Abstract
A single-tube, multiplex, real-time PCR assay with molecular beacons was established in which various probes were used for the simultaneous detection, differentiation, and quantification of human T-cell leukemia virus types 1, 2, and 3 (HTLV-1, HTLV-2, and HTLV-3, respectively) and of simian T-cell leukemia virus types 1 and 3 (STLV-1 and STLV-3, respectively). The quantitative amplification of the standards with MT4 (HTLV-1) and C19 (HTLV-2) cell lines and a molecular clone of HTLV-3 was linear, with the simplex and multiplex methods having similar efficiencies. A maximum difference of 0.9 (mean, 0.4; range, 0.0 to 0.9) was found between threshold cycle values in single and multiplex reactions. The efficiency with each probe in the multiplex reaction was close to 100%, indicating strong linear amplification. The albumin gene was used to standardize the copy number. Comparable results for the detection and quantification of HTLV-1 were obtained with our new methods and with other real-time PCR methods described previously. With our new multiplex assay, however, we were able to detect and quantify HTLV-2 and -3 and STLV-1 and -3 in clinical specimens, with an excellent dynamic range of 106 to 100 copies per assay, which the other assays could not do. Thus, it will be possible to determine a wide range of HTLV types in both standard and clinical samples, with a detection of 1 to 10 HTLV copies in samples containing at least 100 cells. Furthermore, our system can provide evidence for multiple infections with the three HTLV types, with separate proviral load results. Our new method also could be used for epidemiological studies in Africa and in countries where HTLVs and STLVs are endemic.
Human T-cell lymphotropic virus types 1 (HTLV-1), 2 (HTLV-2), and 3 (HTLV-3) belong to a group of primate retroviruses with certain common epidemiological and biological properties, including tropism for T lymphocytes. HTLV-1, isolated in 1980, is the causative agent of adult T-cell leukemia/lymphoma (37) and tropical spastic paraparesis/HTLV-1-associated myelopathy (TSP/HAM) (17). HTLV-2, isolated 2 years after HTLV-1 (24), might be responsible for rare neurological syndromes that are clinically related to TSP/HAM (21, 33). Although no tumors have been linked definitively to such infections (14, 22), frequent lymphocytosis was described recently (1). Two new retroviruses, HTLV-3 and HTLV-4, have been identified in Cameroon, but no diseases have been associated with them yet (5, 45). The finding of these different HTLV types indicates that they are more diverse and polymorphic than previously thought (28). Previous phylogenetic analysis indicated that HTLV-1 can be classified into three major lineages, Cosmopolitan, Central African, and Melanesian, with each lineage being divided into various subtypes (38). Studies of the molecular epidemiology of HTLV-2 have shown at least two major molecular subtypes, designated HTLV-2A and HTLV-2B (19).
HTLV-1 is endemic in southern Japan, some regions of sub-Saharan Africa, and the Caribbean Basin, as well as some parts of South America and the Middle East (16), with an estimated 15 to 20 million infected persons worldwide. HTLV-2 has been shown to be endemic in various American Indian populations (2) and has been endemic for the past 15 to 25 years among intravenous drug users in Europe and North America (34). Furthermore, since 1991, sporadic cases of HTLV-2 infection have been detected in west and central Africa in isolated rural populations, including some Pygmies (18). No information is available yet on the epidemiology of HTLV-3 or -4 (28).
HTLV-1 and -2 are transmitted in three ways: between sexual partners, mainly from men to women; from mothers to infants during prolonged breastfeeding; and through transfusions with HTLV-infected blood cells. It has been reported that 65% of patients who received whole blood or cellular blood components from HTLV-1-seropositive donors seroconverted against HTLV-1 (23). HTLV-1 also has been reported to be transmitted in organ transplants (40).
In Gabon, central Africa, the prevalence of HTLV-1 infection is high (5% of adults in urban areas and 8.5 to 10.5% of those in rural areas) (27). As mother-to-child transmission and blood transfusion appear to be the main routes of transmission in hospitalized children, the prevention of mother-to-child transmission and transfusion- or transplant-associated transmission will require screening, especially of populations in which the infections are endemic (11).
These viruses usually are detected indirectly by enzyme-linked immunosorbent assay (ELISA), and positive samples are confirmed by Western blot analysis. Samples are considered positive when a complete Western blot profile can be obtained; however, no currently available immunological assay can detect acute infection with HTLVs. Furthermore, individuals infected with HTLV-3 and -4 have incomplete Western blot profiles, showing an indeterminate pattern (5, 45). No direct molecular methods are available for the rapid detection of these newly discovered viruses. PCR is a good choice for detecting HTLVs but might be less sensitive for low proviral loads. A rapid, non-labor-intensive amplification technique for the accurate quantification of the viral target is real-time PCR (20), in which internal fluorescent probes provide specificity and allow the detection of amplification products that are directly related to the copy number of the target (36). Real-time PCR already has been used for detecting HTLV-1 and -2 proviral loads (10, 12), including those of subtypes 1A, 1B, 1C, 2A, and 2B; however, the method did not detect HTLV-3 or various simian T-cell leukemia viruses (STLVs).
