) or a -2731 (human) 5′ upstream DYT1 fragment, indicating the presence of a functional promoter within these fragments. Very low levels of reporter gene activity was observed in astrocytes, supporting that the above fragments also contain glial-specific negative regulatory elements. Immunohistochemistry, using an antibody which immunoreacts with only the human torsinA, confirmed promoter function of the -2731fragment and its tissue specificity since no signal was detected in transfected astrocytes. Western Blot analysis further demonstrated that the -2731bp DYT1 promoter is almost as strong as the cytomegalovirus promoter in HEK293 cells, while it is considerably less powerful in N2A cells. This is in agreement with high levels of endogenous torsinA in peripheral tissues compared to neuronal (Shashidharan et al. 2000). Surprisingly, western blot detected low expression of torsinA under the -2731bp DYT1 promoter in transfected astrocytes, which was however, almost equal to the signal observed in the negative control. We reasoned that the primary astrocyte culture might contain low number of other cell types, like fibroblasts or neurons. Indeed, more detailed examination of the transfected astrocytes, co-labeled with GFAP and torsinA, revealed few GFAP-negative cells expressing human torsinA (suppl. Fig. 2). These GFAP-negative cells, could also account for the very low -2731hDYT1-driven-luciferase activity detected in astrocytes.
By serial truncation of the mouse and human 5′ upstream DYT1 fragments, we identified a proximal minimal promoter within -141bp for mouse and -191bp for human, with respect to the DYT1 translational start site. The minimal promoter was able to drive high levels of expression of reporter gene in cultured N2A and HEK293 cell lines. Very low activity was observed in astrocytes by most of the 5′ upstream serial deletion constructs, with the exception of the -243mDYT1, -196mDYT1, -301hDYT1 and -191hDYT1 constructs, exhibiting a slightly increased reporter gene activity. Further analysis is required to test if this is due to loss of glial specific negative regulatory element(s).
Alignment of human, mouse and rat 5′ upstream DYT1 sequences revealed a highly conserved direct repeat of two putative Ets cores, interrupted by a di-nucleotide at -86′GGAA-83′ CC -80′ GGAA -77′ for the mouse and -78′ GGAA-75′ CC -72′ GGAA -69′ for the human DYT1 gene. The -80/-77mBM1 and -72/-69hBM1 cores specifically interacted with brain derived nuclear extracts or mouse Ets1 protein in binding assays. On the contrary, the -86/-83mBM2 and -78/-75hBM2 cores appeared less critical for DNA-protein interaction since probes bearing mutations within the above cores were still efficient competitors in EMSA studies (Fig. 5).
When an in vitro translated mouse Ets1 protein was used, two retarded DNA-mEts1 protein complexes were formed, both specifically interacting with the -80/-77mBM1 core. This complex migrated with mobility similar to the C1 complex formed when using mouse nuclear extracts (Fig. 5A2 and A3); thus the C1 protein-DNA complex could reflect either multimeric forms of endogenous Ets1 factor bound to the -80/-77mEts1 core or the involvement of a higher molecular weight Ets factor.
Using transient transfection assays we showed that mutations within the mBM1 and hBM1 cores, had a concomitant dramatic decrease in DYT1 driven gene reporter expression (Fig. 6). Mutations eliminating the mBM2 and hBM2 cores, shown not very critical in EMSAs, also resulted in reduction of reporter gene expression, although of a lower magnitude compared to the mBM1 and hBM1-dependent decreases (Fig. 6). This could be interpreted as a result of changing the surrounding sequences of the mBM1 and hBM1 cores, since the mBM2- and hBM2-mutations are just 3- and 5-bp away from the mBM1 and hBM1 cores, respectively (Fig. 4). On the other hand, the mutated mBM2 and hBM2 cores might slightly reduce the binding capacity of the wildtype sequence, but still be efficient competitors when used in high excess. To test that, we labeled the primers bearing mutations on mBM1 (-97/-66mDYT1S1), on mBM2 (-97/-66mDYT1S2), or both in mBM1 and 2 cores (-97/-66mDYT1S1S2), and compared their binding capacity (instead of their competition ability) to the wildtype primer. Indeed, the mutated BM2 core slightly affected the binding, the mutated mBM1 core retained minor binding capacity, while combination of the mBM1 and mBM2 within mutations completely abrogated binding (Suppl. Fig. 3).
