Abstract
Identification of major histocompatibility complex (MHC) class II binding peptides is a crucial step in rational vaccine design and immune monitoring. We designed a novel MHC class II molecule-peptide microarray binding assay and evaluated 346 peptides from already identified human immunodeficiency virus (HIV) epitopes and an additional set (n = 206) of 20-mer peptides, overlapping by 15 amino acid residues, from HIV type 1B (HIV-1B) gp160 and Nef as a paradigm. Peptides were attached via the N-terminal part to a linker that covalently binds to the epoxy glass slide. The 552 peptides were printed in triplicate on a single peptide microarray chip and tested for stable formation of MHC class II molecule-peptide complexes using recombinant soluble DRB1*0101(DR1), DRB1*1501(DR2), and DRB1*0401(DR4) molecules. Cluster analysis revealed unique patterns of peptide binding to all three, two, or a single MHC class II molecule. MHC class II binding peptides reside within previously described immunogenic regions of HIV gp160 and Nef, yet we could also identify new MHC class II binding peptides from gp160 and Nef. Peptide microarray chips allow the comprehensive and simultaneous screening of a high number of candidate peptide epitopes for MHC class II binding, guided by subsequent quality data extraction and binding pattern cluster analysis.
Major histocompatibility complex (MHC) class II alleles, cell surface glycoproteins with a high degree of allelic polymorphism (8), are constitutively expressed on professional antigen-presenting cells. The peptide repertoire presented by MHC class II molecules to CD4+ T cells is edited by intracellular, nonclassical MHC molecules, i.e., HLA-DM and -DO (43). MHC class II alleles exhibit four major pockets (13) to accommodate and present a broad peptide repertoire to CD4+ T cells: the same peptide can be presented by different MHC class II alleles due to highly degenerate peptide binding motifs (12, 16, 29, 36). CD4+ T cells play a crucial role in the adaptive immune response: they provide help to B cells and CD8+ T cells and they also display immune effector functions (37, 41).
Different individuals exhibit different immune responses to the same pathogen (4), in part due to the “genetic makeup,” including the MHC class II allelic composition in the individual. Some MHC class II alleles have been associated with “better” immune responses to infectious pathogens (1, 24), while other MHC class II alleles show differential effects: an enhanced or decreased risk of diseases, based on the nature of the antigen and on the T-cell repertoire capable of reacting to a distinct set of peptides. For instance, the MHC class II molecule DQ*0602 is strongly associated with susceptibility to narcolepsy, yet it protects against type 1 diabetes (35).
Only a fraction of a candidate protein may give rise to antigen-specific immune reactivity (3). Definition of the nature and composition of peptides binding to MHC class II molecules is crucial for T-cell-based vaccine design and for the subsequent gauging of the “vaccine take”: proteins which provide epitopes leading to strong CD4+ T-cell responses may be advantageous. Rational design of vaccines considers genetic diversity, including the MHC class II alleles of the target populations, to ensure wide population coverage (9, 17). It would therefore be helpful to determine the following: (i) which set of peptides from a molecularly defined pathogen is able to bind to different MHC class II alleles, (ii) if subtle differences in the MHC class II molecules have an impact on the peptide repertoire selection, and vice versa, (iii) if variations within the candidate peptide have an impact on binding to individual MHC class II alleles. A number of assays are currently available to assess peptide binding to MHC class II alleles. These include computer-based algorithms (5, 20, 27, 40, 47), functional assays (enzyme-linked immunospot and intracellular cytokine staining), mass spectrometric sequencing of peptides eluted from purified HLA alleles (6, 7, 42), and peptide binding assays (12, 25, 34).
The advent of peptide microarray technology now offers a unique platform for studying the full spectrum of peptide attributes in a massively parallel, miniaturized, and automated fashion (23, 31). Using peptide microarrays to explore peptide binding to soluble, recombinant MHC class II molecules represents a departure from traditional one-peptide-at-a-time assays. It allows the simultaneous evaluation of the binding behavior of thousands of peptides for soluble MHC class II molecules, which would require considerable investments of time and patient material if more traditional approaches were implemented. We report here, as a paradigm, the use of a peptide microarray chip to determine the binding of soluble MHC class II molecules to immobilized candidate peptides from human immunodeficiency virus type 1B (HIV-1B). The MHC class II binding groove, unlike that of MHC class I molecules, is open at both ends and thus allows spatial interaction with test peptides linked to a scaffold (32, 33, 38).
DRB1*0101(DR1), DRB1*1501(DR2), and DRB1*0401(DR4) represent the most frequently reported MHC class II alleles (22): DR1 (15.4%), DR2 (32.9%), and DR4 (20.9%) are the alleles most frequently defined for Caucasians (45) by high-resolution typing. Low-resolution MHC class II typing data from Botswana (24) showed that HLA DRB1*01, DRB1*02, and DRB1*04 exhibit population frequencies of 21.7%, 21.3%, and 14.4%, respectively. The association of individual MHC alleles with protection from certain viral infections (21) suggests that “protective alleles” bind more and “better” antigenic peptides than other alleles. The number of candidate peptides cannot be deduced based on allele frequencies; these can only be experimentally determined, since peptide binding reflects the structural constraints of the MHC class II binding molecule (1, 35).
MATERIALS AND METHODS
HIV-1 peptides.
