Abstract
Over recent years, there has been an increasing acknowledgment of the diversity that exists among Mycobacterium tuberculosis clinical isolates. To facilitate comparative studies aimed at deciphering the relevance of this diversity to human disease, an unambiguous and easily interpretable method of strain classification is required. Presently, the most effective means of assigning isolates into a series of unambiguous lineages is the method of Gagneux et al. (S. Gagneux et al., Proc. Natl. Acad. Sci. USA 103:2869-2873, 2006) that involves the PCR-based detection of large sequence polymorphisms (LSPs). In this manner, isolates are classified into six major lineages, the majority of which display a high degree of geographic restriction. Here we describe an independent replicate of the Gagneux study carried out on 798 isolates collected over a 6-year period from mostly foreign-born patients resident on the island of Montreal, Canada. The original trends in terms of bacterial genotype and patient ethnicity are remarkably conserved within this Montreal cohort, even though the patient distributions between the two populations are quite distinct. In parallel with the LSP analysis, we also demonstrate that “clustered” tuberculosis (TB) cases defined through restriction fragment length polymorphism (RFLP) analysis (for isolates with ≥6 IS6110 copies) or RFLP in combination with spoligotyping (for isolates with <6 IS6110 copies) do not stray across the LSP-defined lineage boundaries. However, our data also demonstrate the poor discriminatory power of either RFLP or spoligotyping alone for these low-IS6110-copy-number isolates. We believe that this independent validation of the LSP method should encourage researchers to adopt this system in investigations aimed at elucidating the role of strain variation in TB.
Infection with Mycobacterium tuberculosis, the bacillus responsible for tuberculosis (TB), still results in almost 2 million global deaths each year despite the availability of effective chemotherapy and a partially effective vaccine (bacillus Calmette-Guérin [Mycobacterium bovis BCG]) for well over half a century (43). Even though there have been real advances in our level of understanding of M. tuberculosis metabolic pathways and in the identification of potential virulence-associated molecules since the advent of the “postgenomic era” (8), significant improvements with respect to treatment, diagnosis, and prevention of TB still remain elusive. Recent descriptions of variably expressed virulence factors and disease outcomes that are associated with particular lineages of M. tuberculosis further emphasize the layers of complexity that need to be overcome in our efforts to combat this devastating disease (6, 25, 31, 32, 38).
While the fact that TB phenotypic diversity exists is no longer in dispute, there is heightened interest in epidemiological circles in establishing the relevance of this diversity to human disease. Of particular note are the recent publications suggesting that strains belonging to the M. tuberculosis W/Beijing lineage possess unique attributes that confer an increased ability to cause disease and to be transmitted within certain geographic settings (10, 12, 19, 20) or to cause extrapulmonary TB in certain patient ethnicities (7, 23). There is also the intriguing suggestion of an interaction between host and bacterial genotypes (6). To increase the power of these types of studies, there is a requirement for a robust system of classifying clinical isolates into a genetically defined set of clades or lineages, each member of which would be expected to share a unique set of phenotypic attributes. However, M. tuberculosis genotypes have traditionally been reported in the literature on the basis of rapidly evolving epidemiologic markers, such as IS6110 restriction fragment length polymorphism (RFLP) fingerprints, spoligotypes, or mycobacterial interspersed repeat units (3, 27). The use of relatively nondescript classifications afforded by these techniques often precludes direct comparisons being made across independent studies. Specifically, the boundaries that define frequently cited strain families such as the Haarlem, Latin-American/Mediterranean, or W/Beijing lineages are vague and inconsistent, and moreover, the nature of the evolutionary relationship between these lineages is unclear.
To understand the epidemiological consequences of M. tuberculosis variability, a less ambiguous system of nomenclature is required to demonstrate clearly the genetic and evolutionary relationships among independent clinical isolates. To date, the most effective means of assigning strains into a small number of unambiguous lineages is the method based on the detection of large sequence polymorphisms (LSPs) or regions of difference (RDs) that represent a series of well-characterized unique-event polymorphisms (deletions) (1, 14, 15, 21, 40). Importantly, these LSP-defined lineages are phylogenetically robust and, unlike IS6110 RFLP, spoligotyping, and mycobacterial interspersed repeat unit analysis, they do not suffer from problems associated with convergent evolution (15, 27). Using this approach, M. tuberculosis isolates are currently classified into six major global lineages, the majority of which tend to show a high degree of geographic restriction. Each of the six lineages is defined by a single ancestral LSP common to all isolates within that particular lineage. In some instances, sublineages have also been identified, each possessing its own unique LSP deletion event. Additional laboratories are also reporting on their own LSPs, such as RD(Rio) (RD174), which appear to define sublineages unique to specific locales (16, 29, 30). However, as Alland et al. have recently pointed out, it is important when building phylogenies based on LSPs to distinguish between those that are unique-event LSPs and those that occur repeatedly across multiple lineages because of their association with repetitive genetic elements (1).