Ets proteins form homo- and hetero-oligomers mediated either by their Ets domain or their Pointed-domain (Drewett et al. 2000; Oikawa and Yamada 2003; Buchwalter et al. 2004). To examine the possibility of an Ets:DNA:Ets complex formed on the DYT1 Ets direct repeat motif, we tested the effects of increasing the distance between the two Ets cores. Changing the distance between adjacent binding sites in promoters affects the relative position of a core sequence on the DNA-turn as well as their relative distance along the DNA helix, resulting in decreased promoter activity in the majority of genes (Harvey et al. 1991; Baillat et al. 2002) and in fewer cases in increased activity (Spek et al. 1999). Introducing 3 (1/3 DNA turn) or 6 bases (2/3 DNA turn) changes the relative orientation between the mBM1 and mBM2, locating them on different sides of the helix, thus impairing protein-protein interaction. For some reason, the two Ets sites interact better when they are separated by a 2/3 than 1/3 DNA turn. Introduction of 3G′s at -81 is more deleterious than 3C′s which could reflect increase in steric hindrance between the two Ets sites, further disturbing protein-protein interaction, due to the more bulky structure of the G compare to the C bases. If we hypothesize that the mBM1 and mBM2 sites are optimally aligned in the wildtype DYT1 promoter, then introducing a complete DNA turn (10 bases) between them, brings mBM1 and mBM2 on the same side of the DNA helix, allowing them to still interact but not as well as in the wildtype, thus resulting in more activity compared to addition of 3 or 6 bases, but to decreased activity compared to the wildtype. It can be argued that the drop in transcriptional activity could be due to altering neighboring nucleotides of the critical mBM1 core. However, adding six or ten random bases still alters the mBM1 neighboring bases but the 5′ upstream DYT1 driven gene expression was partially restored. Thus, it is intriguing to hypothesize that the mBM1-mBM2 distance dependent configuration is important for optimal transcriptional activity. Finally, we cannot exclude the possibility that the 6 or 10 randomly added nucleotides introduced a new, undefined binding site, contributing to the activity.
A marked mEts1-DB-depended drop in 5′ upstream DYT1 driven gene expression confirmed the involvement of Ets-like factors in both N2A and HEK293 cells. Mutations introduced in the mBM2 core ameliorated the mEts1-DB effect, further supporting partial contribution of the mBM2 core in promoter activity while mutations within the critical mBM1 core further reduced, but did not abrogate, the dominant negative effect (Fig. 7A). That is probably due to the fact that the mutated mBM1 core retains a very low binding capacity (Suppl. Fig. 3). No mEts1-DB dependent effect was observed when both mBM1 and mBM2 cores were mutated (Fig. 7A). Similar effects of the mEts1-DB were observed using the human 5′ upstream DYT1 region (Fig. 7B).
The Ets family of transcription factors is composed of more than 30 members (Degnan et al. 1993; Oikawa and Yamada 2003), making it difficult to choose appropriate antibodies to demonstrate supershift in EMSAs. Furthermore, results obtained with the mEts1-DB dominant negative do not allow precise identification of the specific Ets paralog mediating the activity in this context. Even though the mEts1-DB has been designed to compete DNA binding of endogenous Ets1 factor, it is considered a general Ets-domain protein inhibitor since high levels of the Ets-domain would inhibit most of the GGAA/T core binding factors (Mattot et al. 2000).
Ubiquitous expression of Ets proteins (Oikawa and Yamada 2003) makes this family an ideal candidate for regulating widespread expression of torsinA. The mEts1-DB dominant negative does not allow precise identification of the specific Ets paralog mediating the activity in this context. Even though the mEts1-DB has been designed to compete DNA binding of endogenous Ets1 factor, it is considered a general Ets-domain protein inhibitor since high levels of the Ets-domain would inhibit most of the GGAA/T core binding factors (Mattot et al. 2000). Thus, further analysis is required to identify the specific Ets factors involved in different tissues. Such studies, will enhance our understanding of DYT1 regulation and potentially shed light on the torsinA-related pathways, since a number of Ets-factors are downstream effectors of distinct biological cascades (Wasylyk et al. 1998; Drewett et al. 2000). Finally, our studies reveal that a single or a two nucleotide changes at -79 of the mouse and -71/-70 of the human 5′ upstream DYT1 region, accounts for ∼90% or 45 to 60% of a -243bp- or -301bp-5′ upstream DYT1 driven activity, respectively. If human polymorphisms prove to exist within the -72/-69hBM1 core, they might provide insights into the decreased EOD penetrance.