Two groups of HIV-1 peptides were printed on peptide microarray slides. One set consisted of 206 20-mer peptides overlapping by 15 amino acids and was generated from the HIV-1B consensus sequences of the gp160 and Nef proteins. The second set consisted of 346 CD4+ T-cell epitopes listed in the Los Alamos HIV Immunology database (http://www.hiv.lanl.gov/content/immunology/index.html) at the end of November 2006.
Peptide microarray printing.
Amino-oxy-acetylated peptides were synthesized on cellulose membranes in a parallel manner using SPOT synthesis technology (11, 30). Following side-chain deprotection, the solid-phase bound peptides were transferred into 96-well microtiter filtration plates (Millipore, Bedford, MA) and treated with 200 μl of aqueous triethylamine (0.5% by volume) in order to cleave the peptides from the cellulose support. The peptide-containing triethylamine solution was filtered off, and the solvent was removed by evaporation under reduced pressure. The resulting peptide derivatives (50 nmol) were redissolved in 25 μl of printing solution (70% dimethyl sulfoxide, 25% 0.2 M sodium acetate [pH 4.5], 5% glycerol [by volume]) and transferred into 384-well microtiter plates. Two droplets of 0.5 nl peptide solution (1 mM) was deposited per spot on epoxy-functionalized glass slides (Corning epoxy no. 40042) using the noncontact printer Nano Plotter of GeSiM (Großerkmannsdorf, Germany) equipped with a piezoelectric NanoTip (GeSiM, Großerkmannsdorf, Germany). The peptides were attached to the glass slide via the N-terminal end, which was attached to a linker that covalently binds to the epoxy glass slide. The peptides and control spots (i.e., empty spots) were printed on positions in each identical subarray, i.e., each peptide microarray slide provided three repeats, and there were three slides prepared for each of the HLA-DR molecules, resulting in nine repeats for each individual peptide species. Printed peptide microarrays were kept at room temperature for 5 h, washed with deionized water, quenched for 1 h with 0.1 mg/ml bovine serum albumin in 75 mM SSC buffer (pH 7.0) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% sodium dodecyl sulfate and 750 mM NaCl at 42°C, washed extensively with 1.5 mM SSC buffer (pH 7.0) followed by five washing steps with water, and dried using a chip centrifuge. Peptide microarrays were then stored at 4°C.
Soluble HLA class II alleles.
Soluble MHC class II molecules, HLA DRB1*0101(DR1), DRB1*1501(DR2), and DRB1*0401(DR4), were used as probes to gauge binding to individual peptide species. Three HLA-DR alleles, HLA DRB1*0101(DR1), DRB1*1501(DR2), and DRB1*0401(DR4), were supplied by Beckman Coulter as described in detail previously (28).
Sample processing.
HLA-DR monomers were diluted to a working concentration of 1 μg/ml using a binding buffer (phosphate [36 mM], citrate [14.4 mM], bovine serum albumin [0.15%], octyl beta-d-glucopyranoside [0.25%], NaN3 [0.02%], with a pH of 5.5). After the surface of the slide was ensured to be clean and dry, the incubation area was circumscribed using a hydrophobic pen (DakoCytomation, Denmark). The diluted HLA-DR monomer was spread evenly across the entire slide, which was covered with a coverslip and incubated for 48 h at 37°C in a humid chamber. After the first incubation step, the coverslip was removed and the slide was placed in a box with a slide holder (VWR International) and washed three times for 5 min each, two times with washing solution (phosphate-buffered saline [PBS] plus Tween 80 [0.05%]), and once with PBS. The washing procedure was performed on a shaker. Without allowing the slide to dry, 300 μl of monoclonal secondary antibody, Cy5-labeled monoclonal antibody (MAb) L243 (Beckman Coulter), diluted to 5 μg/ml in PBS, was pipetted onto the slide, which was then covered with a coverslip and incubated for 1 h at room temperature in a humid chamber, followed by the three washing steps listed above. The slide was then spun dry for 10 s using a slide spinner (Euro Tech, United Kingdom). The optimal incubation time, temperature, and dilution for HLA-DR monomers and for the secondary antibody were determined prior to testing. Negative controls included the following: (i) scanning of the slide without any reagent (blank) and (ii) preparation of slides with only reagent buffer and the secondary antibody (MAb L243; see above) used for detection. Two slides were incubated with buffer and secondary antibody only, and three slides were incubated with each of the three different HLA-DR alleles. Titration of the ligand (peptide) or the HLA-DR molecule would aid in determining HLA-DR-peptide affinity, yet the peptide concentration is fixed (approximately 10,000 individual peptide species/spot); this can be altered only with technical challenges (10,000 binding sites have to be occupied per spot; variation of the numbers of peptides per spot could be achieved by mixing the target peptide with an irrelevant control peptide, with the risk of unspecific interaction with the respective HLA-DR molecule used for testing). Alternate incubation times would provide information concerning the off rate, yet this would be different for each peptide species.
Data acquisition. (i) Scanning and analysis.