Single nucleotide polymorphism (SNP)-based population analyses are also phylogenetically informative due to an inherent paucity of either synonymous or nonsynonymous substitutions within the M. tuberculosis genome (37). This lack of sequence heterogeneity ensures that the chance of encountering recurrent synonymous SNPs within independent lineages is extremely unlikely. To date, combinations of synonymous SNPs have been used to classify many of the same major lineages as those defined through LSP analysis (13, 15, 17). The major advantage that unique-event LSPs have over SNPs lies in the efficiency and technical simplicity with which isolates can be genotyped and assigned to a particular lineage. Using a multiplex PCR approach, large numbers of isolates can be screened for the lineage-defining LSPs in a high-throughput manner very rapidly. In contrast, a minimum of six SNP loci must be screened in order to make the same lineage assignments, and furthermore, as some of the lineages differ from each other by only a single SNP, extreme care needs to be taken in interpreting the results of these assays. In the future, it will prove immensely useful to continually update the broader global classification to incorporate newly identified LSPs and SNPs as they are uncovered.
The description of the six major global lineages was for the most part based on clinical isolates obtained in San Francisco, CA (14, 15). Because the majority of samples analyzed were collected at one site and represent only a tiny fraction of all TB cases diagnosed each year, the possibility exists that the model proposed by Gagneux et al. (14) suffers from sampling bias and, if so, would not be truly representative of the global situation. In the present study, we have carried out an independent replicate using a data bank of 798 isolates collected over a 6-year period from mostly foreign-born TB patients resident on the island of Montreal, Quebec, Canada. While San Francisco and Montreal are both cosmopolitan cities reflecting a large immigrant community, Montreal, unlike San Francisco, has very low rates of ongoing TB transmission (24). In this manner, our strain collection provides a “snapshot” of TB lineages resident in the more than 80 countries represented. With these isolates, we set out to determine (i) whether the framework outlined by Gagneux and colleagues was robust with the use of an independent sample set, (ii) whether the described association between lineages and geography still held true, and (iii) the association between the LSP/RD-defined lineages and RFLP analysis on the same isolates.
MATERIALS AND METHODS
Study population.
The study population consists of Montreal residents diagnosed with active TB over an approximately 6-year period from January 2001 to May 2007. As TB is a notifiable disease in Canada, all diagnosed cases are reported to the local public health authorities (Laboratoire de Santé Publique du Québec [LSPQ]), and clinical and demographic information is collected within a provincial reportable disease registry. For this study, data including patient's country of origin, year of arrival in Canada, age, sex, and primary site of disease (pulmonary or extrapulmonary) were abstracted from the registry database in nonnominal format. Culture confirmation, species determination, and antimicrobial testing were performed at the LSPQ. Bacterial isolates representing culture-positive TB cases (>85% of diagnosed active cases [35]) were received from the LSPQ following inoculation onto Lowenstein-Jensen agar slants. Where multiple samples were collected from the same patient on different dates, only the primary isolate was included in the studies described herein. This study was granted ethics approval by the Biomedical Research Ethics Board of the McGill University Health Center (MUHC study BMC-08-002).
Molecular typing methods.
The RD239, RD750, and RD105 deletions (14) are used to specifically define isolates belonging to the “Indo-Oceanic,” “East African/Indian,” and “East Asian” (or “W/Beijing”) strain lineages, respectively. These regional names are a reference to the fact that, in their original description, each of these lineages appeared to be restricted to patients originating from certain geographic locations. RD9 is absent from all members of the M. tuberculosis complex (MTC) other than M. tuberculosis and M. canetti (5) and was used in the current study to identify potential atypical MTC infections, including those caused by M. africanum (annotated as M. tuberculosis West African-1 and -2 by Gagneux et al. [14]), M. microti, M. caprae, or M. bovis.
