Abstract
A total of 233 isolates of Pasteurella multocida were obtained from 2,912 cases of clinical respiratory disease in pigs in China, giving an isolation rate of 8.0%. Serogroup A P. multocida isolates were isolated from 92 cases (39.5%), and serogroup D isolates were isolated from 128 cases (54.9%); 12 isolates (5.2%) were untypeable. P. multocida was the fourth most frequent pathogenic bacterium recovered from the respiratory tract, after Streptococcus suis, Haemophilus parasuis, and Escherichia coli. All isolates were characterized for their susceptibilities to 20 antibiotics and the presence of 19 genes for virulence factors (VFs). The frequency of antimicrobial resistance among P. multocida isolates from swine in China was higher than that reported among P. multocida isolates from swine in from other countries, and 93.1% of the isolates showed multiple-drug resistance. There was a progressive increase in the rate of multiresistance to more than seven antibiotics, from 16.2% in 2003 to 62.8% in 2007. The resistance profiles suggested that cephalosporins, florfenicol, and fluoroquinolones were the drugs most likely to be active against P. multocida. Use of PCR showed that colonization factors (ptfA, fimA, and hsf-2), iron acquisition factors, sialidases (nanH), and outer membrane proteins occurred in most porcine strains. The VFs pfhA, tadD, toxA, and pmHAS were each present in <50% of strains. The various VFs exhibited distinctive associations with serogroups: concentrated in serogroup A, concentrated in serogroup D, or occurring jointly in serogroups A and D. These findings provide novel insights into the epidemiological characteristics of porcine P. multocida isolates and suggest that the potential threat of such multiresistant bacteria in food-producing animals should not be neglected.
Pasteurella multocida is an important cause of pneumonia and atrophic rhinitis in pigs and is responsible for significant losses on large farms worldwide (11, 16, 30). Strains of P. multocida are grouped into five capsular serogroups (serogroups A, B, D, E, and F) and are further classified into 16 somatic serotypes (serotypes 1 to 16), which are primarily based on lipopolysaccharide antigens (22, 31, 34). To date, only serogroups A, B, and D have been recovered from swine (11, 33). Together with Bordetella bronchiseptica, toxigenic strains of P. multocida serogroups A and D can cause atrophic rhinitis (4, 11). Both toxigenic and nontoxigenic strains of serogroups A and D can cause pneumonic pasteurellosis (15, 30), whereas isolates of serogroup B cause hemorrhagic septicemia and are less frequently associated with pigs (16, 33). Since pasteurellosis was identified as one of the most important zoonoses in 1959 (3), P. multocida has been reported to be the cause of a series of outbreaks, especially in Australia, Vietnam, Canada, and the United States (4, 15, 30, 33). In addition, this organism is typically associated with subacute or chronic pleuritis. Both vertical and horizontal transmissions occur, with the most common route of transmission being nose-to-nose contact. Interspecies transmission of P. multocida is also thought to occur under certain conditions (12).
Although antimicrobial therapy is a widely available tool for the prevention and control of clinical infections (5, 25, 26), antibiotic resistance in pathogenic bacteria from food-producing animals and environmental sources is recognized as a global problem for public health (6, 37). Over the past decade, the high degree of resistance to common antibiotics and the worldwide emergence of multidrug resistant phenotypes have become of increasing concern (8, 36, 37). Previous studies have reported that the imprudent use of antimicrobials bears a high risk for the selection of resistant bacteria and promotes the spread of resistance genes located on plasmids, integrons, and transposons (23, 25). This has resulted in a reduction in the efficacies of the antimicrobial agents that are currently available for the treatment of infections in food-producing animals (25). Indeed, the antimicrobial classes that are commonly used for the treatment of infections in humans may be misused in animals either for therapy or for the prevention of disease, which has a large potential impact on public health (8, 36).
The pathogenicity of P. multocida is associated with various virulence factors (VFs) (18, 20, 24). The key factors that have been identified to date include the capsule and lipopolysaccharide (10, 21). The recognized VFs of this organism also include diverse adhesins (e.g., filamentous hemagglutinin, type 4 fimbriae, and Flp pilin), toxins (dermonecrotic toxin), siderophores (e.g., iron acquisition proteins), sialidases (which may enhance bacterial virulence by unmasking key host receptors and/or reducing the effectiveness of host defenses), and outer membrane proteins (e.g., OmpA, OmpH, Oma87, and PlpB) (2, 17, 18, 23, 35). These VFs facilitate the colonization and invasion of the host, the avoidance or disruption of host defense mechanisms, injury to host tissues, and/or stimulation of a noxious host inflammatory response (20, 23). Thus, the informed selection of the VFs to be targeted for the prevention of P. multocida infections requires knowledge of which VFs are prevalent in specific clinical syndromes, as may be revealed by epidemiological studies. Furthermore, it was reported that there is an obvious correlation between some VFs and capsular serogroups, with the filamentous hemagglutinin gene pfhA being associated with serogroups A, B, E, and F; the iron acquisition gene tbpA being associated with serogroups A and B; and the dermonecrotoxin toxA gene being associated with serogroup D (17). Because pathogenic behavior is predicted both by the VF repertoire and by the serogroup (20), the clonal associations of VFs must be evaluated.
The current investigation is the first large study in China of the prevalence of P. multocida in clinical samples collected from 16 provinces between 2003 and 2007. To obtain more information about the epidemiology of porcine P. multocida infection and to characterize clinical isolates, we investigated a total of 233 isolates of P. multocida that were associated with clinical disease in swine for the distributions of the capsular serogroups, the phenotypic antimicrobial resistance profiles, and the presence of 19 virulence genes.
MATERIALS AND METHODS
Clinical specimens, culture, and P. multocida screening.
Over more than 4 years (from June 2003 to September 2007), 2,912 clinical samples from pigs with clinical respiratory infections which were collected by the Clinical Microbiology Laboratory of the College of Animal Science and Veterinary Medicine, Huazhong Agricultural University, for routine pathogen identification were screened for P. multocida. The samples were plated on tryptic soy agar (Difco, Detroit, MI) containing 10 μg/ml NAD (Sigma, St. Louis, MO) and 5% bovine serum, MacConkey agar, and blood agar (5% fresh sheep blood). All plates were incubated at 37°C in air for a minimum of 48 h. After this isolation stage, the isolates were purified and cultured by standard methods for the identification of strains of bacteria, including Haemophilus parasuis, Streptococcus suis, Actinobacillus pleuropneumoniae, Bordetella bronchiseptica, Escherichia coli, and Staphylococcus aureus (7, 28). Presumptive isolates of P. multocida were confirmed by a PCR assay with primers specific for the amplification of the KMT1 gene (34).