Therefore, a rapid, specific system for evaluating the various primate T-cell leukemia viruses is needed that can be used in epidemiological and pathogenesis investigations. As the HTLV proviral load can indicate the degree of replication and usually is correlated with the onset and progression of TSP/HAM (6, 35), real-time PCR also could provide information about disease progression by the quantification of the HTLV viral load. We report here a one-step (single-tube), multiplex, real-time PCR for the simultaneous detection, differentiation, and quantification of proviral HTLV-1, -2, and -3; the new method also can detect STLV type 1 (STLV-1) and STLV-3. The assay was validated with samples previously described and obtained from blood donors in Franceville (southeast Gabon), where the prevalence of HTLVs, especially HTLV-1B but also 1A, 2A, and 2B, is very high (14, 28).
MATERIALS AND METHODS
Oligonucleotide design.
All of the complete genomes of HTLV-1, -2, and -3 and STLV-1 and -3 available in the GenBank database were included and aligned for the selection and design of various primers and probes (Table 1). Three pairs of primers located in the Tax gene that can detect various strains of HTLV-1, -2, and -3 and STLV-1 and -3 as well as primers for the detection of the albumin gene were designed with Beacon Designer software (Premier Biosoft, Palo Alto, CA) and are presented in Table 1. The four molecular beacon probes allow specific detection, because they are more adaptable than TaqMan probes (Table 2). The primers for the detection of albumin genes (AlbF and AlbR) were reported by Mortreux et al. (32), but we designed a new probe, AlbSd1 (5′-Cy5 and 3′-black hole quencher 2 [3′-BHQ2]) (Table 1).
TABLE 1.
Characteristics of primers and probes
| Name | Sequence and genomic location | Size (bp) | Specificity | Concn in multiplex (μM) |
|---|---|---|---|---|
| H1Z1F3 | 7434-CCGCCTACATCGTCACG-7450 | 0.4 | ||
| H1Z1R5 | 7546-GGAGTCGAGGGATAAGGAA-7528 | 0.4 | ||
| H1Z1Sd5 | 7493-ROX-CGCGATCGGGTCCCAGGTGATCTGATGCTCTGGATCGCG-BHQ1-7517 | 113 | HTLV-1/STLV-1a | 0.3 |
| H2Z2F3 | 7236-GACAGAGCCTCCTATATGG-7254 | 0.4 | ||
| H2Z2R6 | 7424-GGTATTGGAGAGGAGAGC-7407 | 0.4 | ||
| H2Z2Sd3 | 7301-FAM-CGCGATCGACACCAATCGGCCTGTACACAATCGGATCGCG-BHQ1-7276 | 189 | HTLV-2a | 0.3 |
| H3Z1F3 | 6921-TACCCTGTCTACGTTTTCG-6939 | 0.4 | ||
| H3Z1R1 | 7046-CCAGGTAATCTGATGTTCG-7028 | 0.4 | ||
| H3Z1Sd1 | 6983-HEX-CGCGATCTTGTTCCGCTCGGCTACACCGCGATCGCG-BHQ1-7004 | 126 | HTLV-3/STLV-3a | 0.3 |
| AlbF | 16533-GCTGTCATCTCTTGTGGGCTGT-16554 | 0.4 | ||
| AlbR | 16671-ACTCATGGGAGCTGCTGGTTC-16651 | 0.4 | ||
| AlbSd1 | 16616-Cy5-CGCGATCGGGAGAGATTTGTGTGGGCATGACAGGATCGCG-BHQ2-16591 | 139 | Albumin | 0.3 |
The types and subtypes detected (only complete genomes were included) are the following: for HTLV-1, HTLV-1 WHP (AF259264), HTLV-1A ATK1 (J02029), HTLV-1A ATL-YS (U19949), HTLV-1A K30 (L03561), HTLV-1A RKI3A (AF042071), HTLV-1A TSP1 (M86840), HTLV-1B EL (M67514), HTLV-1C Mel5 (L02534), HTLV-1 (AF139170), HTLV-1 (AF033817.1), HTLV-1 Brazil (AY563953.1), STLV-1 Tan90 (AF074966), and STLV-1 TE4 (Z46900); for HTLV-2, HTLV-2 Brazil (AF139382), HTLV-2 Venezuela G2 (AF074965), HTLV-2A AF41231Hsa (AF412314), HTLV-2A K96 (AF326584), HTLV-2A MO (M10060), HTLV-2A RP329 (AF326583), HTLV-2B G12 (L11456), HTLV-2B GAB2 (Y13051), HTLV-2B GU (X89270), and HTLV-2D EFE2 (Y14365); and for HTLV-3: STLV-3 CTO604 (NC_003323), STLV-3 CTONG409 (AY222339), STLV-3 PH969 (Y07616), HTLV-3 Pyl43 (DQ462191), HTLV-3 2026ND (DQ093792), and STLV-3 TGE2117 (AY217650).
TABLE 2.
Comparison of CT values of the simplex and multiplex methods
| Beacon probe | Target(s) | 5′ reporter/3′ quencher fora:
|
CT values for 104 copies/PCR
|
||
|---|---|---|---|---|---|
| Single-dye assay | Multiplex assay | Single-dye assay | Multiplex assay | ||
| H1Z1Sd5 | HTLV-1/STLV-1 | FAM/DABCYL | ROX/BHQ1 | 25.1 ± 0.1 | 25.7 ± 0.1 |
| H2Z2Sd3 | HTLV-2 | FAM/DABCYL | FAM/BHQ1 | 25.2 ± 0.3 | 25.1 ± 0.3 |
| H3Z1Sd1 | HTLV-3/STLV-3 | FAM/DABCYL | HEX/BHQ1 | 25.1 ± 0.1 | 25.7 ± 0.2 |
| AlbSd1 | Albumin | FAM/DABCYL | Cy5/BHQ2 | 25.7 ± 0.3 | 25.7 ± 0.5 |
FAM, 6-carboxyfluorescein: ROX, 6-carboxy-X-rhodamine; DABCYL, 4-(((4-dimethyl)-phenyl)-azo)benzoic acid.