DNA was extracted from Lowenstein-Jensen agar slant-grown bacteria using cetyltrimethylammonium bromide-NaCl according to established protocols (2, 36). Lineage-defining LSPs were detected by multiplex PCR using the oligonucleotide primers specified in Table A1 in the appendix. In addition to a “forward” primer specific for the upstream region of each LSP, each multiplex reaction also included two “reverse” primers—one internal to the deleted region and one located immediately downstream of the LSP (14). The two reverse primers were designed so as to allow an unambiguous visual assignment of strain lineage based on the size of the PCR products obtained. Reactions were carried out using a 96-well format in 25-μl volumes that included 10 ng genomic DNA, 0.4 μM of each oligonucleotide, 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, 1× Fermentas Taq buffer (+KCl), and 0.5 U Taq polymerase (Fermentas). Where necessary, reactions were optimized through the addition of 5% to 10% dimethyl sulfoxide (Sigma). A denaturation step was carried out at 94°C for 2 min, followed by 35 cycles of 94°C for 10 s, 58°C for 10 s, and 68°C for 30 s. PCR products were electrophoresed on 1% agarose gels and visualized under UV light following ethidium bromide staining.
TABLE A1.
Primers used in LSP and DNA sequence analysis
| Lineage | LSP | Primer | Sequence (5′-3′) | Product size (bp)
|
|
|---|---|---|---|---|---|
| Intact | Deleted | ||||
| Indo-Oceanic | RD239 | RD239-F | CGTAGACTGCTCGCATGACC | 531 | |
| RD239-Rint | CAGTGAGATCCCAAATGCTGC | 344 | |||
| RD239-Rdel | CGACGCAATCTGACCGACAG | ||||
| East African/Indian | RD750 | RD750-F | GTCAACTGCCGATGGCTGAC | 556 | |
| RD750-Rint | CGTCAGCGATGATCACCTCG | 371 | |||
| RD750-Rdel | GTGAACTAGGTCGAGCATCG | ||||
| East Asian | RD105 | RD105-F | GGTCATATCACGCGTTCGTG | 547 | |
| RD105-Rint | TGCGGTCAAAGCACGCCTTG | 330 | |||
| RD105-Rdel | GGTGGCCCAGAAACCACCA | ||||
| RD207 | RD207-F | GTCTGACGACTGACAGGGTG | 516 | ||
| RD207-Rint | CACGATGGCCACCTCCATG | 682a | |||
| RD207-Rdel | CGTTGCGTGCTCGACGCTG | ||||
| RD181 | RD181-F | CAACAGCACCAGCATCGGAC | 326 | ||
| RD181-Rint | CTGCCGGTCTTAGTCTGCTC | 537 | |||
| RD181-Rdel | CTTGCTATCGGCGTCGTTGC | ||||
| RD150 | RD150-F | CCATCCTGGCGTTGGTTGG | 505 | ||
| RD150-Rint | GCCATCGCGACGGTCAATG | 300 | |||
| RD150-Rdel | CCGAGGACCTTACTGCGTG | ||||
| RD142 | RD142-F | GGAGGACACATGTCGCAACAC | 400 | ||
| RD142-Rint | CGTGCAGCACGAACACCAC | 577 | |||
| RD142-Rdel | GTCGCAGCGCGAGTAGATC | ||||
| Euro-American/African | pks1-15 | RD1-15-F | GGTGTCCTCCTTTGGGATCA | 561b | |
| RD1-15-R | CTGTCAAGCAAATCCGCATCG | ||||
| Atypical MTC | RD9 | RD9-F | GTGACGGTATCGTCGAGCAG | 501 | |
| RD9-Rint | CGCGCCAACTATGTCTACGG | 368 | |||
| RD9-Rdel | GCTCGAGCTAGACCTGCAC | ||||
| RD711-F | GGTTGGCCACTACCAGAGAC | 580 | |||
| RD711 | RD711-Rint | GAACTCGCCGACTAGGTCG | 361 | ||
| RD711-Rdel | CGACGAAGTGCGTGATTTCG | ||||
| RD702 | RD702-F | CAGCAGCAGGGTGTCATTGC | 553 | ||
| RD702-Rint | GCAGCAGCACGATTCCTTGC | 355 | |||
| RD702-Rdel | GATCGTCGCCGACCAGTGT | ||||
| RD713 | RD713-F | GCACTACCACCATCTGCGCT | 504 | ||
| RD713-Rint | CCCACGCTCTTGAGCTGATC | 316 | |||
| RD713-Rdel | CCCAACCCAGGTATTGGTCC | ||||
| RD10 | RD10-F | CACTGACCATGCCATTTAC | 234 | ||
| RD10-Rint | CGTAACTCACCGGGACCG | 478 | |||
| RD10-Rdel | CATCGACATGTTTACCTTC | ||||
| RD13 | RD13-F | GAAGCAGGTGTCACGATCGG | 526 | ||
| RD13-Rint | CCGCAGATCGGATACTCGAC | 250 | |||
| RD13-Rdel | ATGTGCTGGCTGGCCTGATG | ||||
| RD4 | RD4-F | CAAGCCGTAGTCGTGCAGAAG | |||
| RD4-Rint | CTGATTACACCGGCATCGACG | 368 | |||
| RD4-Rdel | CTGCAGCATCTGTTGAGTACGC | 607 | |||
Product results from the RD207-Rint and RD207-Rdel primers due to a sequence inversion within RD207-deleted strains.