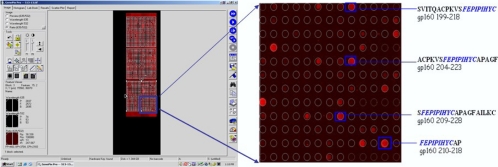
Each slide was scanned with the GenPix 4000B microarray scanner (Axon Instruments) at two wavelengths, 532 and 635 nm, and images were saved in the TIFF and JPG formats (Fig. 1). Image analysis was performed utilizing the circular feature alignment of the GenePix Pro 5.1 software program and the Genepix Array List files supplied by JPT (Berlin, Germany). Spots with a nonuniform foreground or background signal were flagged if they satisfied the following criteria: F635 mean > (1.5 × F635 median) and F635 median > 40 or B635 mean > (1.5 × B635 median) and B635 median > 40.
FIG. 1.
Representative example of MHC class II molecule-peptide formation. Binding of the MHC class II molecule to peptides is visualized using a Cy5 fluorochrome-conjugated anti-human HLA-DR antibody. CD4+ HIV T-cell epitopes, listed in the HIV Los Alamos database, form a complex with the DRB1*101 molecule. The peptide species share a potential MHC class II binding motive (FEPIPIHYC) (http://www.hiv.lanl.gov/content/immunology/index.html) (19, 46).
These and other flags assigned by GenePix resulted in four types of spots: “good” or “nonflagged” spots (labeled “0”), “bad” or “flagged” spots (labeled “−100”), not-found spots (labeled “−50”), and empty spots (labeled “−75”). The image from each subarray was saved as a GPR (GenePix result) file, and the median foreground and background intensities for the 635-nm wavelength from individual peptide spots were used in the analysis of the responses. All GPR files were saved in a common folder and imported into the R/BioConductor software package using the read.GenePix function from the marray R/bioconductor package.
To examine the quality of the acquired data, we examined the distribution of the flags (listed above). We performed this quality-control exercise for each of the four groups of slides: (i) slides incubated with buffer only, (ii) HLA-DRB1*0101 slides, (iii) HLA-DRB1*1501 slides, and (iv) HLA-DRB1*0401 slides, both overall and stratified by the type of feature (control or peptide spots). Visual inspection of the images from the individual subarrays was carried out, using the Image function in Bioconductor, in order to evaluate questionable responses that should be excluded from the analysis. For a measure of the strength of the response, we chose the ratio of median foreground to background (on a log scale). This response index was computed for all spots with a background greater than zero, and any spots with zero background were excluded. The data for each of the four groups of slides were arranged in a large matrix, with columns identifying slide, subarray, and block, and all the analyses described below used these master data sets.
(ii) Data reduction.
Using the distribution of the negative controls to define a cutoff for a “detectable” response, we removed the spots with no detectable response on any slide. The method used to define the cutoff has been described previously (23) and involves three steps: the elimination of extreme outliers, normalization of the responses on all slides in a group using a simple linear model, and computation of the threshold from the mean and standard deviation of the normalized responses as t − mean + 3 standard deviations. In addition to excluding any spots that had nondetectable responses on all slides, we also excluded the “not-found” spots with high intensity, since these were presumably flagged for problems other than a low “not-found” signal.
(iii) False-positive events.
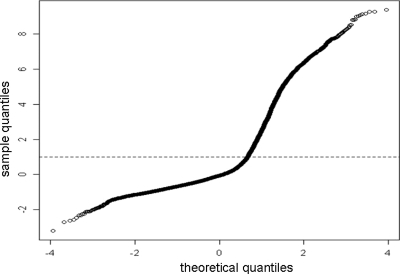
Any peptide with a high response on slides incubated only with buffer and the Cy5-labeled MAb L243 was considered false positive (Fig. 2) and discarded for analysis. After normalizing all valid (i.e., unflagged) peptide responses on the buffer slides using the same linear model as for the negative controls, the cutoff was determined for the definition of a false-positive event (Fig. 3).
FIG. 2.
Strategy to remove false-positive results. A quartile-quartile plot of theoretical quantiles versus sample quantiles for peptide microarray slides incubated with buffer and the secondary MAb (L243) detecting HLA-DR molecules.
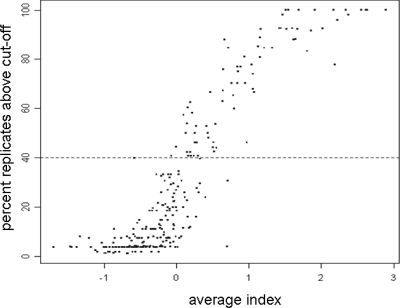
FIG. 3.
Definition of the cutoff in MHC class II molecule-peptide binding experiments. Correlation of the peptide response index and the peptide foreground fluorescence intensity. A representative example for the DRB1*0101 molecule.
(iv) Analysis of peptide responses.
For each group of HLA-DR incubated slides, we used the thresholds defined above to exclude from analysis any peptide that had no detectable response on any slide or had a false-positive response in at least 10% of replicates. The remaining peptide responses were normalized using a linear model to remove artifacts due to slide, subarray, and block: the model was fit using the biglm R software package to accommodate the much larger dimension of the problem, and we estimated the peptide effects as the differences between the observed responses and the responses predicted by the model (i.e., the residuals). Since the systematic effects of slide, subarray, and block were removed, we refer to these as the “normalized responses.” For any peptides that were replicated, the normalized values were averaged. Thus, the preprocessed data consist of a list of unique peptides with their normalized values for each slide.
CD4+ T-cell peptide-specific T-cell expansion.