Standards used for HTLV detection and quantification.
The standard used for HTLV-1 was DNA lysate from the MT4 cell line (kindly provided by Franck Mortreux, Lyon, France). The cell line previously was characterized as having seven copies of the virus per cell (32), and the range of the MT4 standard was 105 to 100 copies per reaction. The standard used for HTLV-2 was DNA lysate from the C19 cell line (kindly provided by Antoine Gessain, Institut Pasteur, Paris, France). It has been reported previously that this cell line carries seven copies of HTLV-2 (15, 26). The range of the C19 standard was 105 to 100 copies per reaction. The standard used for HTLV-3 was a molecular clone of HTLV-3, Pyl 43 (kindly provided by Renaud Mahieux, Institut Pasteur, Paris, France) (7) and was used in the same range as the other standards. The range of the quantitative housekeeping gene standard (albumin) was 104 to 102 cells per reaction.
DNA extraction.
DNA was extracted from 200 μl of infected cell lines or blood samples with the QIAamp DNA mini kit (reference 51304; Qiagen) and eluted in 200 μl of buffer according to the manufacturer's recommendations. For the real-time PCR assays, a 10-fold dilution series was used.
Multiplex real-time PCR.
Real-time PCR was performed with a Quantitect multiplex NoROX kit (catalogue no. 204743; Qiagen) containing primer mix (Table 1). Five microliters of DNA from virus-infected cell lines or from patients, 12.5 μl of 2× master mix, primers and probes at the final concentrations (Table 1), and RNase-free water were added to a final volume of 25 μl. The DNA was amplified at 95°C for 15 min, followed by 50 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. The PCR was stopped by holding the solution at 20°C for 10 min. The iQ5 cycler (Bio-Rad Laboratories, Hercules, CA) was used throughout the study for beacon assays, and the tests were systematically conducted in duplicate. We suggest that all users of this system run their samples in duplicate in order to avoid false-positive or -negative results. Two similar results are highly conclusive.
Clinical samples.
To evaluate the applicability of our new multiplex, real-time PCR assay to clinical specimens, 100 blood samples were collected in Vacutainer tubes with EDTA from the blood transfusion unit at the general hospital in Franceville, Gabon. Furthermore, 19 samples of peripheral blood mononuclear cells were obtained from pregnant women who were either HTLV-1 or HTLV-2 seropositive or had HTLV-indeterminate Western blot profiles (14). We also included six samples from STLV-1-seropositive mandrills (29) and an STLV-3 molecular clone obtained by Chevalier et al. (8). DNA was extracted from 200 μl of collected blood. Before multiplex real-time PCR was conducted, serological tests were performed to confirm the HTLV or STLV status with an enzyme immunoassay (Vironostika HTLV1/2; Biomérieux), and positive or borderline-positive samples were analyzed by Western blotting (HTLV blot 4; Diagnostic Biotechnology, Singapore).
RESULTS
Multiplex assay development and standardization.
The best real-time PCR system for each type of virus was determined, and then each system was adjusted in the same mix to make up the multiplex. After optimization, the results of the multiplex real-time assay were compared to the results achieved in our simplex real-time PCR. Table 2 shows the reporters and quenchers on the different probes. We found a maximum difference in threshold cycle (CT) values of 0.9 (mean, 0.4; range, 0.0 to 0.9) between multiplex and single reactions, which is a weak, acceptable variation.
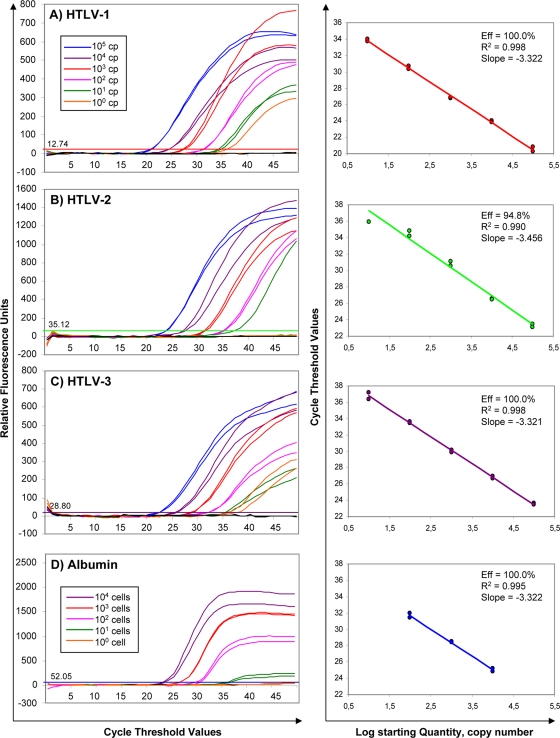
The dynamic range of our multiplex real-time PCR was determined with 10-fold serial dilutions of standards. Standard curves were considered acceptable only when their slope was between −3.1 and −3.5 and the coefficient of correlation, R2, was between 0.990 and 1.000, indicating a strong linear relation (Fig. 1).
FIG. 1.