All isolates of P. multocida were subsequently characterized biochemically by using a MicroStation system (Biolog Inc.) and their capsules were serotyped. For samples in which all isolates were identical with respect to their capsular serotype, only one colony was selected. When one sample yielded colonies with different capsular serotypes, one colony of each serotype was selected for further characterization. All isolates were freeze-dried and kept at −80°C.
Capsule typing.
The capsular types of the isolates were determined by multiplex capsule PCR typing with the capsule-specific primer pairs (primers specific for capA, capB, capD, capE, and capF) described by Townsend et al. (34). All oligonucleotides were synthesized with a DNA synthesizer (with finishing done by Sangon Biological Engineering Technology Inc., Shanghai, China). The primer sequences used in the multiplex capsule PCR typing assay for P. multocida are listed in Table 1.
TABLE 1.
Primers used for the detection of virulence-associated genes in strains of P. multocida
| Gene function and gene | Description | Directiona | Primer sequence (5′-3′) | Amplicon size (bp) |
|---|---|---|---|---|
| Adhesins | ||||
| ptfA | Type 4 fimbriae | s | TGTGGAATTCAGCATTTTAGTGTGTC | 488 |
| a | TCATGAATTCTTATGCGCAAAATCCT GCTGG | |||
| fimA | Fimbriae (from Pm70) | s | CCATCGGATCTAAACGACCTA | 866 |
| a | AGTATTAGTTCCTGCGGGTG | |||
| hsf-1 | Autotransporter adhesion (from Pm70) | s | TTGAGTCGGCTGTAGAGTTCG | 654 |
| a | ACTCTTTAGCAGTGGGGACAACCTC | |||
| hsf-2 | Autotransporter adhesion (from Pm70) | s | ACCGCAACCATGCTCTTAC | 433 |
| a | TGACTGACATCGGCGGTAC | |||
| pfhA | Filamentous hemagglutinin | s | TTCAGAGGGATCAATCTTCG | 286 |
| a | AACTCCAGT TGGTTTGTCG | |||
| tadD | Putative nonspecific tight adherence protein D | s | TCTACCCATTCTCAGCAAGGC | 416 |
| a | ATCATTTCGGGCATTCACC | |||
| Toxins | ||||
| toxA | Dermonecrotic toxin | s | CTTAGATGAGCGACAAGG | 864 |
| a | GAATGCCACACCTCTATAG | |||
| Iron acquisition | ||||
| exbB | Accessory protein Ton-dependent transport of iron | s | TTGGCTTGTGATTGAACGC | 283 |
| compounds | a | TGCAGGAATGGCGACTAA A | ||
| exbD | Accessory protein Ton-dependent transport of iron | s | CGTTCTGATTACAGCCTCTT | 247 |
| compound | a | AACGAAATCTTGGAAACTGG | ||
| tonB | Iron transporters, transport ferric-siderophore complexes | s | CGACGGTGAAACCTGAGCCA | 261 |
| a | CCGAGCGATAAGCATTGACT | |||
| hgbA | A hemoglobin-binding protein | s | TCAACGGCAGATAATCAGGG | 267 |
| a | GCGGGAATGCTGAAGATAAG | |||
| fur | Ferric uptake regulation protein | s | GTTTACCGTGTATTAGACCA | 244 |
| a | CATTACTACATTTGCCATAC | |||
| Sialidases | ||||
| nanB | Outer membrane-associated proteins, an autotransporter protein | s | CATTGCACCTAACACCTCT | 555 |
| a | GGACACTGATTGCCCTGAA | |||
| nanH | Outer membrane-associated proteins, small sialidases | s | GTGGGAACGGGAATTGTGA | 287 |
| a | ACATGCCAAGTTTGCCCTA | |||
| Hyaluronidase | ||||
| pmHAS | Hyaluronan synthase | s | TCAATGTTTGCGATAGTCCGTTAG | 430 |
| a | TGGCGAATGATCGGTGATAGA | |||
| Protectins | ||||
| ompA | Outer membrane protein A | s | CGCATAGCACTCAAGTTTCTCC | 201 |
| a | CATAAACAGATTGACCGAAACG | |||
| ompH | Outer membrane protein H | s | CGCGTATGAAGGTTTAGGT | 438 |
| a | TTTAGATTGTGCGTAGTCAAC | |||
| oma87 | Outer membrane protein 87 | s | GGCAGCGAGCAACAGATAACG | 838 |
| a | TGTTCGTCAAATGTCGGGTGA | |||
| plpB | Lipoprotein B | s | TTTGGTGGTGCGTATGTCTTCT | 282 |
| a | AGTCACTTTAGATTGTGCGTAG | |||
| Capsule serotypes | ||||
| KMT1 | Identification of all P. multocida isolates | s | ATCCGCTATTTACCCAGTGG | 460 |
| a | GCTGTAAACGAACTCGCCAC | |||
| hyaD-hyaC | Serogroup A cap gene | s | GATGCCAAAATCGCAGTCAG | 1048 |
| a | TGTTGCCATCATTGTCAGTG | |||
| bcbD | Serogroup B cap gene | s | CATTTATCCAAGCTCCACC | 758 |
| a | GCCCGAGAGTTTCAATCC | |||
| dcbF | Serogroup D cap gene | s | TTACAAAAGAAAGACTAGGAGCCC | 647 |
| a | CATCTACCCACTCAACCATATCAG | |||
| ecbJ | Serogroup E cap gene | s | TCCGCAGAAAATTATTGACTC | 512 |
| a | GCTTGCTGCTTGATTTTGTC | |||
| fcbD | Serogroup F cap gene | s | AATCGGAGAACGCAGAAATCAG | 852 |
| a | TTCCGCCGTCAATTACTCTG |
s, sense; a, antisense.
