Abstract
The Abbott RealTime human immunodeficiency virus type 1 (HIV-1) assay (ART) and the Cobas AmpliPrep/Cobas TaqMan HIV-1 test (CTM) are commercially available assays for quantification of HIV-1 RNA in plasma. We evaluated performance characteristics, workflow, throughput, reliability, and direct costs of these assays. Both assays yielded good correlation of quantitative results (r = 0.95) among clinical specimens, with a mean difference of −0.34 log10 copies/ml. Testing of healthy donor plasma specimens yielded “target not detected” results by ART, with “HIV-1 RNA detected, <40 copies/ml” results for 3.3% (3 of 90 samples) of these specimens by CTM. Both the m2000sp/m2000rt (ART) and docked CAP/CTM96 (CTM) instrument systems were capable of operating with continuous, uninterrupted workflow. When daily maintenance and cleaning were included, ART and CTM run durations (5 h 52 min and 6 h 4 min, respectively) and hands-on times (53 min and 46 min, respectively) were similar for a run batch size of 24. While ART was more flexible in terms of run batch size, CTM required fewer user interventions and consistently produced higher specimen throughput rates at 8, 16, and 24 h. Assay run failure rates were 6.3% (1 of 16 runs) and 4.2% (1 of 24 runs) for ART and CTM, respectively (P = 1.000), with invalid specimen result rates of 1.0% (5 of 495 specimens) and 2.8% (11 of 399 specimens), respectively (P = 0.073). Direct reagent and consumable costs for each assay were comparable (difference of <10%). In selecting an assay for implementation, laboratories should consider how various assay and instrument features might impact laboratory operation and patient care.
Since quantification of human immunodeficiency virus type 1 (HIV-1) RNA in plasma has become a standard of care in the treatment of HIV-1-infected individuals (10), a number of commercially available assays capable of quantifying HIV-1 RNA in human plasma have been developed and approved for in vitro diagnostic use in the United States. These assays have relied on one of three basic molecular methods, namely, end-point reverse transcription-PCR, nucleic acid sequence-based amplification, and signal amplification (4, 8, 11). While these assays have undoubtedly led to improved patient management, they have limited dynamic ranges and require significant amounts of hands-on time to perform.
Recently, two new assays for the quantification of HIV-1 RNA, the Abbott RealTime HIV-1 assay (ART; Abbott Molecular Inc., Des Plaines, IL) and the Cobas AmpliPrep/Cobas TaqMan HIV-1 test (CTM; Roche Molecular Systems, Inc., Branchburg, NJ), were approved by the U.S. Food and Drug Administration (FDA) for in vitro diagnostic use. ART is performed on an integrated m2000sp and m2000rt instrument system (Abbott Molecular Inc.), while CTM is performed with a Cobas AmpliPrep instrument (CAP instrument; Roche Molecular Systems, Inc.) used in conjunction with either a Cobas TaqMan analyzer (CTM96; Roche Molecular Systems, Inc.) in a docked or undocked instrument configuration or a Cobas TaqMan 48 analyzer (CTM48; Roche Molecular Systems, Inc.) in an undocked configuration.
Both assays are highly automated and utilize real-time PCR methods to provide broad dynamic ranges and closed-tube amplification-detection formats that decrease hands-on time requirements and reduce the potential for contamination with previously amplified reaction products (19, 23). ART offers the option of four different sample extraction volumes (0.2-, 0.5-, 0.6-, and 1.0-ml application protocols), with minimum sample input volumes ranging from 0.7 to 1.8 ml. However, a new assay calibration curve must be generated with each ART application protocol, each reagent lot change, or at a minimum of every 6 months. ART sample preparation and amplification reagents are subdivided into four one-time-use-only sets, with each set containing reagents sufficient for performing 24 tests. CTM performed with the CAP/CTM96 system offers only a single, 0.85-ml sample extraction volume protocol, which requires a sample input volume between 1.0 and 1.05 ml per sample input tube (S tube). Assay calibration is unnecessary with CTM, since kit-specific calibration data and assay control acceptance ranges are encoded in the CTM reagent cassette barcodes. The CTM reagents are packaged in instrument-ready, multiple-use cassettes sufficient for the testing of 48 samples, with multiple aliquots of assay controls.
The availability of these assays and their associated instrument systems provides new options for measuring HIV-1 viral load while challenging clinical laboratories to redefine their current assay selection criteria. Selection of a new diagnostic assay for implementation in a clinical laboratory depends on various factors, such as the performance characteristics of an assay (e.g., analytical sensitivity, analytical specificity, linearity, reproducibility, etc.), reliability (e.g., infrequent instrument and run failures), workflow (e.g., hands-on and hands-off time requirements), flexibility (e.g., variability in sample input volume and assay run batch size), result turnaround time, and the costs of reagents, supplies, and instrumentation. The importance of these various assay characteristics can differ from one laboratory to another, and individual laboratories must determine which factors are relevant and most important in selecting an appropriate assay for their operation and to meet their patient care needs.