Sensitivity and specificity of the one-tube, multiplex, real-time PCR assay for the simultaneous detection, differentiation, and quantification of HTLV-1, -2, and -3. (A) HTLV-1, 10-fold dilutions, 105 to 100 copies per PCR (with standard curve); (B) HTLV-2, 10-fold dilutions, 105 to 100 copies per PCR (with standard curve); (C) HTLV-3, 10-fold dilutions, 105 to 100 copies per PCR (with standard curve); and (D) albumin, 10-fold dilutions, 104 to 100 cells per PCR (with standard curve).
The sensitivity of our multiplex assay for detecting HTLVs and albumin is shown in Fig. 1, and the linearity, sensitivity, and efficiency of PCR are indicated in Table 3 for each probe. Each standard curve was obtained in one multiplex run. For albumin, we wanted a system that could detect and quantify the cell number per sample of a minimum of 100 cells, the linearity required being between 105 and 102 cells. A sample with less than 100 cells (less than 6 ng of DNA) was automatically retested (by DNA extraction and real-time PCR), as the albumin was considered insufficient.
TABLE 3.
Sensitivity and linearity of each beacon probe in the one-step (one-tube) multiplex assay
| Beacon probe | Target(s) | Linearitya | Sensitivityb | Efficiency (%) | Standard correlation |
|---|---|---|---|---|---|
| H1Z1Sd5 | HTLV-1/STLV-1 | 105-101 | 1 | 100.0 | 0.998 |
| H2Z2Sd3 | HTLV-2 | 105-101 | 10 | 94.8 | 0.990 |
| H3Z1Sd1 | HTLV-3/STLV-3 | 105-101 | 1 | 100.0 | 0.998 |
| AlbSd1 | Albumin | 105-102 | 100 | 100.0 | 0.995 |
The range of copy numbers that can be quantified in one assay.
The limit of detection in one assay.
Multiplex detection and quantification in clinical specimens.
The sensitivity and specificity of our multiplex real-time PCR method was tested in clinical specimens and compared to those of another real-time PCR method (Table 4). The efficiency of the detection of each system was greater than 95.0%, and the standard correlations were 0.990 to 0.999.
TABLE 4.
Comparative analysis of results obtained for clinical specimens with the pX primer method and our new multiplex assay
| HTLV status and identity | Sample origin | ELISA result | Western blot pattern | Subtype (by sequencing) | Proviral load (copy no./μg DNA)
|
|
|---|---|---|---|---|---|---|
| pX | Multiplex | |||||
| HTLV-1a | ||||||
| BD DS26* | Blood donor | + | Complete HTLV-1 | NA | 27,602 | 25,277 |
| Gab112LM | Pregnant woman | + | Complete HTLV-1 | A | 22 | 41 |
| Gab683LB | Pregnant woman | + | Complete HTLV-1 | B | 1,200 | 1,924 |
| Gab958LM | Pregnant woman | + | Complete HTLV-1 | B | 36 | 33 |
| Gab70LM | Pregnant woman | + | Complete HTLV-1 | B | 71,100 | 61,061 |
| Gab109LM | Pregnant woman | + | Complete HTLV-1 | B | 76 | 259 |
| Gab826LM | Pregnant woman | + | Complete HTLV-1 | B | 370 | 354 |
| Gab197LM | Pregnant woman | + | Complete HTLV-1 | B | 10 | 6 |
| Gab1014FC | Pregnant woman | + | Complete HTLV-1 | B | 30,800 | 27,454 |
| Gab1058FC | Pregnant woman | + | Complete HTLV-1 | B | 3,450 | 2,920 |
| Gab1144FC | Pregnant woman | + | Complete HTLV-1 | B | 21,260 | 46,123 |
| Gab1123FC | Pregnant woman | + | Complete HTLV-1 | B | 900 | 2,235 |
| Gab1008FC | Pregnant woman | + | Complete HTLV-1 | B | 1,450 | 1,058 |
| Gab1089FC | Pregnant woman | + | Complete HTLV-1 | B | 56,400 | 48,737 |
| HTLV-2a | ||||||
| Gab1080FC | Pregnant woman | + | Complete HTLV-2 | B | Undetectable | 12,259 |
| FE 11355* | Pregnant woman | + | Complete HTLV-2 | NA | Undetectable | 51,800 |
| Indeterminatea | ||||||
| Gab1314PG | Pregnant woman | + | GD21, p19, gp21, p24, p26, p28, p32, p36 | 85 | 108 | |
| Gab1527OY | Pregnant woman | + | p19, gp21, p26, p28, p32, p36 | 0 | 0 | |
| Gab1129FC | Pregnant woman | + | p19, gp21, p24 | 0 | 0 | |
| Gab1004LB | Pregnant woman | + | p19, gp21, p26, p28, p36 | 0 | 0 | |
| STLV-1b | ||||||
| Msp 15G | Wild mandrill | + | Complete HTLV-1 | D | Undetectable | 16,120 |
| Mnd 16B | Wild mandrill | + | Complete HTLV-1 | D | Undetectable | 6,205 |
| Mnd 12 M | Wild mandrill | + | Complete HTLV-1 | D | Undetectable | 9,790 |
| Ms 19 | Wild mandrill | + | Incomplete (no MTA-1) | F | Undetectable | 16,350 |
| Ms 20 | Wild mandrill | + | Complete HTLV-1 | F | Undetectable | 11,556 |
| Ms 25 | Wild mandrill | + | Incomplete (no MTA-1) | F | Undetectable | 3,448 |
| STLV-3 | ||||||
| PPA-F3c | Molecular clone | + | GD21, p24, p36, p53, K55, MTA-1 | NA | Undetectable | 23,504 |
The results for the samples obtained from pregnant women and the sequence data were published previously by Etenna et al. (13). An asterisk indicates that data for the sample are newly reported in this paper. The samples were first tested by ELISA and confirmed by Western blotting. NA, not available.