Size of pks1-15 PCR fragment subject to DNA sequencing.
IS6110 RFLP typing is carried out routinely as part of an ongoing program that monitors TB transmission on the island of Montreal and is described elsewhere (18, 24, 35). Isolates with fewer than six copies of the IS6110 element are also subject to spoligotyping due to the low level of specificity of RFLP for these “low-copy-number” isolates (33, 35, 41). In the present study, clustered isolates were defined as possessing identical IS6110 fingerprints and, in addition, for isolates with less than six IS6110 copies, identical spoligotypes were also required. IS6110 fingerprints were scanned and analyzed using GelCompar II (Applied Maths NV, Belgium). Spoligotype patterns were compared manually in Microsoft Excel (35).
DNA sequence analysis.
Isolates belonging to the “Euro-American/African” lineage were identified through sequence analysis of a portion of the polyketide synthase 1-15 gene (pks1-15) that is known to contain a 7-bp deletion within all isolates of this lineage (14, 26). Oligonucleotides used for both PCR amplification and sequencing of this region are listed in Table A1 in the appendix. PCR conditions were identical to those described above, and sequence analysis of the reaction products was carried out at the McGill University and Genome Québec Innovation Centre. Statistical analysis for age, sex, and site of disease revealed no evident bias in those samples subject to sequencing.
RESULTS
Population structure of Montreal M. tuberculosis isolates.
Eight hundred twenty-seven bacterial isolates representing each of the culture-confirmed TB cases diagnosed on the island of Montreal between January 2001 and May 2007 were obtained from the LSPQ as part of ongoing molecular epidemiology investigations monitoring TB transmission. Of these, genomic DNA was able to be extracted in sufficient quantity for PCR-based LSP genotyping, IS6110 RFLP, and spoligotyping (for isolates with <6 IS6110 bands) for a total of 798 isolates (96% of isolates received). Country-of-origin information was available for 730 of these patient isolates (91%), and 606 (83%) of these originated from foreign-born patients. The latter figure is reflective of previous studies that highlight the fact that immigrant communities are the major risk group for developing active TB within the city of Montreal (24, 34). For the 555 foreign-born patients for whom the year of arrival is known, the median time from entry into Canada to diagnosis of active TB was 5 years (range, 0 to 60 years). Within this group of individuals, 36.4% of all TB cases occurred within 2 years of arrival. This figure is similar to that previously recorded (33.2%) for a 1992-to-1995 Montreal TB patient cohort (34). The age range of the Canadian-born patients is 0.5 to 100 years (median, 52 years), and that of the foreign-born patients is 2 to 95 years (median, 38 years). Finally, the proportions of TB cases associated with males were 63.7% and 51.8% among Canadian-born and foreign-born individuals, respectively.
Using a multiplex PCR-based approach, all 798 isolates were typed for the presence of the RD239, RD750, RD105, and RD9 LSP markers. The proportion of isolates within our Montreal TB data bank belonging to the Indo-Oceanic lineage (RD239 deleted) was determined to be 17.5%. Similarly, 9.0% of isolates were classified as East Asian (RD105 deleted), 5.9% as East African/Indian (RD750), and 0.8% as atypical MTC (RD9 deleted) (see Table S1 in the supplemental material).
Preliminary attempts to design a reliable multiplex PCR typing strategy for identification of strains that fall within the last of the six major strain families described thus far—the Euro-American lineage—were largely unsuccessful. This particular lineage is defined by a short (7-bp) deletion in the pks1-15 gene (14) and consists of strains previously classified as belonging to principal genetic groups II and III (37). In order to confirm that the remainder of the Montreal isolates did, in fact, belong to this large lineage, a representative sample of isolates were chosen among the 533 isolates found to be intact for each of the RD239, RD750, RD105, and RD9 LSPs. The region of pks1-15 containing the deleted segment was PCR amplified and sequenced for 270 (51%) randomly selected samples, and 100% of these were found to contain the expected 7-bp deletion characteristic of the Euro-American lineage (9, 14). Although this obviously does not discount the possibility that an additional, previously undescribed M. tuberculosis lineage exists among the 263 isolates that were not tested for the pks1-15 deletion, it is clear that the frequency of such an isolate(s) is expected to be extremely low in our sample population—certainly not higher than that observed for the atypical MTC lineages. As such, we have made the assumption that all of the isolates that were not classified as Indo-Oceanic, East Asian, East African/Indian, or atypical MTC belong within the Euro-American lineage. Therefore, this lineage accounts for a clear majority (67%) of isolates currently present on the island of Montreal (see Table S1 in the supplemental material). Importantly, none of the DNA samples analyzed throughout the course of this study were found to possess more than one of the markers examined, highlighting both the specificity of the lineage-defining LSP markers and the fact that cross-contamination of bacterial or DNA samples has not occurred to any discernible extent within our isolate data bank.