Peripheral blood mononuclear cells (PBMCs) from HLA class II typed healthy blood donors were cultured in 50% Dulbecco's modified Eagle medium (high glucose) and 50% adoptive immunotherapy (AIM-V) medium supplemented with 1% human serum. Five million cells were added to each well in a 48-well plate, and 1 μg of peptide was added to test wells. On day 3, 10 ng/ml of interleukin 7 (IL-7) and 10 IU/ml of IL-2 were added to PBMC cultures. On day 14 and again on day 21, cells were restimulated with PBMCs which had been radiated with 5,000 rad after being pulsed with 1 μg of the stimulating peptide for 2 h at 24°C in AIM-V medium. After peptide stimulation on day 21, cells rested for 6 days and were then assayed for peptide-specific responses by intracellular cytokine staining, i.e., tumor necrosis factor alpha (TNF-α), gamma interferon (IFN-γ), and IL-2 on a single-cell level as described previously (18). One million events were acquired, and results are reported as absolute numbers of cytokine-producing cells/105 CD4+ T cells. Control PBMCs were cultured as well in medium and cytokines without peptide stimulation.
RESULTS
Visualizing binding of soluble HLA-DR molecules to immobilized peptides.
Two groups of HIV-1 peptides were printed in triplicate on microarray glass slides. One set consisted of 20-mer peptides overlapping by 15 amino acids generated from the gp160 and Nef HIV-1B consensus sequences, resulting in 167 peptides for gp160 and 39 peptides for HIV Nef. The other group of 346 peptides has been listed as CD4+ T-cell epitopes of various lengths in the Los Alamos database. There was a clear distinction between the formation of a peptide-HLA-DR allele complex and peptides to which the soluble MHC class II molecules did not bind. Figure 1 shows a representative picture of a scanned peptide microarray image overlaid with the Genepix Array List file to identify candidate peptide species interacting with soluble MHC class II molecules at each spot. The four peptides depicted in Fig. 3 form complexes with DRB1*0101(DR1), DRB1*1501(DR2), and DRB1*0401(DR4) and show a common peptide core (FEPIPIHYC).
Criteria for determining the formation of HLA-DR peptide complexes.
We identified 208/552 peptides binding to DRB1*0101, 163/552 peptides binding to DRB1*1501(DR2), and 162/552 peptides binding to DRB1*0401(DR4) (summarized in Tables S1 and S2 in the supplemental material). These peptides had to have at least one replicate with an index response above the cutoff defining a detectable response. From these peptides, we chose to focus on those that had an index response above the cutoff in at least 40% of the replicates. Since each peptide was printed three times on the chip and we repeated the experiments three times, we obtained nine data sets for each individual peptide species. After removal of false-positive events (Fig. 2), i.e., binding of the secondary reagent to peptide species (see Materials and Methods for details), we analyzed the complex formation of the soluble MHC class II molecules for each candidate peptide printed on the chip. There was a good correlation between the peptide response index and the percentage of replicates above the cutoff. Using this criterion, we found 78/552 peptides interacting with DRB1*0101(DR1), 91/552 peptides interacting with DRB1*1501(DR2), and 101/552 peptides interacting with DRB1*0401(DR4).
Clustering of HIV peptides by HLA-DR binding.
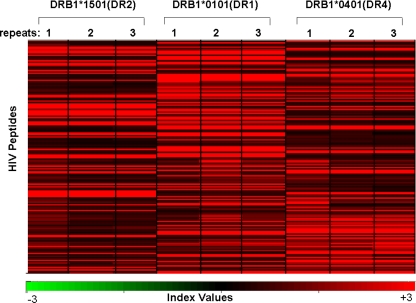
In order to identify clusters of peptides (i.e., using both peptide groups, the CD4+ T-cell epitopes listed in the Los Alamos database and the gp160 and Nef proteins as overlapping peptide stretches) that show differential binding to soluble HLA-DR alleles, a Pearson centered hierarchical clustering analysis was carried out using the software program Acuity (2), using the average of the normalized response indexes on three slides (repeats). Figure 4 shows that the clusters of individual HLA-DR-peptide complexes were unique for each MHC class II allele. There were peptides that bound strongly to all three HLA-DR alleles and peptides which formed complexes with only one or two of the MHC class II alleles.
FIG. 4.
MHC class II molecule-peptide binding pattern. A Pearson centered hierarchical clustering analysis of HIV peptides (n = 206) derived as overlapping peptides from HIV gp160 or Nef and 346 peptides listed as CD4+ T-cell epitopes in the Los Alamos database using the software program Acuity. Note the cluster of the MHC class II molecule-peptide binding pattern in association with the respective HLA-DR molecules. Each experiment was repeated three times. The index values, which represent a measure of the signal intensity, are similar for slides incubated with the same MHC class II monomer; minor differences are attributable to experimental variations.
Complex formation by described CD4+ T-cell epitopes with HLA-DR molecules.
Of the 306 peptides from the HIV immunology database that were printed on the microarray chip, 73 showed interaction with soluble HLA-DR alleles (see Table S1 in the supplemental material). The exact MHC class II-restricting molecule for the majority of the published HIV-1 CD4+ T-cell epitopes has not yet been defined (they have been reported as “DR restricted” or simply as “CD4 epitope”), and it is therefore not surprising that some of the epitopes did not form a complex with the HLA-DR molecules employed in the current assay.
Complex formation by HIV-1B gp160 and Nef peptides with HLA-DR molecules.