The results for the samples obtained from STLV-1-infected monkeys and the corresponding sequences were published by Makuwa et al. (29).
The STLV-3 molecular clone obtained from an STLV-3-infected baboon (Papio anubis) was published by Chevalier et al. (8).
All of the samples were albumin positive, indicating that there was sufficient DNA to perform real-time PCR and to detect any HTLVs. The slopes of all of the curves were similar, and the signals were conclusively either positive or negative despite rare high CT values (>45 was considered a negative result). Samples from blood donors were analyzed first by ELISA. Only one sample was found to be HTLV-1 positive, but HTLV-2 and -3 were not detected (Table 4). Furthermore, in clinical samples obtained from pregnant women, subtypes A and B were detected similarly, and the results obtained with the pX system and our multiplex system were comparable (we considered that the same log scale indicated similar proviral loads). The pX method was unable to detect or quantify HTLV-2, STLV-1, or STLV-3. With our new multiplex method, we were able to detect and quantify HTLV-2 in the two samples from pregnant women and also in the samples obtained from monkeys naturally infected with STLV-1 subtypes D and F and the molecular clone of STLV-3, which was obtained from an infected Papio anubis baboon (Table 4).
We experimentally induced coinfected samples by mixing DNA from HTLV-1 and -2, HTLV-1 and -3, HTLV-2 and -3, or the three types of HTLVs in the same tube. We were able to detect and quantify each type of HTLV independently (data not shown).
DISCUSSION
We have described the first multiplex (one step in one tube) real-time PCR method for the simultaneous detection, differentiation, and quantification of HTLV-1, -2, and -3 proviral loads. The one-tube method was chosen because it requires less work and sample handling, thus reducing the risk for sample cross-contamination and saving time. Like other real-time PCR systems, our method provides the linear quantification of input DNA across a broad range, low DNA consumption, the fast throughput of large numbers of samples, and low risks for contamination and carryover. Although a one-tube method is desirable, it is essential that the assay be highly sensitive. To reach optimal sensitivity in the one-tube assay, many combinations of primers and probes were tested, different reagent concentrations were tried, and different kits were used to find the best combination for all oligonucleotides. Our method was effective for the accurate detection of all of the HTLV subtypes for which complete genome sequences were available in GenBank. We found high sensitivity and specificity, as each dual-labeled probe used in our method could detect each type of HTLV, with no cross-reactivity to other types. Furthermore, we were able to detect and quantify STLV-1 and the STLV-3 strains previously described (8, 9, 31). Our results show that this method can accurately detect more HTLV types and subtypes than other available methods, like the pX system.
In previous real-time PCR methods used for the detection and quantification of HTLVs, linear TaqMan probes were used (10, 12). In our study, we used molecular beacon probes, which are dual-labeled oligonucleotide probes that fluoresce upon hybridization with a complementary target sequence (44). The oligonucleotide is labeled at one end with a fluorescent reporter dye and at the opposite end with a fluorescence quencher. Molecular beacons are designed to form a stem-loop hairpin structure in the absence of a target, forcing the fluorescence reporter group to approach the quencher group (dark state). In the presence of a complementary target molecule, the molecular beacon opens, due to the formation of the more stable probe-target duplex, increasing the distance between the reporter and the quencher and restoring fluorescence (bright state). The competition between hairpin formation and target hybridization makes molecular beacons more sensitive than linear probes (43). The transition between a molecular beacon's dark and bright states allows for differentiation between bound and unbound probes, with signal-to-background ratios that can be greater than 200, making them extremely sensitive probes for nucleic acid detection (3, 42).
The thermodynamics and kinetics of molecular beacons depend on their structure and sequence (3, 25). The beacons are designed such that the loop sequence is complementary to the target, while the stem sequences are self-complementary but unrelated to the target sequence. Therefore, it is necessary to understand the structure-function relations of molecular beacons in order to optimize their performance. This may become critical in certain assays, as the addition (or deletion) of even a single nucleotide can dramatically change the behavior of the molecular beacon (43). We chose a seven-nucleotide stem for each probe (5′-CGCGATC…GATCGCG-3′), which provides a good thermodynamic range for amplification, quantification, and detection. The beacons' efficiency can be increased or decreased by modifying the strength of the stem, with more GC content to limit stem opening and vice versa. Thus, molecular beacon probes remain compliant and flexible. Furthermore, these probes do not undergo hydrolysis during the elongation step, so the baseline after the assay is flat, with no residual fluorescence (41). In addition, molecular beacons allow for the amplification of larger amplicons (100 to 200 or up to 400 bp) than TaqMan probes, which are limited to short sequences. While TaqMan probes must be used at a fixed temperature (about 60°C), the beacons are functional at an annealing temperature between 46 and 62°C. It was shown previously that molecular beacons can be used to screen blood samples for multiple viruses at once by monitoring the color of the fluorescence emission (30, 42), which TaqMan and other linear probes are unable to do.
Our assay was developed for the iQ5 cycler, but it would be easy to transfer to other equipment, because we used four filters (available on different commercial cyclers) and 96-well plates, which are common in research laboratories. Our method also could be adapted to capillaries.