Using some of the practical assay selection criteria that we considered to be most important to clinical laboratories performing HIV-1 viral load testing, we conducted a study comparing the assay performance characteristics, workflow, throughput, reliability, and direct costs of ART and CTM.
MATERIALS AND METHODS
Assays.
ART was performed on a single integrated m2000sp/m2000rt system (Abbott Molecular Inc.), using the m2000 0.6 ml HIV-1 RNA 96 software application, version 1.00, with a quantification range of 40 to 10,000,000 copies/ml. CTM was performed on a single docked CAP/CTM96 system (Roche Molecular Systems, Inc.), using AMPLILINK 3.1.1 software, with a quantification range of 40 to 10,000,000 copies/ml. Both assays were designated for investigational use only at the time of this study, and all testing was performed per the manufacturers’ instructions.
Analytical standards.
OptiQuant HIV-1 RNA quantification panels (AcroMetrix Corp., Benicia, CA), consisting of HIV-1 standards at concentrations of 50, 500, 5,000, 50,000, 500,000, and 5,000,000 copies/ml, were used for analytical evaluation of ART and CTM. Replicate aliquots of additional panel members containing 10, 25, and 100 copies/ml were also prepared from appropriate OptiQuant HIV-1 RNA quantification panel members, using EDTA-normal human plasma (AcroMetrix Corp.) as a diluent, and they were frozen at −70°C prior to testing by ART and CTM.
Representative strains of HIV-1 groups M (subtypes A to H), N, and O were evaluated by ART and CTM. An HIV-1 group M subtype H strain (HIV-1BV5018) was purchased from SeraCare Diagnostics, West Bridgewater, MA, while representative HIV-1 group M subtype A to G strains (HIV-1UG273/GS 001, HIV-1US1/GS 004, HIV-1ZAM18/GS 011, HIV-1SE365/GS 017, HIV-1CM235/GS 020, HIV-1BZ126/GS 030, and HIV-1HH8793/GS 029), one group N strain (HIV-1YBF30), and three representative group O strains (HIV-1BCF07, HIV-1BCF11, and HIV-1BCF13) were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Clinical specimens.
Aliquots of HIV-1 RNA-positive EDTA-plasma specimens obtained from 108 patients and previously submitted to the Hepatitis/HIV Molecular Laboratory at Mayo Clinic Rochester for HIV-1 viral load testing by the Cobas Amplicor HIV-1 Monitor test, version 1.5 (CAM; Roche Molecular Systems, Inc.), were retrospectively selected for testing by ART and CTM. These clinical specimens were selected according to their previous CAM results, with 30 specimens from each of the following HIV-1 RNA viral load ranges, along with 18 specimens with HIV-1 RNA viral loads of >750,000 copies/ml: 50 to 999, 1,000 to 9,999, and 10,000 to 750,000. HIV-1 RNA-negative EDTA-plasma specimens from 90 healthy blood donors (previously testing negative by the Cobas AmpliScreen HIV-1 test, version 1.5 [Roche Molecular Systems, Inc.]) were also selected for testing by ART and CTM. An additional group of 528 residual clinical EDTA-plasma specimens previously submitted to the Hepatitis/HIV Molecular Laboratory at Mayo Clinic Rochester for HIV-1 viral load testing were randomly selected (regardless of HIV-1 RNA viral load) and tested by either ART or CTM for comparison of workflow, throughput, and reliability. All plasma specimens were held frozen at −70°C prior to testing by both ART and CTM.
Assay performance characteristics.
The analytical sensitivities of ART and CTM were estimated by testing replicate HIV-1 RNA standards at concentrations of 0, 10, 25, 50, and 100 copies/ml, while replicate aliquots of HIV-1 RNA standards at concentrations of 500, 5,000, 50,000, 500,000, and 5,000,000 copies/ml were tested to assess assay precision. Testing was performed over eight separate ART and CTM assay runs, yielding a total of 16 replicate results per standard. Amplification efficiencies (i.e., equivalent quantification) among HIV-1 groups M (subtypes A to H), N, and O were evaluated by testing three 10-fold dilutions of each representative HIV-1 strain with each assay.
To assess the correlation and agreement of HIV-1 RNA quantification among ART, CTM, and CAM, the 108 HIV-1 RNA-positive specimens were tested in 10 parallel ART and CTM assay runs. To also assess the potential for sample-to-sample contamination during sample processing, 48 of the 90 HIV-1 RNA-negative specimens from healthy blood donors were interspersed at regular intervals among 72 of the 108 HIV-1 RNA-positive specimens and tested in six of these parallel ART and CTM assay runs. The remaining 36 HIV-1 RNA-positive specimens were tested in two separate assay runs, while the remaining 42 HIV-1 RNA-negative specimens from healthy blood donors were tested in two additional assay runs.