Antimicrobial susceptibility testing.
All determinations of antimicrobial MICs for the P. multocida strains were performed by the broth microdilution methods recommended by the Clinical and Laboratory Standards Institute (CLSI; formerly the NCCLS) (29). The antimicrobial agents tested included amoxicillin, cefazolin, ceftiofur, spectinomycin, kanamycin, neomycin, gentamicin, amikacin, chloramphenicol, florfenicol, erythromycin, tilmicosin, lincomycin, clindamycin, chlortetracycline, tetracycline, ciprofloxacin, sulfamethazine (sulfadimidine), trimethoprim-sulfamethoxazole, and polymyxin B. They were supplied by the National Institute for the Control of Pharmaceutical and Biological Products in Beijing, China. Determination of the MICs and their evaluation were done by use of the interpretive criteria of the CLSI (29). The MIC was defined as the lowest concentration that prevented visible growth. Ranges of susceptibility, along with the MIC50s and the MIC90s of the isolates, were recorded. The breakpoint used for ciprofloxacin was that previously used by Aarestrup et al. (1) for Danish strains of H. parasuis. For the other antimicrobials, the breakpoint values were taken from the CLSI guidelines (29). Reference strains Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 29213 were used as quality control organisms in all antimicrobial susceptibility tests. Isolates that were nonsusceptible to at least three different antibiotic classes were classified as multidrug resistant.
Detection of virulence genes by PCR.
Nineteen pairs of oligonucleotide primers to be used for the detection of VFs were designed by using the software program Prime (version 5.0; Premier Biosoft International, Palo Alto, CA) and were synthesized commercially (with finishing done by Sangon Biological Engineering Technology Inc.). The base sequences and the predicted sizes of the amplified products for the specific oligonucleotide primers used in this study are shown in Table 1. The bacterial lysates used as templates for the PCR were prepared as follows. A loopful of bacteria from a fresh overnight culture on a tryptic soy agar plate was resuspended homogeneously in 200 μl of sterile water, and the mixture was boiled at 100°C for 5 min to release the DNA and centrifuged. A 4-μl volume of the supernatant was used as a template for each 25-μl PCR mixture. The appropriate positive and negative controls for amplification were generated from clinical isolates of P. multocida by PCR carried out with a GeneAmp PCR system 9700 instrument (Applied Biosystems, Foster City, CA) and were confirmed by sequencing. The amplified products were analyzed in 0.8% agarose gels by electrophoresis, and the results were recorded with a gel documentation system. All tests were repeated three times in parallel with the relevant positive and negative controls. Discrepant results for each VF were investigated further, and samples were sequenced for gene verification.
Clinical data and statistical analysis.
Clinical data were collected by retrospective analysis of the protocols. Statistical testing was performed with SPSS software (version 12.0; SPSS Inc., Chicago, IL). Comparisons of proportions were made by two-tailed Fisher's exact test or the χ2 test. Comparisons of the prevalence of different traits within the same population were made by McNemar's test. Aggregate VF scores were compared by the Mann-Whitney U test. P values of <0.05 were considered statistically significant.
RESULTS
Prevalence of P. multocida in porcine clinical samples.
We analyzed clinical specimens from diseased pigs with pneumonia or atrophic rhinitis from 16 provinces in China for the presence of P. multocida. Strains of P. multocida were detected in 233 (8.0%) of the 2,912 cases investigated, and the isolation rate at different time points (years) ranged from 6.4 to 10.2% (Table 2). Isolates of P. multocida of capsular type A were obtained from 92 cases (39.5%), capsular type D strains were isolated from 128 cases (54.9%), and 1 isolate was identified as capsular type B, whereas 12 isolates were untypeable. Capsular types E and F were not detected in the population sampled. All these isolates gave positive results by the PCR assays with primers specific for P. multocida (34).
TABLE 2.
Isolation of bacterial species and the prevalence of P. multocida in clinical samples from China from June 2003 to September 2007
| Yr | No. of samples analyzed | No. of samples from which the following bacterial species were isolateda:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P. multocida | S. suis | H. parasuis | E. coli | B. bronchiseptica | S. aureus | A. pleuropneumoniae | Othersb | Nonec | ||
| 2007 | 594 | 49 (33) | 167 (15) | 132 (15) | 87 (11) | 42 (5) | 26 (2) | 2 | 112 (9) | 61 |
| 2006 | 726 | 74 (46) | 214 (22) | 156 (23) | 95 (11) | 69 (5) | 31 (1) | 2 | 137 (6) | 86 |
| 2005 | 493 | 35 (17) | 126 (9) | 91 (13) | 68 (3) | 35 (3) | 17 | 5 (3) | 83 (7) | 55 |
| 2004 | 502 | 32 (14) | 97 (7) | 64 (8) | 55 (5) | 25 (4) | 9 | 11 (3) | 59 (3) | 48 |
| 2003 | 597 | 43 (15) | 101 (8) | 55 (8) | 62 (4) | 39 (6) | 10 (1) | 6 (3) | 64 (3) | 52 |
| Total no. (%) of samples from which bacterial species were isolated | 2,912 | 233 (8.0) | 705 (24.2) | 498 (17.1) | 367 (12.6) | 210 (7.2) | 93 (3.2) | 26 (0.9) | 455 (15.6) | 302 (10.3) |
| Total no. (%) of samples containing other pathogenic bacteria as well as P. multocida | 125 | 61 (48.8) | 67 (53.6) | 34 (27.2) | 23 (18.4) | 4 (3.2) | 9 (7.2) | 28 (22.4) | ||
Values in parentheses in the rows for the years 2003 to 2007 indicate the number of samples containing other pathogenic bacteria among 125 samples coinfected with P. multocida.
Others include, for example, Salmonella enterica, P. aeruginosa, Staphylococcus epidermidis, Actinobaculum suis, Streptococcus equinus, and Erysipelothrix rhusiopathiae.
None, no bacteria were isolated from the clinical samples.