Table S2 in the supplemental material lists 20-mer peptides, generated in a systematic way as overlapping peptides from the HIV-1B consensus gp160 and Nef sequences, that showed complex formation with any of the HLA-DR alleles tested in the current report. Thirty-two of 167 peptides from gp160 formed complexes with DRB1*0101, 34/167 with DRB1*1501, and 38/167 with DRB1*0401; 10/39 peptides from Nef formed complexes with DRB1*0101, 8/39 with DRB1*1501, and 6/39 with DRB1*0401, respectively. The comparison of these peptides to the CD4+ T-cell epitopes listed in the Los Alamos HIV-1 Immunology database indicates a good overlap of the protein regions which provide immunogenic peptide epitopes (see Table S2 in the supplemental material). In a parallel approach, we selected overlapping peptides from HIV-1 gp160 and Nef and evaluated MHC class II complex formation; Fig. 5 shows the compilation of HLA-DR binding epitopes for HIV Nef and Fig. 6 the HLA-DR binding patterns for HIV gp160 as a paradigm. The newly identified peptides reside within already described immunodominant regions of the HIV proteins, which further lends support to our approach (Fig. 5 and 6). Although the MHC class II molecule-peptide binding was reproducible, we lacked information on whether HLA-DR binding peptides would indeed be able to expand peptide-specific CD4+ T cells. We therefore used some of the identified MHC class II binding peptides to expand CD4+ T cells in vitro defined by the production of IFN-γ, TNF-α, and IL-2 (Fig. 7).
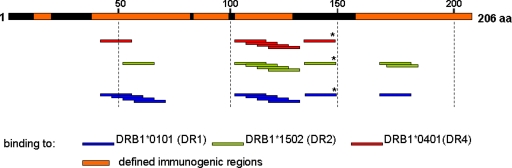
FIG. 5.
HLA-DR epitopes on HIV Nef. Compilation of the DRB1*0101, DRB1*1501, and DRB1*0401 interactions with HIV-1B Nef defined by the peptide array platform. Areas with CD4+ T-cell epitopes that have been listed in the Los Alamos HIV database are highlighted in orange. All HLA-DR binding epitopes have been described except an epitope in position 130 to 150 (*).
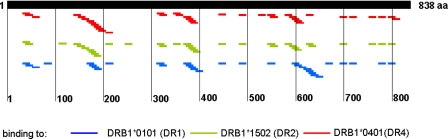
FIG. 6.
HLA-DR epitopes on HIV gp160. Compilation of the DRB1*0101, DRB1*1501, and DRB1*0401 interactions with HIV-1B gp160 defined by the peptide array platform.
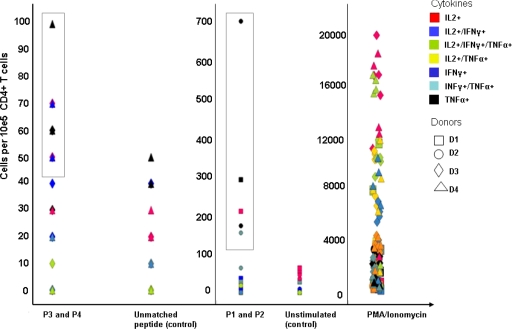
FIG. 7.
MHC class II-restricted expansion of peptide-specific CD4+ T cells. PBMCs from HIV-negative and MHC class II-genotyped blood donors were stimulated with the following peptides: for donors 1 to 3, peptides SLYVTVATLYCVHQRIEV and FRKQNPDIVIYQYMDDLYVG, representing HIV Gag (amino acids 77 to 94) and HIV reverse transcriptase (amino acids 171 to 190), respectively (peptides 1 and 2 [P1 and P2]). PBMCs from donor 4 were stimulated with ALFYKLDVVPINDNTSYRL from HIV gp160 (amino acids 174 to 193) and EKLWVTVYYGVPVWKEATTT from HIV gp160 (amino acids 32 to 51) (peptides 3 and 4 [P3 and P4]). After three rounds of peptide stimulation, PBMCs were tested for peptide-specific reactivity defined by intracellular production of TNF-α, IFN-γ, and IL-2 in intracellular cytokine staining. Phorbol myristate acetate/ionomycin served as the positive control; negative controls included the incubation in medium or stimulation with an irrelevant HIV peptide which was not used for stimulation (EKLWVTVYYGVPVWKEATTT for donors 1 to 3 and peptide FRKQNPDIVIYQYMDDLYVG for donor 4). Results are reported as absolute numbers of cytokine-producing cells/105 CD4+ T cells.
DISCUSSION
We describe here, to our knowledge for the first time, an MHC class II binding assay implementing a peptide microarray and recombinant synthetic soluble MHC class II molecules, using HIV-derived peptides as a paradigm. Already described CD4+ T-cell epitopes were printed on the chip to act as positive controls, and 73/306 peptides formed detectable MHC class II complexes. Of note, the peptides listed in the Los Alamos database show various lengths and have been selected based on different criteria, particularly functional T-cell assays; most of the epitopes have been listed as “CD4+ T-cell epitope” or “DR-restricted.” Of the 346 peptides listed as CD4+ T-cell epitopes in the Los Alamos HIV Immunology database, only 280 were defined for humans. Other peptides were identified in different species, e.g., mice, chimpanzees, and macaques. Out of these 280, only 70 have been mapped to a specific MHC class II allele or an allele group. It could very well be that specific MHC class II alleles were not covered in the current experiments due to failure to employ the correct MHC class II alleles for binding.