Workflow, throughput, and reliability.
Hands-on and hands-off time requirements were determined for each of the various assay run batch sizes (24, 48, and 96 samples for ART and 12 and 24 samples for CTM) by calculating the average time requirements for each assay performed in duplicate by a single operator. Hands-on time for the first ART and CTM run in a given workday included all daily instrument maintenance and reagent preparation. For ART, hands-on time also included the manual transfer of specimen aliquots from their primary specimen tubes to uniform sample tubes necessary for centrifugation prior to m2000sp sample processing. For CTM, hands-on time was expanded to also include the time required to perform the manufacturer's supplemental “best practices” recommendations for periodic maintenance and cleaning of the CAP/CTM96 system issued in May 2007 (14), shortly after completion of our initial workflow studies.
For each assay, run and specimen throughput at 8, 16, and 24 h were calculated from projected assay run timelines, based on the efforts of one dedicated, experienced operator using a single instrument system. Only completed runs were included in the cumulative throughput estimates for each assay. The individual run and specimen result data from assay runs with clinical specimens used to evaluate assay performance and workflow (108 HIV-1 RNA-positive and 90 healthy donor specimens were used for method comparison and 528 clinical specimens were tested by either ART or CTM for workflow analyses) were also used to determine the assay run failure and invalid result rates for each assay, based on failed or invalid assay control results and invalid specimen results, respectively.
Costs.
Reagent, supply, and total direct costs (excluding labor costs) were calculated on a per specimen basis for ART and CTM, using the manufacturers’ list prices and appropriate assay run batch sizes for each assay. For ART, the costs associated with assay calibration (required every 6 months or with any change in reagent lots) were not included in the cost estimates.
Statistical analyses.
Assay precision was evaluated by calculating the percent coefficient of variation for log10-transformed data from replicate aliquots at each of the five different HIV-1 RNA concentrations (2.7 to 6.7 log10 copies/ml) tested over eight separate runs. Probit analysis (with a 95% hit rate) was used to estimate the analytical sensitivities of ART and CTM (5). For each assay, quantification equivalency among HIV-1 group M (subtypes A to H), N, and O strains was evaluated by comparing the slope estimates for individual linear regression lines constructed from test results for the three 10-fold dilutions of each of the representative HIV-1 groups and subtypes. Individual slope estimates, along with their 95% confidence intervals (95% CI), were plotted and examined for the presence of overlapping 95% CI (i.e., the slope estimate for one group falling within the confidence interval of another), which would indicate a lack of statistically significant differences in target amplification efficiency among the genotypes and subtypes tested. Quantitative results for HIV-1 RNA-positive specimens tested by ART and CTM were compared to each other and to CAM results by Deming regression analysis (9) and Bland-Altman plots (1). As a measure of assay and system reliability, assay run failure and invalid specimen result rates, along with 95% CI, were estimated for each assay. Two-sided Fisher's exact test was used to compare these rates for ART and CTM, with P values of <0.05 considered statistically significant.
RESULTS
Assay performance characteristics.
Our study yielded percent coefficients of variation ranging from 2.7% (at 2.7 log10 IU/ml) to 0.5% (at 6.7 log10 IU/ml) for ART and from 6.2% (at 4.7 log10 IU/ml) to 1.6% (at 6.7 log10 IU/ml) for CTM, which are within the ranges of reproducibility data provided in the respective product inserts. The analytical sensitivities of ART and CTM were also determined to be 32.2 copies/ml (95% CI, 24.7 to 51.8) and 45.3 copies/ml (95% CI, 36.3 to 64.5), respectively, closely approximating the lower limits of quantification (40 copies/ml) set by the manufacturers of both assays.
All serial dilutions of the HIV-1 strains of groups M (subtypes A to H), N, and O were quantifiable by ART, while only group M and N strains were quantifiable by CTM. Comparison of the 95% CI for individual slope estimates derived from linear regression analyses showed no statistically significant differences among the slope estimates obtained for the quantifiable strains relevant to ART and CTM (data not shown). These findings suggest equivalency of quantification among HIV-1 groups M (subtypes A to H) and N by both assays, with further equivalency of quantification for HIV-1 group O by ART.