After Streptococcus suis, Haemophilus parasuis, and Escherichia coli, P. multocida was the bacterial pathogen that was the fourth most frequently isolated from clinical porcine specimens in this study (Table 2). Streptococcus suis was isolated from 24.2% of the study samples, Haemophilus parasuis was isolated from 17.1%, Escherichia coli was isolated from 12.6%, Bordetella bronchiseptica was isolated from 7.2%, Staphylococcus aureus was isolated from 3.2%, and Actinobacillus pleuropneumoniae was isolated from 0.9%. The simultaneous detection of P. multocida and other species of bacteria pathogenic for pigs occurred in 125 specimens. Haemophilus parasuis (53.6%), Streptococcus suis (48.8%), Escherichia coli (27.2%) and Bordetella bronchiseptica (18.4%) were the agents that were the most frequently found in coinfections with P. multocida. Eleven toxigenic strains of P. multocida of serogroup D were isolated from 37 samples of nasal swabs and lungs lesions from pigs with typical clinical signs of atrophic rhinitis. Six strains of Bordetella bronchiseptica and five strains of Pseudomonas aeruginosa were also cultured from the same samples.
All infections with P. multocida detected in this study were from territorial outbreaks on pig farms, and the sources of infection were not identified in most cases. Strains of P. multocida were isolated throughout the year without seasonal variation, and 62.2% of the organisms were detected from growing pigs 60 to 110 days old (145 cases). Eleven toxigenic strains of P. multocida were collected from growing pigs 80 to 100 days old from June to August.
Antimicrobial susceptibility.
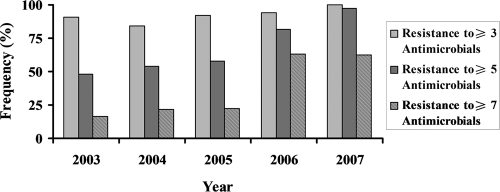
Two hundred thirty-three isolates of P. multocida recovered from diseased swine were tested for resistance to 20 antibiotics (Table 3). The most prevalent phenotypes detected were resistance to lincomycin (96.6%), sulfamethazine (85.4%), amoxicillin (80.3%), clindamycin (80.3%), trimethoprim-sulfamethoxazole (74.2%), chlortetracycline (65.2%), and tetracycline (58.0%), followed by tilmicosin (28.3%), amikacin (14.2%), gentamicin (13.7%), kanamycin (12.8%), and spectinomycin (12.0%). Less than 10% of the isolates were resistant to erythromycin or chloramphenicol (6.0 and 2.6%, respectively). No resistance to cefazolin, ceftiofur, florfenicol, or ciprofloxacin could be detected. The MIC90s of neomycin and polymyxin B for the P. multocida isolates tested were 32 μg/ml and 4 μg/ml, respectively. The proportion of P. multocida isolates categorized as resistant could not be evaluated in this study because the breakpoints of neomycin and polymyxin B for veterinary use have not been determined according to the CLSI criteria (29). In addition, it was observed that 98.6% of the isolates were resistant to at least one antibiotic and 93.1% were multiresistant (resistant to from 3 to 10 antibiotics). Multiresistance was predominant in isolates of serogroup D, including toxigenic P. multocida strains. Resistance to amoxicillin, chlortetracycline and tetracycline, lincomycin and clindamycin, and sulfamethazine and trimethoprim-sulfamethoxazole was the common feature of these multiresistant isolates. The percentage of isolates with resistant to at least three antimicrobials was equally high in all years from 2003 to 2007 (Fig. 1). Isolates resistant to more than five antimicrobials became more frequent over time. The prevalence increased from 47.8% in 2003, 54.1% in 2004, and 57.6% in 2005 to 81.6% in 2006 and 97.1% in 2007. It is important to note that the proportion of isolates resistant to more than seven antimicrobials increased approximately fourfold between 2003 and 2007, from 16.2% to 62.8% (P < 0.05).
TABLE 3.
MICs for 20 antimicrobial agents against 233 strains of P. multocida
| Antimicrobiala | No. of isolates with MIC of (μg/ml):
|
Breakpoint MIC (μg/ml)b | MIC50 (μg/ml) | MIC90 (μg/ml) | % Resistancec | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | ≥1,024 | |||||
| AMX | 4 | 1 | 2 | 1 | 38 | 49 | 62 | 54 | 12 | 7 | 3 | 32 | 64 | 128 | 80.3 | |||||
| FAM | 1 | 1 | 8 | 101 | 96 | 18 | 6 | 1 | 1 | 32 | 0.5 | 0.5 | 0 | |||||||
| XLN | 165 | 58 | 6 | 1 | 3 | 8 | ≤0.03 | 0.06 | 0 | |||||||||||
| SPT | 14 | 36 | 71 | 84 | 15 | 7 | 4 | 2 | 128 | 32 | 128 | 12.0 | ||||||||
| KAN | 17 | 34 | 66 | 58 | 21 | 7 | 18 | 10 | 2 | 64 | 4 | 64 | 12.8 | |||||||
| NEO | 14 | 18 | 62 | 48 | 45 | 26 | 9 | 11 | NDc | 8 | 32 | ND | ||||||||
| GEN | 2 | 11 | 30 | 62 | 56 | 40 | 23 | 7 | 2 | 16 | 4 | 16 | 13.7 | |||||||
| AMK | 2 | 2 | 23 | 76 | 49 | 48 | 17 | 14 | 2 | 64 | 16 | 64 | 14.2 | |||||||
| CHL | 13 | 99 | 85 | 12 | 9 | 3 | 2 | 4 | 5 | 1 | 32 | 0.5 | 2 | 2.6 | ||||||
| FFC | 8 | 48 | 107 | 54 | 9 | 3 | 4 | 8 | 0.25 | 0.5 | 0 | |||||||||
| ERY | 16 | 39 | 18 | 36 | 54 | 56 | 12 | 1 | 1 | 8 | 2 | 4 | 6.0 | |||||||
| TYL | 2 | 19 | 17 | 10 | 33 | 36 | 28 | 22 | 24 | 17 | 12 | 9 | 4 | 32 | 4 | 128 | 28.3 | |||
| LIN | 1 | 7 | 22 | 102 | 71 | 25 | 5 | 4 | 8 | 32 | 96.6 | |||||||||
| CLI | 1 | 1 | 12 | 32 | 61 | 61 | 42 | 18 | 5 | 4 | 4 | 16 | 80.3 | |||||||
| CTET | 5 | 14 | 28 | 34 | 36 | 34 | 33 | 25 | 20 | 2 | 2 | 16 | 16 | 128 | 65.2 | |||||
| TET | 1 | 13 | 25 | 41 | 18 | 22 | 36 | 32 | 27 | 8 | 10 | 16 | 16 | 128 | 58.0 | |||||
| CIP | 91 | 84 | 31 | 22 | 3 | 1 | 1 | 4 | 0.06 | 0.25 | 0 | |||||||||
| SDM | 2 | 9 | 12 | 11 | 67 | 132 | 512 | ≥1,024 | ≥1,024 | 85.4 | ||||||||||
| SXT | 1 | 2 | 5 | 7 | 24 | 21 | 43 | 130 | 512 | ≥1,024 | ≥1,024 | 74.2 | ||||||||
| PB | 2 | 5 | 12 | 26 | 39 | 51 | 37 | 39 | 16 | 3 | 3 | ND | 1 | 4 | ND | |||||
AMX, amoxicillin; FAM, cefazolin; XLN, ceftiofur; SPT, spectinomycin; KAN, kanamycin; NEO, neomycin; GEN, gentamicin; AMK, amikacin; CHL, chloramphenicol; FFC, florfenicol; ERY, erythromycin; TYL, tilmicosin; LIN, lincomycin; CLI, clindamycin; CTET, chlortetracycline; TET, tetracycline; CIP, ciprofloxacin; SDM, sulfamethazine; SXT, trimethoprim-sulfamethoxazole; PB, polymyxin B.