The limitations of the assay described may be the following: (i) structural constraints imposed by the immobilization of peptide on the glass slide via the N-terminal end, possibly precluding spatial interaction with soluble MHC class II molecules; (ii) the current 20-mer peptides may not represent the optimal length facilitating binding to a specific MHC class II allele; or (iii) glycopeptides cannot currently be evaluated for MHC class II interaction (7).
A number of assays to identify peptides binding to MHC class II molecules (5, 20, 27, 40, 47) have been described, but only a very limited number of peptides could be tested simultaneously. This makes it difficult to comprehensively screen an entire proteome from a pathogen or a comprehensive library from target epitopes in cancer or autoimmune diseases. A number of reports addressed a multidimensional approach to enhance prediction of MHC class II molecule-peptide interactions (26, 44). If T-cell-based assays are employed, it is not only the turnaround time that is limited but also the availability of biological material. Computational predictions of MHC class II binding peptides offer the advantage of a short turnaround time (44), yet it may be challenging to rely solely on computational analysis without selected biological readouts (14). This demand has been addressed by Stone and coworkers, who developed an MHC class II-peptide array supplemented with costimulatory molecules to identify MHC class II-restricted epitopes defined by functional T-cell responses (39). This assay requires peptides that are known to bind to certain MHC class II molecules. Peptides identified in the current assay were linked to biological activity: some peptides, previously shown to be functional HIV-specific CD4+ T-cell epitopes (10, 15, 39), showed good binding to soluble MHC class II molecules employed in the current assay (see Table S1 in the supplemental material), and newly identified MHC class II-binding peptide epitopes resided in “immunogenic regions” of HIV-1B Nef (see Fig. 5) or gp160 and showed the capacity to expand MHC class II-restricted and peptide-specific CD4+ T cells defined by cytokine production.
The assay described in the current report offers the unique advantage of screening thousands of peptides within a single experiment using high-density peptide spotting using SPOT synthesis technology (11, 30). This enables the printing of entire viral proteomes as linear peptide stretches (our unpublished data) on glass slides. In addition, the orientation of the candidate peptide is defined and can also be used for a more detailed structural analysis of MHC class II molecule-peptide interactions: the crystal structure of MHC class II molecules shows that the peptide binding groove is open at both ends (32, 33, 38), which enables complex formation with peptides immobilized on the glass scaffold.
In conclusion, we developed a robust assay using a peptide microarray and recombinant soluble MHC class II molecules to identify HIV-1 peptides which form complexes with different HLA-DR alleles. The assay can be implemented to screen for MHC class II binding of peptide libraries with a short turnaround time and may also be used to probe for differences in MHC class II molecule-peptide complex formation associated with variant peptides or with differences in MHC class II alleles. Peptide microarrays, such as those described in the current report, will help to speed up the process of CD4+ T-cell epitope identification and advance efforts targeting a rational platform for vaccine design and immunomonitoring.
Supplementary Material
Acknowledgments
This work was supported by a grant from the EU Marie Curie Early Training program to A.E. and grants from Cancerfonden, Vetenkapsrådet, Söderberg-foundation, and Karolinska Institutet, Sweden, to M.M.
Footnotes
Published ahead of print on 18 February 2009.
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1.Angyalosi, G., R. Neveu, I. Wolowczuk, A. Delanoye, J. Herno, C. Auriault, and V. Pancre. 2001. HLA class II polymorphism influences onset and severity of pathology in Schistosoma mansoni-infected transgenic mice. Infect. Immun. 695874-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axon Instruments Inc. 2005. Acuity 4.0 microarray informatics software user's guide, 2005. Axon Instruments/Molecular Devices Corp., Sunnyvale, CA.