Only strictly consensus regions in our sequences' alignments were selected for detecting each HTLV type (HTLV-1, -2, and -3 and all the corresponding subtypes). After having designed primers and probes (located on the Tax gene), we confirmed the sequence homology with all of the subtypes listed in Table 1. During multiplex optimization, we determined whether the different subtypes (for example, HTLV-1A and -1B) could be detected with the same accuracy. The results were conclusive, and there is no reason that the probes would behave differently for other subtypes, as they target exactly the same sequence (no mismatches were accepted). For some other subtypes, such as HTLV-1E and -1F, only partial sequences on the Env gene were available; however, as the sequence variation among HTLV subtypes is very low and our primers and probes have 100% homology with all subtypes, including HTLV-1A, -1B, -1C, and -1D and STLV-1 Tan90 and STLV-1 TE4, our new assay should be able to detect all of them. A similar procedure was used for the primers and probes designed for HTLV-2 (subtypes 2A and 2B) and HTLV-3/STLV-3. Furthermore, by experimentally inducing coinfection with various HTLV types, we showed that our multiplex method can identify multiple HTLV infections by quantifying the proviral load of each HTLV, which no other available system currently is able to do. Consequently, the method allows simultaneous but differential detection and quantification.
In our clinical specimens, no new HTLV-3 or STLV-3 infection could be detected. There are few patients positive for HTLV-3 in the world, and all have been found in Cameroon (4, 5, 39). Thus, there was very little probability that this new virus would be found in our clinical samples. Nevertheless, a large epidemiological study is ongoing in our laboratory, with more than 4,000 samples collected from rural Africa. Our method will help us to evaluate the epidemiology of this newly discovered virus in central Africa.
In conclusion, a one-tube, highly sensitive, specific, fast, multiplex, real-time PCR method for measuring HTLV-1, -2, and -3 and STLV-1 and -3 proviral loads has been developed. This method could be used in studies of HTLV-related disease progression and in epidemiological studies in Africa and in countries where HTLVs and STLVs are endemic.
Acknowledgments
We thank F. Mortreux (Centre Léon Bérard, Lyon, France) for providing MT4 cells and A. Gessain and R. Mahieux (Institut Pasteur, Paris, France) for providing the C19 cell line and HTLV-3 and STLV-3 molecular clones.
Guillaume Besson is the recipient of a postdoctoral fellowship from the European Community. The Centre International de Recherches Médicales de Franceville, Gabon, is funded by the Gabonese Government, Total-Gabon, and the French Foreign Ministry.
Footnotes
Published ahead of print on 11 February 2009.
REFERENCES
- 1.Bartman, M. T., Z. Kaidarova, D. Hirschkorn, R. A. Sacher, J. Fridey, G. Garratty, J. Gibble, J. W. Smith, B. Newman, A. E. Yeo, and E. L. Murphy. 2008. Long-term increases in lymphocytes and platelets in human T-lymphotropic virus type II infection. Blood 1123995-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biglione, M., O. Vidan, R. Mahieux, M. de Colombo, M. de Los Angeles, A. de Basualdo, M. Bonnet, G. Pankow, M. Avila de Efron, A. Zorrilla, F. Tekaia, E. Murphy, G. de Thé, and A. Gessain. 1999. Seroepidemiological and molecular studies of HTLV-II, subtype b, in isolated groups of Mataco and Toba indians of northern Argentina. AIDS Res. Hum. Retrovir. 15407-417. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet, G., S. Tyagi, A. Libchaber, and F. R. Kramer. 1999. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. USA 966171-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calattini, S., E. Betsem, S. Bassot, S. A. Chevalier, R. Mahieux, A. Froment, and A. Gessain. 19 December 2008. New strain of human T lymphotropic virus (HTLV) type 3 in a Pygmy from Cameroon with peculiar HTLV serologic results. J. Infect. Dis. [Epub ahead of print.] [DOI] [PubMed]
- 5.Calattini, S., S. A. Chevalier, R. Duprez, S. Bassot, A. Froment, R. Mahieux, and A. Gessain. 2005. Discovery of a new human T-cell lymphotropic virus (HTLV-3) in central Africa. Retrovirology 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Césaire, R., A. Dehee, A. Lezin, N. Desire, O. Bourdonne, F. Dantin, O. Bera, D. Smadja, S. Abel, A. Cabie, G. Sobesky, and J. C. Nicolas. 2001. Quantification of HTLV type I and HIV type I DNA load in coinfected patients: HIV type 1 infection does not alter HTLV type I proviral amount in the peripheral blood compartment. AIDS Res. Hum. Retrovir. 17799-805. [DOI] [PubMed] [Google Scholar]