“Phylogeographic” associations of major M. tuberculosis lineages.
Country-of-origin information was available for 730 patient isolates, and 606 (83%) of these were collected from patients who were self-identified as being foreign born. These foreign-born patients represented 80 different countries that were well distributed over 16 diverse geographic regions. Only the Pacific region was poorly represented in our patient cohort. Aside from Canada (17% of patients in the cohort for whom the country of origin was recorded), 19.3% of patients were from the Americas or the Caribbean, 8.6% from Europe, 21.8% from Africa and the Middle East, 12.5% from the Indian subcontinent, and 20.7% from Asia (Table 1).
TABLE 1.
LSP deletion analysis of Montreal M. tuberculosis isolates organized by geographic region
| Region | No. of isolates
|
|||||
|---|---|---|---|---|---|---|
| By region | By lineage:
|
|||||
| Indo-Oceanic (RD239) | East African/Indian (RD750) | East Asian (RD105) | Euro-American/African (pks1−15) | Atypical MTC (RD9) | ||
| North America | 127 | 4 | 2 | 6 | 115 | |
| Central America | 7 | 0 | 0 | 0 | 7 | |
| South America | 27 | 0 | 0 | 0 | 27 | |
| Caribbean | 104 | 2 | 1 | 0 | 101 | |
| Eastern Europe | 27 | 0 | 0 | 2 | 24 | 1 |
| Western Europe | 36 | 1 | 0 | 0 | 35 | |
| Middle East | 19 | 0 | 5 | 1 | 13 | |
| North Africa | 33 | 1 | 0 | 0 | 32 | |
| South Africa | 14 | 2 | 0 | 0 | 12 | |
| West Africa | 15 | 0 | 0 | 0 | 12 | 3 |
| East Africa | 13 | 0 | 2 | 1 | 10 | |
| Central Africa | 65 | 1 | 0 | 0 | 63 | 1 |
| Indian subcontinent | 91 | 33 | 33 | 2 | 23 | |
| Southeast Asia | 107 | 78 | 0 | 19 | 10 | |
| East Asia | 44 | 2 | 0 | 32 | 10 | |
| Pacific | 1 | 0 | 0 | 0 | 1 | |
| Total (% of total) | 730 | 124 (17.0%) | 43 (5.9%) | 63 (8.6%) | 495 (67.8%) | 5 (0.7%) |
| No. (%) of clustered casesa | 134 (18.4%) | 19 | 6 | 109 | ||
Number of cases involving clustered (nonunique) isolates as determined by RFLP and spoligotype analysis. Where all patients in a particular cluster were born in the same country, the number of “clustered (secondary) cases” was assumed to equal n − 1, where n represents the total number of cases attributed to that cluster.