The values, with the exception of those for ciprofloxacin, are based on CLSI standards.
ND, not determined.
FIG. 1.
Distribution by year of P. multocida isolates that showed phenotypes of multidrug resistance to the antimicrobial agents tested. All determinations of antimicrobial MICs with the P. multocida isolates were performed by broth microdilution methods recommended by the CLSI (29). The breakpoint used for ciprofloxacin was those previously used by Aarestrup et al. (1) for Danish strains of H. parasuis. For the other antimicrobials tested, breakpoint values were taken from the CLSI guidelines (29). Isolates that were nonsusceptible to at least three different antibiotic classes were classified as multidrug resistant. The percentage of isolates resistant to at least three antimicrobials was equally high between 2003 and 2007. The proportion of isolates resistant to more than five antimicrobials became more frequent, with a progressive increase from 47.8% in 2003 to 97.1% in 2007. The proportion of isolates resistant to more than seven antimicrobials increased approximately fourfold between the years 2003 and 2007, from 16.2% to 62.8% (P < 0.05).
Distribution of virulence genes.
Among the 233 porcine P. multocida isolates, the 19 virulence gene regions ranged in prevalence from 4.7% (toxA) to 100% (ompA). Multiple adhesins (including ptfA, fimA, and hsf-2), all iron acquisition factors (exbB, exbD, tonB, hgbA, and fur), nanH, and various outer membrane proteins (ompA, ompH, oma87, and plpB) were each found to occur in over 90% of the strains (Table 4). This shows that these virulence genes are highly prevalent in porcine isolates of P. multocida. Of the adhesin-encoding genes studied, hsf-2 (99.1%) was more prevalent than hsf-1 (67.0%; P < 0.001, McNemar's test), and tadD was more prevalent than pfhA (P < 0.001, McNemar's test). However, there was no statistically significant difference in the prevalence of hsf-1 and tadD (67.0% and 43.3%, respectively; P > 0.05, McNemar's test). Of the sialidase-encoding genes studied, nanH (97.0%) was more prevalent than nanB (81.5%; P < 0.001, McNemar's test). Of note was the substantial prevalence of the gene for hyaluronan synthase (pmHAS) and the low prevalence of the virulence genes toxA (4.7%) and pfhA (15%). The tbpA determinant was not detected in any of the 233 clinical strains studied.
TABLE 4.
Distribution of VFs according to capsule serotypes among 233 porcine isolates of P. multocida
| Associated VF genea | Total no. (% of 233) with trait | No. (%) of VFs within the following capsule serotypes:
|
P value | ||
|---|---|---|---|---|---|
| capA (n = 92) | capD (n = 128) | Other (n = 13) | |||
| ptfA | 218 (93.6) | 82 (89.1) | 123 (96.1) | 13 (100) | |
| fimA | 231 (99.1) | 90 (97.8) | 128 (100) | 13 (100) | |
| hsf-1 | 156 (67.0) | 35 (38.0) | 119 (93.0)d | 2 (15.4) | <0.001 |
| hsf-2 | 231 (99.1) | 91 (98.9) | 127 (99.2) | 13 (100) | |
| pfhA | 35 (15.0) | 23 (25.0)d | 4 (3.1) | 8 (61.5)d | <0.001 |
| tadD | 101 (43.3) | 85 (92.4)d | 13 (10.2) | 3 (23) | <0.001 |
| toxA | 11 (4.7) | 0 (0) | 11 (8.9)d | 0 (0) | 0.009 |
| exbB | 231 (99.1) | 91 (98.9) | 127 (99.2) | 13 (100) | |
| exbD | 231 (99.1) | 91 (98.9) | 127 (99.2) | 13 (100) | |
| tonB | 228 (97.9) | 87 (94.6) | 128 (100) | 13 (100) | |
| hgbA | 225 (96.6) | 90 (97.8) | 125 (97.7) | 10 (76.9)b | <0.001 |
| fur | 216 (92.7) | 89 (96.7) | 118 (92.2) | 9 (69.2)c | 0.002 |
| nanB | 190 (81.5) | 60 (65.2) | 122 (95.3)d | 8 (61.5) | <0.001 |
| nanH | 226 (97.0) | 91 (98.9) | 123 (96.1) | 12 (92.3) | |
| pmHAS | 105 (45.1) | 77 (83.7)d | 19 (14.8) | 9 (69.2)d | <0.001 |
| ompA | 233 (100) | 92 (100) | 128 (100) | 13 (100) | |
| ompH | 217 (93.1) | 88 (95.7) | 121 (94.5) | 12 (92.3) | |
| oma87 | 220 (94.4) | 83 (90.2) | 125 (97.7) | 12 (92.3) | |
| plpB | 231 (99.1) | 91 (98.9) | 128 (100) | 12 (92.3) | |
All genes were detected by PCR.