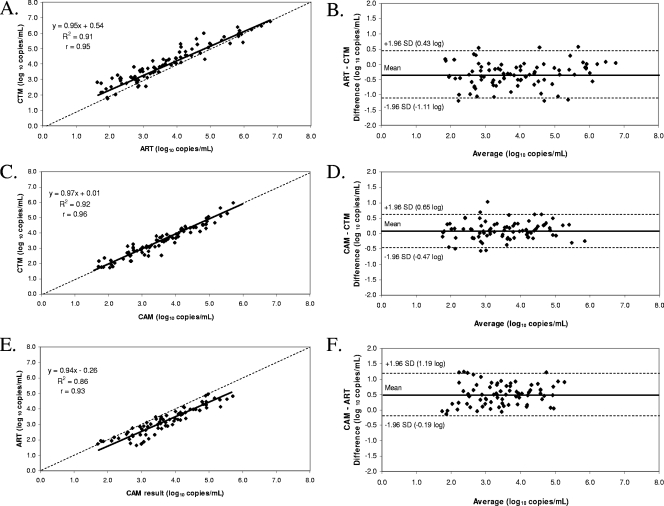
Among the 108 clinical specimens previously tested by CAM, 87, 81, and 75 yielded valid, quantifiable results by both ART and CTM, both CAM and CTM, and both CAM and ART, respectively (Fig. 1). While good correlation was observed for ART versus CTM (r = 0.95), CAM versus CTM (r = 0.96), and CAM versus ART (r = 0.93), viral load differences of >0.5 log10 copies/ml occurred in 36.8% (32 of 87), 9.9% (8 of 81), and 48.0% (36 of 75) of paired results, respectively. Bland-Altman plotting of the ART versus CTM data yielded a mean difference of −0.34 log10 copies/ml in viral load, with individual differences ranging from 0.57 to −1.20 log10 copies/ml. Similar analyses of the data for CAM versus CTM and CAM versus ART resulted in mean differences of 0.09 log10 copies/ml (range, 1.03 to −0.57 log10 copies/ml) and 0.50 log10 copies/ml (range, 1.25 to −0.14 log10 copies/ml), respectively. Of note, no clinically significant bias was observed for any assay over the relevant mean quantification ranges shown in the Bland-Altman plots.
FIG. 1.
Deming regression analyses and Bland-Altman plots of valid quantitative results for clinical plasma specimens. Graphs show comparisons of ART to CTM (A and B) (n = 87), CAM to CTM (C and D) (n = 81), and CAM to ART (E and F) (n = 75).
Among the healthy donor plasma specimens tested by ART and CTM, all 90 yielded “target not detected” results by ART, while a single healthy donor specimen yielded an invalid result and 3.3% (3 of 90 samples) of specimens yielded HIV-1 RNA-positive results below the limit of quantification (i.e., “HIV-1 RNA detected, <40 copies/ml”) by CTM. Two of the specimens yielding “HIV-1 RNA detected, <40 copies/ml” results by CTM were tested in an assay run containing both HIV-1 RNA-negative and -positive specimens, while the remaining specimen was tested in an assay run consisting of only healthy donor specimens. Thus, these unexpected findings were unlikely to be due to sample-to-sample contamination during automated processing by CAP. These discordant specimens could not be retested due to insufficient volume.
Workflow, throughput, and reliability.
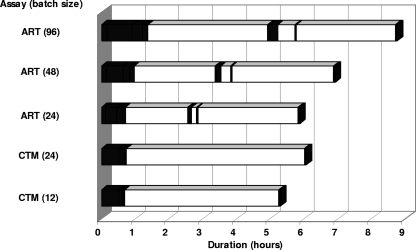
As shown in Fig. 2, larger run batch sizes generally increased the hands-on time requirement (especially during initial specimen processing) along with total assay run duration for both assays. Run durations for ART and CTM (5 h 52 min and 6 h 4 min, respectively) and total hands-on time requirements for both assays (53 min and 46 min, respectively) were similar for the first run of the day with a run batch size of 24. However, daily maintenance procedures for ART performed on the m2000sp/m2000rt system required ∼8 min to perform, while a substantial portion of the hands-on time required to perform the initial CTM run (30 of 46 min) was spent performing daily maintenance procedures on the CAP/CTM system. On subsequent assay runs, hands-on time for ART was greater than that for CTM. In addition, the hands-on time for ART included several minor manual manipulations separated by relatively long periods of hands-off time during each run, whereas virtually all of the hands-on time for CTM occurred at the beginning of each run.
FIG. 2.
Assay workflow comparison between ART and CTM. Averaged hands-on and hands-off times are represented by the filled and open bars, respectively. Hands-on time for both assays includes all currently recommended instrument maintenance procedures necessary to start the first assay run of the day. For CTM, the hands-on time does not include a manual review of amplification growth curves for all clinical specimens in the assay run (see reference 16).
The cumulative assay run and specimen throughput for a batch size of 24 was consistently lower with ART than with CTM after 8, 16, and 24 h of continuous operation (Table 1). While throughput was consistently lower for ART than for CTM with a batch size of 24 (21 specimens and 3 controls), CTM had a higher projected throughput of only 21 specimens (i.e., one run) at each time point. Importantly, ART exhibited substantial increases in specimen throughput for batch sizes of >24.