- 7.Chevalier, S. A., N. L. Ko, S. Calattini, A. Mallet, M. C. Prevost, K. Kehn, J. N. Brady, F. Kashanchi, A. Gessain, and R. Mahieux. 2008. Construction and characterization of a human T-cell lymphotropic virus type 3 infectious molecular clone. J. Virol. 826747-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevalier, S. A., M. Walic, S. Calattini, A. Mallet, M. C. Prevost, A. Gessain, and R. Mahieux. 2007. Construction and characterization of a full-length infectious simian T-cell lymphotropic virus type 3 molecular clone. J. Virol. 816276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courgnaud, V., S. Van Dooren, F. Liegeois, X. Pourrut, B. Abela, S. Loul, E. Mpoudi-Ngole, A. Vandamme, E. Delaporte, and M. Peeters. 2004. Simian T-cell leukemia virus (STLV) infection in wild primate populations in Cameroon: evidence for dual STLV type 1 and type 3 infection in agile mangabeys (Cercocebus agilis). J. Virol. 784700-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dehée, A., R. Cesaire, N. Desire, A. Lezin, O. Bourdonne, O. Bera, Y. Plumelle, D. Smadja, and J. C. Nicolas. 2002. Quantitation of HTLV-I proviral load by a TaqMan real-time PCR assay. J. Virol. Methods 10237-51. [DOI] [PubMed] [Google Scholar]
- 11.Delaporte, E., M. Peeters, J. L. Bardy, Y. Ville, L. Placca, I. Bedjabaga, B. Larouze, and P. Piot. 1993. Blood transfusion as a major risk factor for HTLV-I infection among hospitalized children in Gabon (equatorial Africa). J. Acquir. Immune Defic. Syndr. 6424-428. [PubMed] [Google Scholar]
- 12.Estes, M. C., and J. S. Sevall. 2003. Multiplex PCR using real time DNA amplification for the rapid detection and quantitation of HTLV I or II. Mol. Cell. Probes 1759-68. [DOI] [PubMed] [Google Scholar]
- 13.Etenna, S. L., M. Caron, G. Besson, M. Makuwa, A. Gessain, A. Mahé, and M. Kazanji. 2008. New insights into the prevalence, genetic diversity and proviral load of human T-cell leukemia viruses types 1 and 2 in pregnant women in Gabon, equatorial central Africa. J. Clin. Microbiol. 463607-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouchard, N., B. Flageul, M. Bagot, M. F. Avril, O. Hermine, F. Sigaux, H. Merle-Beral, X. Troussard, J. F. Delfraissy, G. de Thé, and A. Gessain. 1995. Lack of evidence of HTLV-I/II infection in T CD8 malignant or reactive lymphoproliferative disorders in France: a serological and/or molecular study of 169 cases. Leukemia 92087-2092. [PubMed] [Google Scholar]
- 15.Gallo, D., L. M. Penning, and C. V. Hanson. 1991. Detection and differentiation of antibodies to human T-cell lymphotropic virus types I and II by the immunofluorescence method. J. Clin. Microbiol. 292345-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gessain, A. 1996. Epidemiology of HTLV-I and associated diseases, p. 33-64. In P. Höllsberg and D. A. Hafler (ed.), Human T-cell lymphotropic virus type 1. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 17.Gessain, A., F. Barin, J. C. Vernant, O. Gout, L. Maurs, A. Calender, and G. de Thé. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii407-410. [DOI] [PubMed] [Google Scholar]
- 18.Gessain, A., P. Mauclère, A. Froment, M. Biglione, J. Y. Le Hesran, F. Tekaia, J. Millan, and G. de Thé. 1995. Isolation and molecular characterization of a human T-cell lymphotropic virus type II (HTLV-II), subtype B, from a healthy Pygmy living in a remote area of Cameroon: an ancient origin for HTLV-II in Africa. Proc. Natl. Acad. Sci. USA 254041-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall, W. W., R. Ishak, S. W. Zhu, P. Novoa, N. Eiraku, H. Takahashi, M. d. C. Ferreira, V. Azevedo, M. O. Ishak, O. D. C. Ferreira, C. Monken, and T. Kurata. 1996. Human T lymphotropic virus type II (HTLV-II): epidemiology, molecular properties, and clinical features of infection. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13S204-S214. [DOI] [PubMed] [Google Scholar]
- 20.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6986-994. [DOI] [PubMed] [Google Scholar]
- 21.Hjelle, B., O. Appenzeller, R. Mills, S. Alexander, N. Torrezmartinez, R. Jahnke, and G. Ross. 1992. Chronic neurodegenerative disease associated with HTLV-II infection. Lancet 339645-646. [DOI] [PubMed] [Google Scholar]
- 22.Hjelle, B., R. Mills, S. Swenson, G. Mertz, C. Key, and S. Allen. 1991. Incidence of hairy cell leukemia, mycosis fungoides, and chronic lymphocytic leukemia in first known HTLV-II-endemic population. J. Infect. Dis. 163435-440. [DOI] [PubMed] [Google Scholar]
- 23.Inaba, S., K. Okochi, H. Sato, K. Fukada, N. Kinukawa, H. Nakata, K. Kinjyo, F. Fujii, and Y. Maeda. 1999. Efficacy of donor screening for HTLV-I and the natural history of transfusion-transmitted infection. Transfusion 391104-1110. [DOI] [PubMed] [Google Scholar]
- 24.Kalyanaraman, V. S., M. G. Sarngadharan, M. Robert-Guroff, I. Miyoshi, D. Golde, and R. C. Gallo. 1982. A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science 218571-573. [DOI] [PubMed] [Google Scholar]
- 25.Kuhn, H., V. V. Demidov, J. M. Coull, M. J. Fiandaca, B. D. Gildea, and M. D. Frank-Kamenetskii. 2002. Hybridization of DNA and PNA molecular beacons to single-stranded and double-stranded DNA targets. J. Am. Chem. Soc. 1241097-1103. [DOI] [PubMed] [Google Scholar]