P < 0.05 for the indicated group compared with the results for all other strains (negative association).
P <0.01 for the indicated group compared with the results for all other strains (negative association).
P < 0.001 for the indicated group compared with the results for all other strains.
The distribution of the virulence-associated genes among capsular serogroups, which was compared by Fisher's exact test or the χ2 test, is presented in Table 4. As anticipated, the great majority of VFs, including ptfA, fimA, hsf-2, exbB, exbD, tonB, nanH, ompA, ompH, oma87, and plpB, were equally distributed in each capsular serogroup. However, when each serogroup was compared with all other capsular serogroups combined, tadD was significantly associated with serogroup A, hsf-1 and nanB were significantly associated with serogroup D, and hgbA and fur were significantly associated with serogroups A and D. The pfhA and pmHAS genes were less common in isolates of serogroup D than in isolates of the other serogroups. It was noted that the toxA gene, which is involved in the pathogenesis of progressive atrophic rhinitis in pigs, was found in only 11 strains, and it was strictly restricted to strains belonging to capsular serogroup D. The different capsular serogroups exhibited disparate median aggregate VF scores: serogroup A, 15.6 (range, 12 to 18); serogroup D, 15.0 (range, 9 to 17); serogroup B, 14.0 (only one strain); and the nonaligned strains (nonserogroup strains), 14.1 (range, 11 to 16). These results did not differ significantly (for all comparisons, P > 0.05, Mann-Whitney U test).
DISCUSSION
Pasteurellosis is one of the most common diseases of grower and finisher pigs worldwide. It is widely accepted that specific serotypes and pathotypes of P. multocida strains are responsible for most respiratory disease syndromes in pigs that are associated with pneumonia, atrophic rhinitis, and/or mycoplasma infection (4, 11, 30). However, the distribution and prevalence of serotypes and pathotypes can vary considerably from region to region and over time in a given region. This is the first study in China of a large collection of isolates of P. multocida obtained from pigs with clinical signs of respiratory infection. Our findings suggest that strains of P. multocida are widely prevalent on pig farms, and we have confirmed that on the Chinese mainland, infections caused by P. multocida strains of serogroup D are more common than those caused by strains of serogroup A (P < 0.01). Similar results were reported by Ewers et al. (17) in Germany (58.1% versus 34.9%) and Chandrasekaran and Yeap (9) in Malaysia (45% versus 20%). In contrast, in the United States (30) and England and Wales (11), the prevalence of strains of capsular serogroup D is lower than that of serogroup A. It is interesting that although untypeable strains are generally uncommon in pigs (11), only 12 untypeable isolates associated with pneumonic pigs were isolated during the period of investigation. We do not know whether the presence of untypeable isolates is attributable to a lack of available tests.
Atrophic rhinitis is seldom reported in China; but our investigations have confirmed the presence of atrophic rhinitis in Henan, Shandong, Fujian, Hainan, and Hubei Provinces by the isolation of P. multocida from clinical cases and the use of assays for toxigenicity. In agreement with the findings of previous studies (4, 11), toxigenic P. multocida strains of serogroup D may play a more important role in atrophic rhinitis in China than toxigenic strains of serogroup A. During the study period, other bacterial species, including Haemophilus parasuis and Streptococcus suis, were often coisolated with pathogenic P. multocida strains from the same sample. Although it is not easy to distinguish whether P. multocida is a primary or a secondary pathogen in herds with mixed infections, the fact that various bacterial species may coexist in a given herd should be considered when attempts are made to control disease outbreaks (7).
Treatment for infections with P. multocida commonly includes broad-spectrum antimicrobials (5, 25, 26). The findings of our antibiotic susceptibility studies, like the findings of Kehrenberg et al. (25) in France, Salmon et al. (32) in North America, and Yoshimura et al. (38) in Japan, indicated that cephalosporins (cefazolin, ceftiofur), florfenicol, and fluoroquinolones (ciprofloxacin) were the most active drugs. The aminoglycoside antibiotics usually showed poor activity against P. multocida, as reported by Gutiérrez Martin and Rodríguez Ferri (19) in Spain and Yoshimura et al. (38) in Japan; however, in the present study, spectinomycin, kanamycin, gentamicin, and amikacin exhibited moderate activity against all strains tested. The average prevalence of resistance to conventional antibiotics, including amoxicillin, lincomycin, clindamycin, chlortetracycline, tetracycline, sulfamethazine, and trimethoprim-sulfamethoxazole, among the P. multocida isolates was found to be in excess of 60% for each antibiotic. Therefore, preventive and therapeutic effects on porcine P. multocida strains should no longer be expected from these antibiotics. Furthermore, the increased incidence of multidrug-resistant pathogenic bacteria has been widely reported in the last decade (8, 25, 37). This is presumably attributable at least in part to the use of antibiotic additives in animal feed and the extensive use of antimicrobial agents in veterinary medicine. Here we have shown that P. multocida exhibited a rapid increase in the rate of resistance to a large number of antimicrobial agents. This revealed that a high prevalence of multiple-drug resistance exists among isolates of P. multocida from pigs. If this situation continues, there will be no effective antibiotic therapeutic reserve for some bacterial infections. The implications of a large reservoir of multiresistant organisms, particularly P. multocida, which is not host specific, with resistance that is potentially transferable among livestock species are obvious (3, 12, 24). Therefore, the use of antimicrobial agents in food animals in ways that minimize the emergence of resistance not only in target pathogens but also in zoonotic bacteria is warranted in the future for the protection of public health.