TABLE 1.
ART and CTM throughput over 8, 16, and 24 h with various assay run batch sizesa
| Assay | Duration (h) | Cumulative run/specimen throughput (no. of runs/no. of specimens) for batch size of:
|
|||
|---|---|---|---|---|---|
| 12 | 24 | 48 | 96 | ||
| ART | 8 | 1/21 | 1/45 | 0/0 | |
| 16 | 4/84 | 3/135 | 2/186 | ||
| 24 | 7/147 | 6/270 | 4/372 | ||
| CTM | 8 | 3/27 | 2/42 | ||
| 16 | 9/81 | 5/105 | |||
| 24 | 16/144 | 8/168 | |||
Only those assay runs that could be completed within the allotted time were included.
Assay run failure rates among ART and CTM assay runs containing clinical specimens were 1 of 16 runs (6.3%; 95% CI, 0.2% to 30.2%) and 1 of 24 runs (4.2%; 95% CI, 0.1% to 21.1%), respectively. The invalid specimen result rates for ART and CTM among these runs were 5 of 495 specimens (1.0%; 95% CI, 0.3% to 2.3%) and 11 of 399 specimens (2.8%; 95% CI, 1.4% to 4.9%), respectively. Of note, none of the 108 clinical or 90 healthy blood donor specimens used for method comparison yielded invalid specimen results by both assays. There were no significant differences between the two assays in either the run failure rate (P = 1.000) or the invalid specimen result rate (P = 0.073).
Costs.
Reagent, consumable, and total direct costs per specimen for ART and CTM are shown in Table 2. For a 24-sample batch size, despite the higher cost of consumables for CTM, the total direct costs were comparable (difference of <10%) between the two assays, and the total direct cost per specimen was reduced with increasing assay run batch size.
TABLE 2.
Comparison of reagent, consumable, and total direct costs for ART and CTM
| Assay | Batch size | Reagent cost/specimen (US $) | Consumable cost/specimen (US $) | Total direct cost/specimen (US $) |
|---|---|---|---|---|
| ART | 24 | 145 | 2 | 147 |
| 48 | 135 | 1 | 136 | |
| 96 | 130 | 1 | 131 | |
| CTM | 12 | 144 | 12 | 156 |
| 24 | 124 | 10 | 134 |
DISCUSSION
The general assay performance characteristics (e.g., analytical sensitivity, analytical specificity, linearity, reproducibility, correlation, and agreement) of both ART and CTM have been evaluated extensively by other investigators (2, 3, 6, 7, 12, 19-21, 23, 24). Our data for ART versus CTM (R2 = 0.91, differences of >0.5 log10 copies/ml among paired results from 36.8% of specimens, and a mean difference of −0.34 log10 copies/ml) were consistent with previously published studies that reported R2 values ranging from 0.76 to 0.94, differences of >0.5 log10 copies/ml among paired results from 39.8% of specimens, and mean differences ranging from −0.24 to 0.51 log10 copies/ml (2, 6, 7, 20, 24). For CAM versus CTM, our data (R2 = 0.92, differences of >0.5 log10 copies/ml among paired results from 9.9% of specimens, and a mean difference of 0.09 log10 copies/ml) were also consistent with the findings of studies showing R2 values ranging from 0.85 to 0.97, differences of >0.5 log10 copies/ml among paired results from 6.8% to 26.1% of specimens, and mean differences ranging from 0.10 to 0.28 log10 copies/ml (2, 7, 12, 19, 24). Finally, our study data comparing CAM versus ART (R2 = 0.86, differences of >0.5 log10 copies/ml among paired results from 48.0% of specimens, and a mean difference of 0.50 log10 copies/ml) were also generally comparable to those reported in previous studies (2, 7, 20, 23, 24), which showed R2 values ranging from 0.83 to 0.95, differences of >0.5 log10 copies/ml among paired results from 8.5% to 17.0% of specimens, and mean differences ranging from −0.15 to 0.40 log10 copies/ml.