Supplementary Material
Fig.1- Primer extension analysis for the mouse DYT1 gene. Mouse brain (lane 1) or liver (lane 2) total RNA was subjected to primer extension analysis with an oligonucleotide complementary to nucleotides +61 to +80 of the mouse DYT1 gene, Multiple initiation transcription sites were detected, point out with an arrowhead, corresponding to -35, -48, -61, -220, -233 and -289 from the ATG (arrowheads). Yeast t-RNA was used as negative control (lane 3). Sequence including the lower extension products is shown. The first nucleotide of the ATG translation start codon is indicated by an asterisk.
Fig.-2. Low levels of torsinA expression under the -2731 DYT1 promoter, detected in astrocyte cultures, arises from non glial-cells. Primary astrocyte culture, showing GFAP-negative cells expressing torsinA (red) under the -2731bp-5′ upstream DYT1 fragment. GFAP-positive cells (green) are devoid of torsinA expression under this promoter. Nuclei were stained with bis-benzimide. Bar indicates 20μm.
Fig.-3-.Binding properties of the -97/-66m probe and effects of within introduced mutations. The wildtype probe -97/-66 and the mutant versions -97/-66mS1, -97/-66mS2 and -97/-66mS1S2, were radiolabelled and incubated with mouse brain nuclear extract (MBNE). The S2 mutation (lane 2) reduces the binding capacity of the wildtype probe (lane 1), while very minimal binding capacity is retained when the S1 mutation is introduced (lane 3). Binding is completely eliminated when both S1 and S2 mutations are introduced (lane 4).
Acknowledgments
* We thank Dr J. Bieker (Mount Sinai School of Medicine, NY, USA) for critical reading of the manuscript. We thank: Dr B. Vandenbunder (Institute of Biology, Lille, France) who provided us with the mEts1-DB dominant negative expression construct as well as the wildtype pcDNA3-mEts1, Dr. G. John (Mount Sinai School of Medicine, NY, USA) for providing us with human primary astrocytes, Dr P. Gonzalez-Alegre for the polyclonal human torsinA antibody (University of Iowa, Neurology department), and M. Singh for technical assistance. This study was supported by grants from National Institute of Neurological disorders and Stroke (RO1 NS43038, P.S.) and Bachmann-Strauss Parkinson and Dystonia Foundation (P.S.).
REFERENCES
- Augood SJ, Keller-McGandy CE, Siriani A, Hewett J, Ramesh V, Sapp E, DiFiglia M, Breakefield XO, Standaert DG. Distribution and ultrastructural localization of torsinA immunoreactivity in the human brain. Brain Res. 2003;986:12–21. doi: 10.1016/s0006-8993(03)03164-0. [DOI] [PubMed] [Google Scholar]
- Baillat D, Begue A, Stehelin D, Aumercier M. ETS-1 transcription factor binds cooperatively to the palindromic head to head ETS-binding sites of the stromelysin-1 promoter by counteracting autoinhibition. J Biol Chem. 2002;277:29386–29398. doi: 10.1074/jbc.M200088200. [DOI] [PubMed] [Google Scholar]
- Bressman SB, Sabatti C, Raymond D, de Leon D, Klein C, Kramer PL, Brin MF, Fahn S, Breakefield X, Ozelius LJ, Risch NJ. The DYT1 phenotype and guidelines for diagnostic testing. Neurology. 2000;54:1746–1752. doi: 10.1212/wnl.54.9.1746. [DOI] [PubMed] [Google Scholar]
- Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Degnan BM, Degnan SM, Naganuma T, Morse DE. The ets multigene family is conserved throughout the Metazoa. Nucleic Acids Res. 1993;21:3479–3484. doi: 10.1093/nar/21.15.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewett V, Muller S, Goodall J, Shaw PE. Dimer formation by ternary complex factor ELK-1. J Biol Chem. 2000;275:1757–1762. doi: 10.1074/jbc.275.3.1757. [DOI] [PubMed] [Google Scholar]
- Fahn S. Concept and classification of dystonia. Adv Neurol. 1988;50:1–8. [PubMed] [Google Scholar]
- Goodchild RE, Kim CE, Dauer WT. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–932. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Harvey C, Jackson SM, Siddiqui SK, Gutierrez-Hartmann A. Structure-function analysis of the rat prolactin promoter: phasing requirements of proximal cell-specific elements. Mol Endocrinol. 1991;5:836–843. doi: 10.1210/mend-5-6-836. [DOI] [PubMed] [Google Scholar]
- Kustedjo K, Bracey MH, Cravatt BF. Torsin A and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J Biol Chem. 2000;275:27933–27939. doi: 10.1074/jbc.M910025199. [DOI] [PubMed] [Google Scholar]