- 26.Lacoste, V., J. G. Judde, G. Bestett, J. Cadranel, M. Antoine, F. Valensi, E. Delabesse, E. Macintyre, and A. Gessain. 2000. Virological and molecular characterisation of a new B lymphoid cell line, established from an AIDS patient with primary effusion lymphoma, harbouring both KSHV/HHV8 and EBV viruses. Leuk. Lymphoma 38401-409. [DOI] [PubMed] [Google Scholar]
- 27.Le Hesran, J. Y., E. Delaporte, C. Gaudebout, A. Trebuck, D. Schrijvers, R. Josse, M. Peeters, H. Cheringou, A. Dupont, and B. Larouze. 1994. Demographic factors associated with HTLV-1 infection in a Gabonese community. Int. J. Epidemiol. 23812-817. [DOI] [PubMed] [Google Scholar]
- 28.Mahieux, R., and A. Gessain. 2 May 2008. The human HTLV-3 and HTLV-4 retroviruses: new members of the HTLV family. Pathol. Biol. (Paris) [Epub ahead of print.] [DOI] [PubMed]
- 29.Makuwa, M., S. Souquière, S. L. Clifford, P. T. Telfer, B. Sallé, O. Bourry, R. Onanga, A. Mouinga-Ondeme, E. Wickings, K. Abernethy, P. Rouquet, F. Simon, and P. Roques. 2004. Two distinct STLV-1 subtypes infecting Mandrillus sphinx follow the geographic distribution of their hosts. AIDS Res. Hum. Retrovir. 201137-1143. [DOI] [PubMed] [Google Scholar]
- 30.Marras, S. A., F. R. Kramer, and S. Tyagi. 1999. Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal. 14151-156. [DOI] [PubMed] [Google Scholar]
- 31.Meertens, L., V. Shanmugam, A. Gessain, B. E. Beer, Z. Tooze, W. Heneine, and W. M. Switzer. 2003. A novel, divergent simian T-cell lymphotropic virus type 3 in a wild-caught red-capped mangabey (Cercocebus torquatus torquatus) from Nigeria. J. Gen. Virol. 842723-2727. [DOI] [PubMed] [Google Scholar]
- 32.Mortreux, F., M. Kazanji, A. S. Gabet, B. de Thoisy, and E. Wattel. 2001. Two-step nature of human T-cell leukemia virus type 1 replication in experimentally infected squirrel monkeys (Saimiri sciureus). J. Virol. 751083-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy, E. L., J. Fridey, J. W. Smith, J. Engstrom, R. A. Sacher, K. Miller, J. Gibble, J. Stevens, R. Thomson, D. Hansma, J. Kaplan, R. Khabbaz, G. Nemo, et al. 1997. HTLV-associated myelopathy in a cohort of HTLV-I and HTLV-II-infected blood donors. Neurology 48315-320. [DOI] [PubMed] [Google Scholar]
- 34.Murphy, E. L., R. Mahieux, G. de Thé, F. Tekaia, D. Ameti, J. Horton, and A. Gessain. 1998. Molecular epidemiology of HTLV-II among United States blood donors and intravenous drug users: an age-cohort effect for HTLV-II RFLP type aO. Virology 242425-434. [DOI] [PubMed] [Google Scholar]
- 35.Nagai, M., K. Usuku, W. Matsumoto, D. Kodama, N. Takenouchi, T. Moritoyo, S. Hashiguchi, M. Ichinose, C. R. Bangham, S. Izumo, and M. Osame. 1998. Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J. Neurovirol. 4586-593. [DOI] [PubMed] [Google Scholar]
- 36.Niesters, H. G. 2001. Quantitation of viral load using real-time amplification techniques. Methods 25419-429. [DOI] [PubMed] [Google Scholar]
- 37.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type-C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 777415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slattery, J. P., G. Franchini, and A. Gessain. 1999. Genomic evolution, patterns of global dissemination, and interspecies transmission of human and simian T-cell leukemia/lymphotropic viruses. Gen. Res. 9525-540. [PubMed] [Google Scholar]
- 39.Switzer, W. M., S. H. Qari, N. D. Wolfe, D. S. Burke, T. M. Folks, and W. Heneine. 2006. Ancient origin and molecular features of the novel human T-lymphotropic virus type 3 revealed by complete genome analysis. J. Virol. 807427-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toro, C., B. Rodes, E. Poveda, and V. Soriano. 2003. Rapid development of subacute myelopathy in three organ transplant recipients after transmission of human T-cell lymphotropic virus type I from a single donor. Transplantation 75102-104. [DOI] [PubMed] [Google Scholar]
- 41.Tse, C., and J. Capeau. 2003. Real time PCR methodology for quantification of nucleic acids. Ann. Biol. Clin. (Paris) 61279-293. [PubMed] [Google Scholar]
- 42.Tsourkas, A., and G. Bao. 2003. Shedding light on health and disease using molecular beacons. Brief. Funct. Genomic Proteomic 1372-384. [DOI] [PubMed] [Google Scholar]
- 43.Tsourkas, A., M. A. Behlke, and G. Bao. 2002. Structure-function relationships of shared-stem and conventional molecular beacons. Nucleic Acids Res. 304208-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14303-308. [DOI] [PubMed] [Google Scholar]
- 45.Wolfe, N. D., W. Heneine, J. K. Carr, A. D. Garcia, V. Shanmugam, U. Tamoufe, J. N. Torimiro, A. T. Prosser, M. Lebreton, E. Mpoudi-Ngole, F. E. McCutchan, D. L. Birx, T. M. Folks, D. S. Burke, and W. M. Switzer. 2005. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc. Natl. Acad. Sci. USA 1027994-7999. [DOI] [PMC free article] [PubMed] [Google Scholar]