Although the m2000sp instrument is capable of accepting uncapped primary specimen collection tubes for processing, the ART assay protocol requires centrifugation of all specimen tubes prior to processing by the m2000sp instrument, thus making it impractical for clinical laboratories receiving specimen collection tubes of differing sizes and fill volumes to take full advantage of this instrument feature. Additionally, among the four different ART application protocols with various input volumes, there appears to be no practical advantage to using the 0.5-ml and 1.0-ml application protocols because they require essentially the same sample input volume and yield the same lower limit of quantification (40 copies/ml), respectively, as the 0.6-ml application protocol. Sample preparation, amplification, calibration, and control reagents for ART are packaged separately, thereby potentially complicating purchasing, inventory management, and assay calibration. Reagents also require some manual preparation (e.g., mixing, transfer into vessels, and barcode labeling of vessels) prior to use by the m2000sp instrument. As a result of the reagent packaging format, the m2000sp/m2000rt system offers optimal assay run batch sizes of 24, 48, 72, or 96 samples (including assay controls). Because only a single set of assay controls is required per assay run, regardless of assay run batch size, ART may be appealing to cost-conscious clinical laboratories wishing to test samples in run batch sizes larger than 24 samples/run. However, with larger run batch sizes, the hands-on time requirements increase (especially during initial specimen processing), along with the total assay run duration. Furthermore, since laboratory personnel must be available to seal and transfer the 96-well assay reaction plate from the m2000sp instrument to the m2000rt instrument following sample processing, increases in assay run batch size may also necessitate longer work shift duration, multiple work shifts, or limitations in assay run start time within a given work shift.
Despite the lack of a more highly automated instrument platform and “instrument-ready” reagents, ART offers several unique advantages over CTM. When rapid technical assistance or troubleshooting is required, remote diagnostic access to the m2000sp/m2000rt system by the manufacturer is available via modem connection. ART also has FDA approval for the quantification of HIV-1 groups M, N, and O, and it may offer improved quantification of atypical strains and circulating recombinant forms, particularly CRF02 (3, 7, 21, 23; Abbott RealTime HIV-1 test package insert [Abbott Molecular Inc., 2007]).
While the 96-well assay reaction plates are manually sealed after processing on the m2000sp instrument, there are currently no additional safeguards (e.g., uracil-N-glycosylase) incorporated into the assay to prevent carryover contamination. Therefore, great care should be taken to avoid release of previously amplified products into the laboratory environment, and the manufacturer's recommended cleaning procedures should be followed carefully. Periodic monitoring for the presence of laboratory contamination by amplification products is also recommended by the manufacturer (Abbott RealTime HIV-1 test package insert; Abbott Molecular, Inc., 2007).
In contrast to the m2000sp instrument, the CAP instrument is incapable of accepting primary specimen collection tubes for processing. However, CTM does offer some labor-saving advantages over ART. All sample preparation, amplification, and control reagents for CTM are packaged in a single, complete kit, thus simplifying purchasing and inventory management. Additionally, the CTM reagent cassettes are “instrument ready,” requiring no further preparation prior to use. Since the assay permits an optimal run batch size of either 12 or 24 samples (including assay controls), CTM may be preferred by clinical laboratories wishing to perform small run batch sizes (e.g., ≤24 samples/run) with a continuous workflow approach, whereby up to 24 samples (including controls) can be placed in each sample rack, with up to three sample racks loaded on the CAP instrument at any one time. Use of the CAP/CTM96 system in the docked configuration also eliminates hands-on transfer steps in the midst of each assay run. This “walk-away” feature may reduce the need for extended work shift duration or multiple work shifts and thereby result in fewer limitations in assay run start time within a given work shift.
The CAP/CTM96 system software (AMPLILINK 3.1 series) used in this evaluation did not allow remote diagnostic access to the instrument system. However, this feature will be available with future software releases (AMPLILINK 3.2 series). When our evaluation was conducted in early 2007, the manufacturer's recommendations for routine, manual review of all assay growth curves had not yet been issued (16). Implementation of this manual review process was later estimated to add <5 min of additional hands-on time to each assay run consisting of 24 samples, and this brief step was not included in the workflow analysis (Fig. 1). Of note, CTM has FDA approval for the quantification of HIV-1 group M (subtypes A to H) strains only.
While the CTM reaction tubes (K tubes) are capped individually, our experience revealed that they can open unexpectedly during the final K tube discard process or instrument malfunctions. With the docked CAP/CTM96 system in which reusable K carriers are moved back and forth across a mechanical bridge between the two instruments, the K carriers can potentially be contaminated by amplified products inadvertently released from K tubes by the CTM96 instrument. As the empty (but potentially contaminated) K carriers are moved back to the CAP instrument for use in subsequent assay runs, open K tubes placed in these K carriers have the potential to be contaminated by previously amplified products, resulting in carryover contamination of subsequent assay reaction mixtures. Therefore, the manufacturer's recommended instrument cleaning procedures should be followed carefully to reduce the potential for contamination of the laboratory environment with previously amplified products (13, 17). The incorporation of dUTP and uracil-N-glycosylase (AmpErase) into the CTM reagents may also provide an additional safeguard against carryover contamination by previously amplified products. However, under the conditions encountered in routine use of the docked CAP/CTM96 system in a single room, AmpErase activity capable of destroying 103 or even 107 copies of previously amplified target sequence per reaction could be insufficient to reliably prevent false-positive assay results.