- Laudet V, Niel C, Duterque-Coquillaud M, Leprince D, Stehelin D. Evolution of the ets gene family. Biochem Biophys Res Commun. 1993;190:8–14. doi: 10.1006/bbrc.1993.1002. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zolkiewska A, Zolkiewski M. Characterization of human torsinA and its dystonia-associated mutant form. Biochem J. 2003;374:117–122. doi: 10.1042/BJ20030258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattot V, Vercamer C, Soncin F, Calmels T, Huguet C, Fafeur V, Vandenbunder B. Constitutive expression of the DNA-binding domain of Ets1 increases endothelial cell adhesion and stimulates their organization into capillary-like structures. Oncogene. 2000;19:762–772. doi: 10.1038/sj.onc.1203248. [DOI] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- Ozelius LJ, Hewett JW, Page CE, Bressman SB, Kramer PL, Shalish C, de Leon D, Brin MF, Raymond D, Corey DP, Fahn S, Risch NJ, Buckler AJ, Gusella JF, Breakefield XO. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet. 1997;17:40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD, Brown AL, Ling Y, Yates PR. The ETS-domain transcription factor family. Int J Biochem Cell Biol. 1997;29:1371–1387. doi: 10.1016/s1357-2725(97)00086-1. [DOI] [PubMed] [Google Scholar]
- Shashidharan P, Kramer BC, Walker RH, Olanow CW, Brin MF. Immunohistochemical localization and distribution of torsinA in normal human and rat brain. Brain Res. 2000;853:197–206. doi: 10.1016/s0006-8993(99)02232-5. [DOI] [PubMed] [Google Scholar]
- Singer VL, Wobbe CR, Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990;4:636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- Spek CA, Bertina RM, Reitsma PH. Unique distance- and DNA-turn-dependent interactions in the human protein C gene promoter confer submaximal transcriptional activity. Biochem J. 1999;340(Pt 2):513–518. [PMC free article] [PubMed] [Google Scholar]
- Swick AG, Blake MC, Kahn JW, Azizkhan JC. Functional analysis of GC element binding and transcription in the hamster dihydrofolate reductase gene promoter. Nucleic Acids Res. 1989;17:9291–9304. doi: 10.1093/nar/17.22.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RH, Brin MF, Sandu D, Good PF, Shashidharan P. TorsinA immunoreactivity in brains of patients with DYT1 and non-DYT1 dystonia. Neurology. 2002;58:120–124. doi: 10.1212/wnl.58.1.120. [DOI] [PubMed] [Google Scholar]
- Wasylyk B, Hagman J, Gutierrez-Hartmann A. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem Sci. 1998;23:213–216. doi: 10.1016/s0968-0004(98)01211-0. [DOI] [PubMed] [Google Scholar]
- Xiao J, Gong S, Zhao Y, LeDoux MS. Developmental expression of rat torsinA transcript and protein. Brain Res Dev Brain Res. 2004;152:47–60. doi: 10.1016/j.devbrainres.2004.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig.1- Primer extension analysis for the mouse DYT1 gene. Mouse brain (lane 1) or liver (lane 2) total RNA was subjected to primer extension analysis with an oligonucleotide complementary to nucleotides +61 to +80 of the mouse DYT1 gene, Multiple initiation transcription sites were detected, point out with an arrowhead, corresponding to -35, -48, -61, -220, -233 and -289 from the ATG (arrowheads). Yeast t-RNA was used as negative control (lane 3). Sequence including the lower extension products is shown. The first nucleotide of the ATG translation start codon is indicated by an asterisk.
Fig.-2. Low levels of torsinA expression under the -2731 DYT1 promoter, detected in astrocyte cultures, arises from non glial-cells. Primary astrocyte culture, showing GFAP-negative cells expressing torsinA (red) under the -2731bp-5′ upstream DYT1 fragment. GFAP-positive cells (green) are devoid of torsinA expression under this promoter. Nuclei were stained with bis-benzimide. Bar indicates 20μm.
Fig.-3-.Binding properties of the -97/-66m probe and effects of within introduced mutations. The wildtype probe -97/-66 and the mutant versions -97/-66mS1, -97/-66mS2 and -97/-66mS1S2, were radiolabelled and incubated with mouse brain nuclear extract (MBNE). The S2 mutation (lane 2) reduces the binding capacity of the wildtype probe (lane 1), while very minimal binding capacity is retained when the S1 mutation is introduced (lane 3). Binding is completely eliminated when both S1 and S2 mutations are introduced (lane 4).