- 3.Castellino, F., G. Zhong, and R. N. Germain. 1997. Antigen presentation by MHC class II molecules: invariant chain function, protein trafficking, and the molecular basis of diverse determinant capture. Hum. Immunol. 54159-169. [DOI] [PubMed] [Google Scholar]
- 4.Deeks, S. G., and B. D. Walker. 2007. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27406-416. [DOI] [PubMed] [Google Scholar]
- 5.DeLuca, D. S., B. Khattab, and R. Blasczyk. 2007. A modular concept of HLA for comprehensive peptide binding prediction. Immunogenetics 5925-35. [DOI] [PubMed] [Google Scholar]
- 6.Dengjel, J., M. D. Nastke, C. Gouttefangeas, G. Gitsioudis, O. Schoor, F. Altenberend, M. Muller, B. Kramer, A. Missiou, M. Sauter, J. Hennenlotter, D. Wernet, A. Stenzl, H. G. Rammensee, K. Klingel, and S. Stevanovic. 2006. Unexpected abundance of HLA class II presented peptides in primary renal cell carcinomas. Clin. Cancer Res. 124163-4170. [DOI] [PubMed] [Google Scholar]
- 7.Dengjel, J., H. G. Rammensee, and S. Stevanovic. 2005. Glycan side chains on naturally presented MHC class II ligands. J. Mass Spectrom. 40100-104. [DOI] [PubMed] [Google Scholar]
- 8.Erlich, H. A., and U. B. Gyllensten. 1991. The evolution of allelic diversity at the primate major histocompatibility complex class II loci. Hum. Immunol. 30110-118. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, W., S. Perkins, J. Theiler, T. Bhattacharya, K. Yusim, R. Funkhouser, C. Kuiken, B. Haynes, N. L. Letvin, B. D. Walker, B. H. Hahn, and B. T. Korber. 2007. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 13100-106. [DOI] [PubMed] [Google Scholar]
- 10.Fonseca, S. G., A. Coutinho-Silva, L. A. Fonseca, A. C. Segurado, S. L. Moraes, H. Rodrigues, J. Hammer, E. G. Kallas, J. Sidney, A. Sette, J. Kalil, and E. Cunha-Neto. 2006. Identification of novel consensus CD4 T-cell epitopes from clade B HIV-1 whole genome that are frequently recognized by HIV-1 infected patients. AIDS 202263-2273. [DOI] [PubMed] [Google Scholar]
- 11.Frank, R. 2002. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports—-principles and applications. J. Immunol. Methods 26713-26. [DOI] [PubMed] [Google Scholar]
- 12.Gaudebout, P., D. Zeliszewski, J. J. Golvano, C. Pignal, S. Le Gac, F. Borras-Cuesta, and G. Sterkers. 1997. Binding analysis of 95 HIV gp120 peptides to HLA-DR1101 and -DR0401 evidenced many HLA-class II binding regions on gp120 and suggested several promiscuous regions. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1491-101. [DOI] [PubMed] [Google Scholar]
- 13.Godkin, A. J., K. J. Smith, A. Willis, M. V. Tejada-Simon, J. Zhang, T. Elliott, and A. V. Hill. 2001. Naturally processed HLA class II peptides reveal highly conserved immunogenic flanking region sequence preferences that reflect antigen processing rather than peptide-MHC interactions. J. Immunol. 1666720-6727. [DOI] [PubMed] [Google Scholar]
- 14.Gowthaman, U., and J. N. Agrewala. 2008. In silico tools for predicting peptides binding to HLA-class II molecules: more confusion than conclusion. J. Proteome Res. 7154-163. [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann, D. E., P. M. Bailey, J. Sidney, B. Wagner, P. J. Norris, M. N. Johnston, L. A. Cosimi, M. M. Addo, M. Lichterfeld, M. Altfeld, N. Frahm, C. Brander, A. Sette, B. D. Walker, and E. S. Rosenberg. 2004. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J. Virol. 784463-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi, H., M. Wood, Y. Song, E. Appella, and E. Celis. 2000. Defining promiscuous MHC class II helper T-cell epitopes for the HER2/neu tumor antigen. Cancer Res. 605228-5236. [PubMed] [Google Scholar]
- 17.Letourneau, S., E. J. Im, T. Mashishi, C. Brereton, A. Bridgeman, H. Yang, L. Dorrell, T. Dong, B. Korber, A. J. McMichael, and T. Hanke. 2007. Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS One 2e984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magalhaes, I., N. K. Vudattu, E. Jäger, and M. J. Maeurer. 2008. Tumor antigen-specific T-cells are present in the CD8alpha/alpha+ T-cell effector-memory pool. J. Immunother. 31840-848. [DOI] [PubMed] [Google Scholar]
- 19.Malhotra, U., S. Holte, T. Zhu, E. Delpit, C. Huntsberry, A. Sette, R. Shankarappa, J. Maenza, L. Corey, and M. J. McElrath. 2003. Early induction and maintenance of Env-specific T-helper cells following human immunodeficiency virus type 1 infection. J. Virol. 772663-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallios, R. R. 2003. A consensus strategy for combining HLA-DR binding algorithms. Hum. Immunol. 64852-856. [DOI] [PubMed] [Google Scholar]
- 21.Martin, M. P., and M. Carrington. 2005. Immunogenetics of viral infections. Curr. Opin. Immunol. 17510-516. [DOI] [PubMed] [Google Scholar]
- 22.Middleton, D., L. Menchaca, H. Rood, and R. Komerofsky. 2003. New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens 61403-407. [DOI] [PubMed] [Google Scholar]
- 23.Nahtman, T., A. Jernberg, S. Mahdavifar, J. Zerweck, M. Schutkowski, M. Maeurer, and M. Reilly. 2007. Validation of peptide epitope microarray experiments and extraction of quality data. J. Immunol. Methods 3281-13. [DOI] [PubMed] [Google Scholar]
- 24.Ndung'u, T., S. Gaseitsiwe, E. Sepako, F. Doualla-Bell, T. Peter, S. Kim, I. Thior, V. A. Novitsky, and M. Essex. 2005. Major histocompatibility complex class II (HLA-DRB and -DQB) allele frequencies in Botswana: association with human immunodeficiency virus type 1 infection. Clin. Diagn. Lab. Immunol. 121020-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman, M. J., B. Livingston, D. M. McKinney, R. W. Chesnut, and A. Sette. 2002. T-lymphocyte epitope identification and their use in vaccine development for HIV-1. Front. Biosci. 7d1503-d1515. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen, M., C. Lundegaard, T. Blicher, B. Peters, A. Sette, S. Justesen, S. Buus, and O. Lund. 2008. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput. Biol. 4e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen, M., C. Lundegaard, and O. Lund. 2007. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinform. 8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novak, E. J., A. W. Liu, G. T. Nepom, and W. W. Kwok. 1999. MHC class II tetramers identify peptide-specific human CD4(+) T cells proliferating in response to influenza A antigen. J. Clin. Investig. 104R63-R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panina-Bordignon, P., A. Tan, A. Termijtelen, S. Demotz, G. Corradin, and A. Lanzavecchia. 1989. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur. J. Immunol. 192237-2242. [DOI] [PubMed] [Google Scholar]
- 30.Scharn, D., H. Wenschuh, U. Reineke, J. Schneider-Mergener, and L. Germeroth. 2000. Spatially addressed synthesis of amino- and amino-oxy-substituted 1,3,5-triazine arrays on polymeric membranes. J. Comb. Chem. 2361-369. [DOI] [PubMed] [Google Scholar]
- 31.Schena, M. 2005. Protein microarray, vol. 1. Jones and Bartlett, Sudbury, MA.