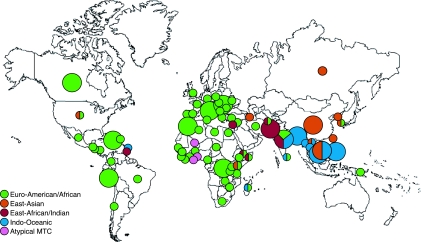
When the isolate LSP genotypes were stratified according to patient country of origin, it was immediately apparent that the striking regional trends with respect to bacterial lineage described in the San Francisco study were very precisely mirrored within the Montreal population (Table 1; see also Table S1 in the supplemental material). These “phylogeographical” trends can be summarized as follows: the Indo-Oceanic lineage (RD239) was almost entirely restricted to TB patients originating from either the Indian subcontinent or Southeast Asia. Indeed, 90% of the 124 cases due to the Indo-Oceanic lineage were from patients from these two regions, and in particular from Bangladesh, India, Vietnam, and the Philippines (Fig. 1 and 2). Seventy-seven percent of isolates within the East African/Indian lineage also originated from the Indian subcontinent, with only a small number of cases identified from the East Africa region (2 out of 13 patients from the region). The Middle East region accounted for 12% of the East African/Indian TB cases. The RD105-deleted East Asian or W/Beijing lineage was largely restricted to patients with Eastern or Southeast Asian heritage (82%), while the predominant lineage in our study sample, the Euro-American lineage, was identified in patients from all 16 geographic regions and, indeed, from all but 12 of the 81 countries represented. The regions that were predominantly Euro-American include Europe and the Americas, the Caribbean, the Middle East, and all subregions of Africa. Together, these regions accounted for 91% of all TB cases due to Euro-American isolates. Given the preponderance of this lineage throughout Africa both in our study and in the Gagneux study (14), we feel that the Euro-American tag is a misnomer and may well be misleading with respect to the origins of this lineage. As such we will refer to this lineage as “Euro-American/African” in order to reflect its broad global distribution (Fig. 1 and 2). Finally, 80% of RD9-deleted (atypical MTC) samples were from patients from either western or central Africa. Despite the fact that there are 68 samples listed as “country unknown” in our database, we do not believe that this has created a bias in our analysis. Indeed, we observe no significant difference in the ratios of the five major lineages between isolates associated with a particular country of origin and those of unknown origin (χ2 = 5.22, P = 0.27).
FIG. 1.
Global distribution of countries and lineages represented by Montreal TB patients. Each color represents a distinct LSP-defined TB lineage. Large circles indicate where ≥10 cases were detected in patients originating from a single country. Small circles indicate <10 cases. Symbols incorporating multiple colors indicate that more than one lineage accounts for ≥25% of all cases originating from that particular country. Only those cases identified as being “unique” through IS6110 RFLP or spoligotype analysis (i.e., isolates not involved in forming a cluster of recent transmission) have been included. (The world outline map used in the preparation of the figure was obtained from WorldAtlas.com and is used with permission.)
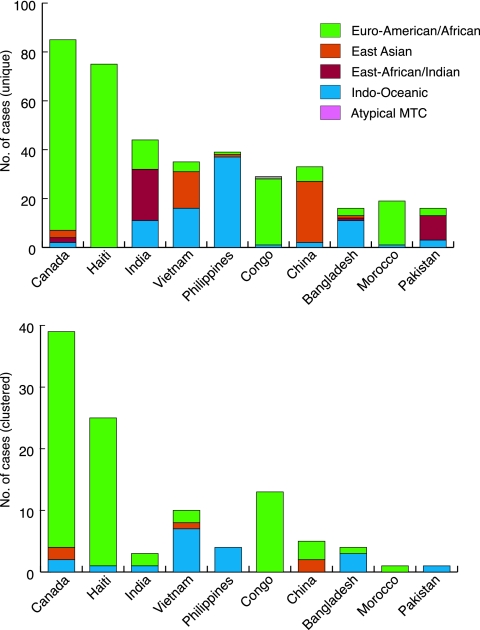
FIG. 2.
Major countries and lineages represented among Montreal TB cases. The “top 10” countries of origin with the highest numbers of TB cases are shown and are stratified according to LSP-defined TB lineage. The upper panel consists of only the isolates identified as being IS6110 RFLP or spoligotype “unique.” The lower panel consists of isolates involved in chains of recent transmission (“clustered”). Note that the relative scales of the two panels differ.
Information regarding the sublineage classification of the RD105 and RD9 isolates is provided in the appendix.
Comparative assessment of LSP, RFLP, and spoligotyping methods.
One would predict that isolates bearing identical IS6110 fingerprints would be classified within the same major lineage through LSP analysis. To confirm this, IS6110 patterns were compared for all isolates within the Montreal cohort. For the isolates with ≥6 IS6110 copies, 114 samples were identified within 44 clusters (data not shown). An additional 11 clusters involving 41 low-copy-number isolates were subsequently identified through spoligotyping. In both instances, the individual clusters ranged in size from two to nine patients/cluster (median = 2). The fact that the great majority of clusters involve ≤3 individuals (87%) is again indicative of the low rates of ongoing TB transmission in Montreal.
Where the country of origin is known, the total number of isolates associated with recent transmission was 134 (18.4% of all cases), and aside from East Africa, all geographic regions represented in the Montreal patient database were involved in one or more clusters (Table 1; see also Table S1 in the supplemental material). More importantly, all 55 clusters were composed of isolates within the same LSP lineage, thereby adding to the validity of using LSPs as lineage-defining markers. Of clustered cases, 81.3% involved the Euro-American/African lineage, 14.2% involved the Indo-Oceanic lineage, and 4.5% involved the East Asian lineage. Neither the RD750-deleted East African/Indian nor the RD9-deleted atypical MTC families were involved in clustered TB cases in Montreal (Fig. 2; see also Table S1 in the supplemental material).