Although the molecular basis of the pathogenicity and host specificity of P. multocida is not well understood, several studies have reported that a number of VFs are correlated with the pathogenic mechanisms (20, 23). The present study has provided novel epidemiological information on the prevalence and distribution of the various VFs of porcine strains of P. multocida. Consistent with previous observations (17), the prevalence of 11 of the 19 VF genes examined, which encode colonization factors (ptfA, fimA, and hsf-2), iron acquisition factors, sialidases (nanH), and outer membrane proteins, were broadly characteristic of the three categories of isolates (serogroup A, serogroup D, and others [1 serogroup B isolate and 12 untypeable isolates]). These distribution patterns would support some lines of evidence that suggest that factors involved in cross-protection may potentially serve as vaccine candidates that can elicit homologous protective immunity against all serotypes of P. multocida (2, 20, 35). However, certain VFs varied significantly among the different serogroups. For example, hsf-1, which has been described to be an autotransporter adhesin in a common avian clone, Pm70 (27), was more frequently seen in serogroup D, whereas tadD, which has been described as putative nonspecific tight adherence protein D in Pm70 (27), was concentrated significantly in serogroup A. pfhA, which governs the adherence of Bordetella pertussis to host cells and which plays a role in the virulence of P. multocida (18, 20, 27), showed a low prevalence in strains of serogroup D compared with its prevalence in serogroup A or untypeable isolates. Various hyaluronan synthases have been described in the last 5 years (13, 14). Preliminary data from a Southern blot analysis suggested that the P. multocida serogroup A hyaluronan synthase PmHAS and the P. multocida serogroup D hyaluronan synthase PmHS1 were not similar at the DNA level (13). However, our study showed that PmHAS not only was prevalent in serogroup A strains but also was found in other serogroups of porcine P. multocida. It seems probable that different VFs have entered P. multocida strains independently at multiple different times in the evolutionary history of the species and at multiple positions within the phylogenetic tree. Moreover, the observed distribution pattern suggests that it is likely that the acquisition of certain VFs has led to divergent patterns of vertical inheritance and horizontal transmission (via pathogenicity-associated islands, plasmids, and transposons) within the P. multocida population.
In conclusion, given that it is a pathogenic microorganism that is not host specific, we believe that the occurrence of P. multocida in food-producing animals should not be forgotten. In China, as in many other countries, strains of P. multocida have frequently been isolated from pigs, and they represent a significant cause of territorial outbreaks of respiratory infections. The high prevalence of multiresistant strains of P. multocida in pigs and the association of such strains with serious disease strongly suggest that more attention should be paid to the prudent use of antimicrobials and to vaccination. Nowadays, many key VFs of P. multocida are slowly being identified. Further work is required to elucidate the mechanisms of pathogenesis and to determine unequivocally the role of these factors in immunity to pasteurellosis.
Acknowledgments
We thank Xiangdong Liu (Wuhan, China) for support with the statistical analyses and Feng Liu for help with the capsule typing and biochemical assays.
This study was supported by grants from the National Basic Research Program (973 Program; no. 2006CB504404) and the Science and Technology Brainstorm Project of Hubei Province, China (Emergency Program, 2006-2008).
Footnotes
Published ahead of print on 21 January 2009.
REFERENCES
- 1.Aarestrup, F. M., A. M. Seyfarth, and Ø. Angen. 2004. Antimicrobial susceptibility of Haemophilus parasuis and Histophilus somni from pigs and cattle in Denmark. Vet. Microbiol. 101:143-146. [DOI] [PubMed] [Google Scholar]
- 2.Adler, B., D. Bulach, J. Chung, S. Doughty, M. Hunt, K. Rajakumar, M. Serrano, A. van Zanden, Y. Zhang, and C. Ruffolo. 1999. Candidate vaccine antigens and genes in Pasteurella multocida. J. Biotechnol. 7383-90. [DOI] [PubMed] [Google Scholar]
- 3.Arashima, Y., and K. Kumasaka. 2005. Pasteurellosis as zoonosis. Intern. Med. 44692-693. [DOI] [PubMed] [Google Scholar]
- 4.Bäckström, L. R., T. A. Brim, and M. T. Collins. 1988. Development of turbinate lesions and nasal colonization of Bordetella bronchiseptica and Pasteurella multocida during long term exposure of healthy pigs affected by atrophic rhinitis. Can. J. Vet. Res. 5223-29. [PMC free article] [PubMed] [Google Scholar]
- 5.Brogden, K. A., G. Nordholm, and M. Ackermann. 2007. Antimicrobial activity of cathelicidins BMAP28, SMAP28, SMAP29, and PMAP23 against Pasteurella multocida is more broad-spectrum than host species specific. Vet. Microbiol. 11976-81. [DOI] [PubMed] [Google Scholar]
- 6.Bronzwaer, S. L., O. Cars, U. Buchholz, S. Mölstad, W. Goettsch, I. K. Veldhuijzen, J. L. Kool, M. J. Sprenger, and J. E. Degener. 2002. A European study on the relationship between antimicrobial use and antimicrobial resistance. Emerg. Infect. Dis. 8278-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, X., H. Chen, P. J. Blackall, Z. Yin, L. Wang, Z. Liu, and M. Jin. 2005. Serological characterization of Haemophilus parasuis isolates from China. Vet. Microbiol. 111231-236. [DOI] [PubMed] [Google Scholar]
- 8.Caprioli, A., L. Busani, J. L. Martel, and R. Helmuth. 2000. Monitoring of antibiotic resistance in bacteria of animal origin: epidemiological and microbiological methodologies. Int. J. Antimicrob. Agents 14295-301. [DOI] [PubMed] [Google Scholar]
- 9.Chandrasekaran, S., and P. C. Yeap. 1982. Pasteurella multocida in pigs: the serotypes and the assessment of their virulence in mice. Br. Vet. J. 138332-336. [DOI] [PubMed] [Google Scholar]