At the time that our study was conducted in early 2007, the manufacturer's suggested daily maintenance and cleaning procedures for the docked CAP/CTM96 system included recommendations for cleaning with deionized or distilled water and 70% ethanol. The manufacturer subsequently issued their supplemental “best practices” recommendations for periodic maintenance and cleaning of CAP/CTM96 systems in May 2007 (14), with several more recent updates (15, 17). More specific recommendations for additional cleaning procedures to be performed following sample spills or suspected contamination from amplified products were issued in August 2007 (13). Of note, these updated maintenance and cleaning procedures now include the use of DNA Away (Molecular BioProducts, Inc., San Diego, CA), a proprietary alkaline solution for cleaning DNA-contaminated surfaces, followed by cleaning with distilled or deionized water and 70% ethanol.
Our finding of 3 false-positive results (all with a result of “HIV-1 RNA detected, <40 copies/ml”) among 89 valid test results from the healthy blood donor plasma specimens included in our study is consistent with data presented in the FDA-approved package insert for this assay, which showed an analytical specificity of 99% (95% CI, 97% to 100%; 1 false-positive result among 179 replicate aliquots of an HIV-1-negative panel member) and a clinical specificity of 99.4% (3 false-positive results among 519 EDTA-plasma specimens from healthy, HIV-1 antibody-negative donors) for this assay (Cobas AmpliPrep/Cobas TaqMan HIV-1 test package insert, rev. 2.0.; Roche Molecular Systems, Inc., 2008).
In summary, while ART was more flexible in terms of batch size, CTM required less hands-on time with fewer user interventions and consistently produced higher assay and specimen throughput at 8, 16, and 24 h. However, both the m2000sp/m2000rt and docked CAP/CTM96 systems were capable of reliable operation with a continuous, uninterrupted workflow, and the direct costs (reagents and consumables) were also comparable between ART and CTM. Clinical laboratories selecting the most suitable assay for test implementation must carefully consider how various features of each assay and instrument system might best meet the needs of their laboratory operation and clinical practice.
Acknowledgments
We thank Abbott Molecular Inc. and Roche Diagnostics Corp. for providing the instruments, assay reagents, supplies, and funding to support this study.
The following HIV-1 strains were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1UG273/GS 001, HIV-1US1/GS 004, HIV-1ZAM18/GS 011, HIV-1SE365/GS 017, HIV-1CM235/GS 020, HIV-1BZ126/GS 030, and HIV-1HH8793/GS 029, submitted by Nelson Michael; and HIV-1BCF07, HIV-1BCF11, HIV-1BCF13, and HIV-1YBF30, submitted by Sentob Saragosti, Francoise Brun-Vézinet, and Francois Simon.
Footnotes
Published ahead of print on 4 February 2009.
REFERENCES
- 1.Bland, J. M., and D. G. Altman. 1999. Measuring agreement in method comparison studies. Stat. Methods Med. Res. 8135-160. [DOI] [PubMed] [Google Scholar]
- 2.Braun, P., R. Ehret, F. Wiesmann, F. Zabbai, M. Knickmann, R. Kuhn, S. Thamm, G. Warnat, and H. Knechten. 2007. Comparison of four commercial quantitative HIV-1 assays for viral load monitoring in clinical daily routine. Clin. Chem. Lab. Med. 4593-99. [DOI] [PubMed] [Google Scholar]
- 3.Damond, F., B. Roquebert, A. Benard, G. Collin, M. Miceli, P. Yeni, F. Brun-Vezinet, and D. Descamps. 2007. Human immunodeficiency virus type 1 (HIV-1) plasma load discrepancies between the Roche Cobas Amplicor HIV-1 Monitor, version 1.5, and the Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 assays. J. Clin. Microbiol. 453436-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dewar, R. L., H. C. Highbarger, M. D. Sarmiento, J. A. Todd, M. B. Vasudevachari, R. T. Davey, Jr., J. A. Kovacs, N. P. Salzman, H. C. Lane, and M. S. Urdea. 1994. Application of branched DNA signal amplification to monitor human immunodeficiency virus type 1 burden in human plasma. J. Infect. Dis. 1701172-1179. [DOI] [PubMed] [Google Scholar]
- 5.Finney, D. J. 1971. Probit analysis, 3rd ed. Cambridge University Press, Cambridge, United Kingdom.