For the low-IS6110-copy-number samples, it is interesting that when spoligotyping is not included as part of the analysis, a total of 92 isolates appear to form 20 clusters of cases. Of these, five are composed of isolates from different LSP lineages (Euro-American/African and Indo-Oceanic; data not shown). This result serves as a direct demonstration of the poor specificity of using IS6110 RFLP for epidemiological investigations where low-copy-number isolates are involved (27, 33). A similar phenomenon arises when spoligotyping is carried out independently of the IS6110 fingerprint for these same low-copy-number samples. In this case, 78 isolates appear to form clusters, of which three are composed of mixed lineages.
Epidemiological features of major M. tuberculosis lineages.
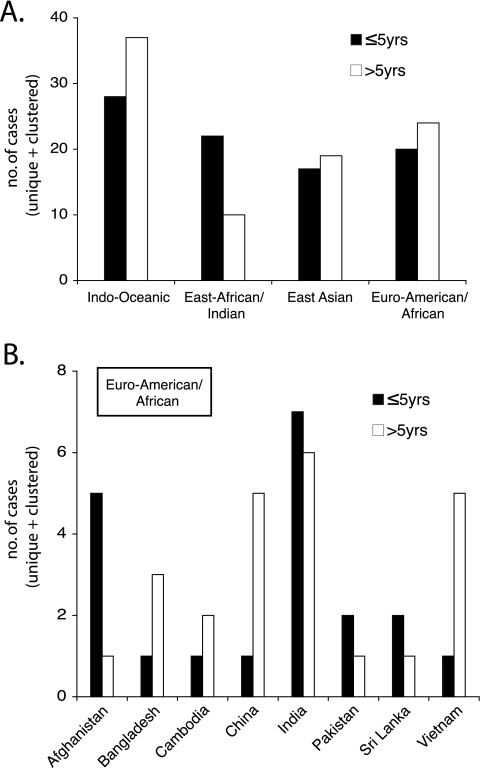
In Montreal, where transmission and incidence rates of TB are low, it is difficult to undertake a comparative assessment of virulence or transmissibility for each of the major lineages—particularly when almost 70% of active cases in Montreal are due to a single lineage. However, for the cohort of patients originating from countries where a mixture of lineages was found to cause infection, we assessed whether or not there were any observable lineage-specific trends in regards to causing early (i.e., patients diagnosed with active TB ≤5 years following immigration to Canada) or late (>5 years following immigration) disease. As TB transmission in Montreal occurs relatively infrequently, the “early” group most likely represents those patients originally infected outside Canada. The “late” group could also include patients infected while living in Montreal, where exposure in the general community is most likely to be due to the Euro-American/African lineage. We would predict that among ethnically related individuals infected with a range of strains from distinct lineages, if a particular lineage is more virulent and therefore more prone to cause active disease, then it should appear more frequently among the cases occurring during the early time frame. Similarly, if a particular lineage is more transmissible, then we would predict an increase in the frequency of cases due to this lineage over time. However, from Fig. 3A, it can be seen that the Indo-Oceanic, East Asian, and Euro-American/African lineages appear equally likely to cause disease during both the early and late time frames. There was also no indication that the lineage that predominates in Montreal, the Euro-American/African lineage, was becoming established over time within these ethnic communities (Fig. 3B). Only the East African/Indian lineage (RD750) demonstrated a trend toward causing active disease more often among newly arrived (68.8%) than among established immigrants.
FIG. 3.
Lineage-related trends over time for patients from “mixed-lineage” countries. (A) For foreign-born patients with either “early” (≤5 years following arrival in Canada) or “late” (>5 years after arrival in Canada) TB, the sum of the lineage-specific trends for each of the countries listed in panel B is presented. The former are most likely to represent patients infected outside Canada; the latter could also include patients infected within Montreal, where general exposure is most likely to be due to the Euro-American/African lineage. (B) The Euro-American/African lineage data presented in panel A, arranged by country. Only those countries of origin where multiple lineages are present (each of which represents a significant proportion of cases within the countries involved) have been included. For both panels, all TB cases are presented (unique plus clustered).