- 10.Chung, J. Y., I. Wilkie, J. D. Boyce, K. M. Townsend, A. J. Frost, M. Ghoddusi, and B. Adler. 2001. Role of capsule in the pathogenesis of fowl cholera caused by Pasteurella multocida serogroup A. Infect. Immun. 692487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, R. L., R. MacCorquodale, S. Baillie, and B. Caffrey. 2003. Characterization and comparison of Pasteurella multocida strains associated with porcine pneumonia and atrophic rhinitis. J. Med. Microbiol. 5259-67. [DOI] [PubMed] [Google Scholar]
- 12.Davies, R. L., R. MacCorquodale, and S. Reilly. 2004. Characterisation of bovine strains of Pasteurella multocida and comparison with isolates of avian, ovine and porcine origin. Vet. Microbiol. 99145-158. [DOI] [PubMed] [Google Scholar]
- 13.DeAngelis, P. L., and C. L. White. 2002. Identification and molecular cloning of a heparosan synthase from Pasteurella multocida type D. J. Biol. Chem. 2777209-7213. [DOI] [PubMed] [Google Scholar]
- 14.DeAngelis, P. L., and C. L. White. 2004. Identification of a distinct, cryptic heparosan synthase from Pasteurella multocida types A, D, and F. J. Bacteriol. 1868529-8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djordjevic, S. P., G. J. Eamens, H. Ha, M. J. Walker, and J. C. Chin. 1998. Demonstration that Australian Pasteurella multocida isolates from sporadic outbreaks of porcine pneumonia are non-toxigenic (toxA−) and display heterogeneous DNA restriction endonuclease profiles compared with toxigenic isolates from herds with progressive atrophic rhinitis. J. Med. Microbiol. 47679-688. [DOI] [PubMed] [Google Scholar]
- 16.Dziva, F., A. P. Muhairwa, M. Bisgaard, and H. Christensen. 2008. Diagnostic and typing options for investigating diseases associated with Pasteurella multocida. Vet. Microbiol. 1281-22. [DOI] [PubMed] [Google Scholar]
- 17.Ewers, C., A. Lübke-Becker, A. Bethe, S. Kiebling, M. Filter, and L. H. Wieler. 2006. Virulence genotype of Pasteurella multocida strains isolated from different hosts with various disease status. Vet. Microbiol. 114304-317. [DOI] [PubMed] [Google Scholar]
- 18.Fuller, T. E., M. J. Kennedy, and D. E. Lowery. 2000. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb. Pathog. 2925-38. [DOI] [PubMed] [Google Scholar]
- 19.Gutiérrez Martin, C. B., and E. F. Rodríguez Ferri. 1993. In vitro susceptibility of Pasteurella multocida subspecies multocida strains isolated from swine to 42 antimicrobial agents. Zentralbl. Bakteriol. Parasitenkd. Infektkrankh. Hyg. Abt. 1 Orig. 279387-393. [DOI] [PubMed] [Google Scholar]
- 20.Harper, M., J. D. Boyce, and B. Adler. 2006. Pasteurella multocida pathogenesis: 125 years after Pasteur. FEMS Microbiol. Lett. 2651-10. [DOI] [PubMed] [Google Scholar]
- 21.Harper, M., A. Cox, F. St. Michael, H. Parnas, I. Wilkie, P. J. Blackall, B. Adler, and J. D. Boyce. 2007. Decoration of Pasteurella multocida lipopolysaccharide with phosphocholine is important for virulence. J. Bacteriol. 1897384-7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heddleston, K. H., J. E. Gallagher, and P. A. Rebers. 1972. Fowl cholera: gel diffusion precipitin test for serotyping Pasteurella multocida from avian species. Avian. Dis. 16925-936. [PubMed] [Google Scholar]
- 23.Hunt, M. L., B. Adler, and K. M. Townsend. 2000. The molecular biology of Pasteurella multocida. Vet. Microbiol. 723-25. [DOI] [PubMed] [Google Scholar]
- 24.Hunt, M. L., D. J. Boucher, J. D. Boyce, and B. Adler. 2001. In vivo-expressed genes of Pasteurella multocida. Infect. Immun. 693004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kehrenberg, C., G. Schulze-Tanzil, J. L. Martel, E. Chaslus-Dancla, and S. Schwarz. 2001. Antimicrobial resistance in Pasteurella and Mannheimia: epidemiology and genetic basis. Vet. Res. 32323-339. [DOI] [PubMed] [Google Scholar]
- 26.Lion, C., M. C. Conroy, A. M. Carpentier, and A. Lozniewski. 2006. Antimicrobial susceptibilities of Pasteurella strains isolated from humans. Int. J. Antimicrob. Agents 27290-293. [DOI] [PubMed] [Google Scholar]
- 27.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 983460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray, P. R., E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken. 2003. Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, DC.
- 29.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard, 2nd ed. NCCLS document M31-A2. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 30.Pijoan, C., R. B. Morrison, and H. D. Hilley. 1983. Serotyping of Pasteurella multocida isolated from swine lungs collected at slaughter. J. Clin. Microbiol. 171074-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhoades, K. R., and R. B. Rimler. 1987. Capsular groups of Pasteurella multocida isolated from avian hosts. Avian Dis. 31895-898. [PubMed] [Google Scholar]
- 32.Salmon, S. A., J. L. Watts, C. A. Case, L. J. Hoffman, H. C. Wegener, and R. J. Yancey, Jr. 1995. Comparison of MICs of ceftiofur and other antimicrobial agents against bacterial pathogens of swine from the United States, Canada, and Denmark. J. Clin. Microbiol. 33:2435-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Townsend, K. M., D. O'Boyle, T. T. Phan, T. X. Hanh, T. G. Wijewardana, I. Wilkie, N. T. Trung, and A. J. Frost. 1998. Acute septicaemic pasteurellosis in Vietnamese pigs. Vet. Microbiol. 63205-215. [DOI] [PubMed] [Google Scholar]
- 34.Townsend, K. M., J. D. Boyce, J. Y. Chung, A. J. Frost, and B. Adler. 2001. Genetic organization of Pasteurella multocida cap loci and development of a multiplex capsular PCR typing system. J. Clin. Microbiol. 39924-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasfi Marandi, M., and K. R. Mittal. 1997. Role of outer membrane protein H (OmpH)- and OmpA-specific monoclonal antibodies from hybridoma tumors in protection of mice against Pasteurella multocida. Infect. Immun. 654502-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh, C., and S. Fanning. 2008. Antimicrobial resistance in foodborne pathogens—a cause for concern? Curr. Drug. Targets 9808-815. [DOI] [PubMed] [Google Scholar]
- 37.White, D. G., S. Zhao, S. Simjee, D. D. Wagner, and P. F. McDermott. 2002. Antimicrobial resistance of foodborne pathogens. Microbes Infect. 4405-412. [DOI] [PubMed] [Google Scholar]
- 38.Yoshimura, H., M. Ishimaru, Y. S. Endoh, and A. Kojima. 2001. Antimicrobial susceptibility of Pasteurella multocida isolated from cattle and pigs. J. Vet. Med. B 48555-560. [DOI] [PubMed] [Google Scholar]