- 6.Foulongne, V., B. Montes, M. N. Didelot-Rousseau, and M. Segondy. 2006. Comparison of the LCx human immunodeficiency virus (HIV) RNA quantitative, RealTime HIV, and COBAS AmpliPrep-COBAS TaqMan assays for quantitation of HIV type 1 RNA in plasma. J. Clin. Microbiol. 442963-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gueudin, M., J. C. Plantier, V. Lemee, M. P. Schmitt, L. Chartier, T. Bourlet, A. Ruffault, F. Damond, M. Vray, and F. Simon. 2007. Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J. Acquir. Immune Defic. Syndr. 44500-505. [DOI] [PubMed] [Google Scholar]
- 8.Kievits, T., B. van Gemen, D. van Strijp, R. Schukkink, M. Dircks, H. Adriaanse, L. Malek, R. Sooknanan, and P. Lens. 1991. NASBA isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. J. Virol. Methods 35273-286. [DOI] [PubMed] [Google Scholar]
- 9.Linnet, K. 1993. Evaluation of regression procedures for methods comparison studies. Clin. Chem. 39424-432. [PubMed] [Google Scholar]
- 10.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 2721167-1170. [DOI] [PubMed] [Google Scholar]
- 11.Mulder, J., N. McKinney, C. Christopherson, J. Sninsky, L. Greenfield, and S. Kwok. 1994. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J. Clin. Microbiol. 32292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver, A. R., S. F. Pereira, and D. A. Clark. 2007. Comparative evaluation of the automated Roche TaqMan real-time quantitative human immunodeficiency virus type 1 RNA PCR assay and the Roche Amplicor, version 1.5, conventional PCR assay. J. Clin. Microbiol. 453616-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roche Diagnostics Corporation. 2007. Additional cleaning procedures for the COBAS AmpliPrep/COBAS TaqMan systems. Analyzer bulletin 07-192. Roche Diagnostics Corporation, Indianapolis, IN.
- 14.Roche Diagnostics Corporation. 2007. Best practices for periodic maintenance of the COBAS AmpliPrep/COBAS TaqMan test. Analyzer bulletin 07-135. Roche Diagnostics Corporation, Indianapolis, IN.
- 15.Roche Diagnostics Corporation. 2007. Best practices for periodic maintenance of the COBAS AmpliPrep/COBAS TaqMan test. Analyzer bulletin 07-135R. Roche Diagnostics Corporation, Indianapolis, IN.
- 16.Roche Diagnostics Corporation. 2007. Growth curve review for the COBAS AmpliPrep/COBAS TaqMan HIV-1 test. Software bulletin 07-233. Roche Diagnostics Corporation, Indianapolis, IN.
- 17.Roche Diagnostics Corporation. 2008. Best practices for periodic maintenance of the COBAS AmpliPrep/COBAS TaqMan test. Analyzer bulletin 07-135RR. Roche Diagnostics Corporation, Indianapolis, IN.
- 18.Reference deleted.
- 19.Schumacher, W., E. Frick, M. Kauselmann, V. Maier-Hoyle, R. van der Vliet, and R. Babiel. 2007. Fully automated quantification of human immunodeficiency virus (HIV) type 1 RNA in human plasma by the COBAS AmpliPrep/COBAS TaqMan system. J. Clin. Virol. 38304-312. [DOI] [PubMed] [Google Scholar]
- 20.Schutten, M., D. Peters, N. K. Back, M. Beld, K. Beuselinck, V. Foulongne, A. M. Geretti, L. Pandiani, C. Tiemann, and H. G. Niesters. 2007. Multicenter evaluation of the new Abbott RealTime assays for quantitative detection of human immunodeficiency virus type 1 and hepatitis C virus RNA. J. Clin. Microbiol. 451712-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swanson, P., S. Huang, K. Abravaya, C. de Mendoza, V. Soriano, S. G. Devare, and J. Hackett, Jr. 2007. Evaluation of performance across the dynamic range of the Abbott RealTime HIV-1 assay as compared to VERSANT HIV-1 RNA 3.0 and AMPLICOR HIV-1 MONITOR v1.5 using serial dilutions of 39 group M and O viruses. J. Virol. Methods 14149-57. [DOI] [PubMed] [Google Scholar]
- 22.Reference deleted.
- 23.Tang, N., S. Huang, J. Salituro, W. B. Mak, G. Cloherty, J. Johanson, Y. H. Li, G. Schneider, J. Robinson, J. Hackett, Jr., P. Swanson, and K. Abravaya. 2007. A RealTime HIV-1 viral load assay for automated quantitation of HIV-1 RNA in genetically diverse group M subtypes A-H, group O and group N samples. J. Virol. Methods 146236-245. [DOI] [PubMed] [Google Scholar]
- 24.Wolff, D., and A. Gerritzen. 2007. Comparison of the Roche COBAS Amplicor Monitor, Roche COBAS Ampliprep/COBAS Taqman and Abbott RealTime Test assays for quantification of hepatitis C virus and HIV RNA. Clin. Chem. Lab. Med. 45917-922. [DOI] [PubMed] [Google Scholar]