- 32.Sercarz, E. E., and E. Maverakis. 2003. Mhc-guided processing: binding of large antigen fragments. Nat. Rev. Immunol. 3621-629. [DOI] [PubMed] [Google Scholar]
- 33.Sette, A., L. Adorini, S. M. Colon, S. Buus, and H. M. Grey. 1989. Capacity of intact proteins to bind to MHC class II molecules. J. Immunol. 1431265-1267. [PubMed] [Google Scholar]
- 34.Sidney, J., S. Southwood, C. Oseroff, M. F. del Guercio, A. Sette, and H. M. Grey. 2001. Measurement of MHC/peptide interactions by gel filtration, chapter 18, unit 18.3. Current protocols in immunology. John Wiley & Sons, Hoboken, NJ. [DOI] [PubMed]
- 35.Siebold, C., B. E. Hansen, J. R. Wyer, K. Harlos, R. E. Esnouf, A. Svejgaard, J. I. Bell, J. L. Strominger, E. Y. Jones, and L. Fugger. 2004. Crystal structure of HLA-DQ0602 that protects against type 1 diabetes and confers strong susceptibility to narcolepsy. Proc. Natl. Acad. Sci. USA 1011999-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinigaglia, F., M. Guttinger, J. Kilgus, D. M. Doran, H. Matile, H. Etlinger, A. Trzeciak, D. Gillessen, and J. R. Pink. 1988. A malaria T-cell epitope recognized in association with most mouse and human MHC class II molecules. Nature 336778-780. [DOI] [PubMed] [Google Scholar]
- 37.Southwood, S., J. Sidney, A. Kondo, M. F. del Guercio, E. Appella, S. Hoffman, R. T. Kubo, R. W. Chesnut, H. M. Grey, and A. Sette. 1998. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 1603363-3373. [PubMed] [Google Scholar]
- 38.Stern, L. J., J. H. Brown, T. S. Jardetzky, J. C. Gorga, R. G. Urban, J. L. Strominger, and D. C. Wiley. 1994. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature 368215-221. [DOI] [PubMed] [Google Scholar]
- 39.Stone, J. D., W. E. Demkowicz, Jr., and L. J. Stern. 2005. HLA-restricted epitope identification and detection of functional T cell responses by using MHC-peptide and costimulatory microarrays. Proc. Natl. Acad. Sci. USA 1023744-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong, J. C., T. W. Tan, and S. Ranganathan. 2007. Methods and protocols for prediction of immunogenic epitopes. Brief. Bioinform. 896-108. [DOI] [PubMed] [Google Scholar]
- 41.Topalian, S. L. 1994. MHC class II restricted tumor antigens and the role of CD4+ T cells in cancer immunotherapy. Curr. Opin. Immunol. 6741-745. [DOI] [PubMed] [Google Scholar]
- 42.Wahlstrom, J., J. Dengjel, B. Persson, H. Duyar, H. G. Rammensee, S. Stevanovideltac, A. Eklund, R. Weissert, and J. Grunewald. 2007. Identification of HLA-DR-bound peptides presented by human bronchoalveolar lavage cells in sarcoidosis. J. Clin. Investig. 1173576-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walter, W., C. Scheuer, M. Loos, T. E. Reichert, and M. J. Maeurer. 2001. H2-Mbeta 1 and H2-Mbeta 2 heterodimers equally promote clip removal in I-A(q) molecules from autoimmune-prone DBA/1 mice. J. Biol. Chem. 27611086-11091. [DOI] [PubMed] [Google Scholar]
- 44.Wang, P., J. Sidney, C. Dow, B. Mothe, A. Sette, and B. Peters. 4 April 2008, posting date. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 4e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams, F., A. Meenagh, R. Single, M. McNally, P. Kelly, M. P. Nelson, D. Meyer, A. Lancaster, G. Thomson, and D. Middleton. 2004. High resolution HLA-DRB1 identification of a Caucasian population. Hum. Immunol. 6566-77. [DOI] [PubMed] [Google Scholar]
- 46.Zhan, X., K. S. Slobod, S. Surman, S. A. Brown, T. D. Lockey, C. Coleclough, P. C. Doherty, and J. L. Hurwitz. 2003. Limited breadth of a T-helper cell response to a human immunodeficiency virus envelope protein. J. Virol. 774231-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, S., K. Udaka, J. Sidney, A. Sette, K. F. Aoki-Kinoshita, and H. Mamitsuka. 2006. Improving MHC binding peptide prediction by incorporating binding data of auxiliary MHC molecules. Bioinformatics 221648-1655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.