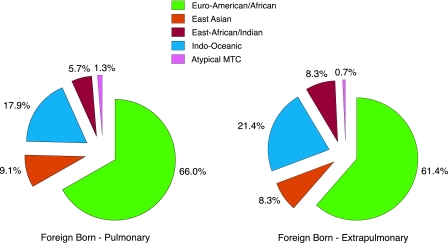
To examine for a possible effect of lineage on disease presentation, both pulmonary and extrapulmonary TB cases were stratified according to lineage. While 17.5% of Canadian-born patients exhibited nonrespiratory symptoms, this cohort was largely uninformative due to the fact that the predominant Euro-American/African lineage accounted for 89% and 94% of pulmonary and extrapulmonary cases, respectively (data not shown). Within the foreign-born cohort, 31% of patients were diagnosed with extrapulmonary TB. As demonstrated by Fig. 4, no significant difference was observed between lineages with respect to their association with either pulmonary or extrapulmonary disease (χ2 = 2.39, P = 0.66).
FIG. 4.
Major M. tuberculosis lineages and primary site of disease among foreign-born TB patients residing in Montreal. The proportions of foreign-born patients infected with each of the major LSP-defined lineages and displaying either pulmonary or extrapulmonary disease are presented. Both unique and clustered TB cases have been included in the analysis.
DISCUSSION
A remarkable degree of concordance exists between our study and the San Francisco study with respect to the phylogeographic associations of the major strain lineages. This finding is even more striking when it is considered that the patient distributions between the two populations are quite different. For example, 71% of the San Francisco patients were born in just five countries or regions (the United States, China, the Philippines, Vietnam, and Central America) and of the original 875 samples subject to LSP analysis, 85% were from the Americas (24%), Southeast Asia (31%), or East Asia (30%). In comparison, 43% of Montreal TB patients with a known country of origin were born outside these three regions and only 21% of isolates originated from East Asian or Southeast Asian patients. In this regard, the Montreal cohort is much more geographically diverse than the corresponding San Francisco cohort, with a much greater representation from Africa (19% of isolates), the Indian subcontinent (12.5%), and Eastern and Western European countries (9% of isolates). Nevertheless, despite the absence of significant numbers of patients from these regions within the Gagneux study, the results were highly predictive of what we found in Montreal, and as such, the lineage distributions between the two studies are virtually superimposable (14, 15). The fact that the lineage-country associations are so highly conserved for the Montreal isolates that were collected in the 6 years following the San Francisco cohort study (1991 to 2001) is strong evidence in support of the fact that the strain distributions within the various regions and countries represented by the two studies have remained largely constant for at least the past 2 decades.
For both the Montreal and San Francisco patient cohorts, the majority of isolates were determined to be part of the highly ubiquitous Euro-American/African lineage that comprises 67% and 48% of Montreal and San Francisco isolates, respectively. In both cases, this particular lineage was isolated from TB patients originating from each of the geographic regions represented by the studies. In this respect, the Euro-American/African lineage is unique, and its peculiar distribution pattern is presumably a function of multiple migration routes involving the individuals infected with these strains. For both cohorts, this lineage is clearly predominant throughout the Americas, Europe, and Africa, and together with Canadian-born patients, patients from Haiti and the Democratic Republic of Congo comprise more than half of all Montreal cases involving this lineage. Interestingly, the Haitian community continues to be a “high-risk” group for developing active TB within Montreal, as has been noted previously in two retrospective studies carried out on patient samples collected between 1992 and 1998 (24, 34). Although not attempted in the present study, it is possible to classify isolates of the Euro-American/African lineage into a number of distinct sublineages that show a degree of geographic restriction through the use of additional LSP or SNP markers (13, 14, 17).
Although the Indo-Oceanic clade appeared to be largely restricted to Southeast Asia within the San Francisco patient population, a significant proportion of these isolates (27%) were also found in Montreal patients originating from the Indian subcontinent. As this lineage retains the TbD1 region, it may be tempting to speculate that this lineage arose in the Indian subcontinent or Southeast Asia. However, a small number of these isolates also occur throughout much of Africa. It is therefore possible that this lineage has its origins in Africa and was transported east along human migration routes into regions where, for reasons of host-pathogen adaptation, it has continued to flourish. A recent publication by Wirth et al. has also suggested an African origin for this particular lineage (42). As in the Gagneux et al. analysis, the RD105-deleted East Asian or W/Beijing lineage was largely restricted to patients of Asian (East or Southeast) origin. The last of the major lineages, namely, the East African/Indian lineage, occurs most frequently among Montreal patients originating from countries within the Indian subcontinent, once again reflecting the San Francisco data. However, unlike the latter study, we identified very few East African patients harboring this lineage. In Montreal, one-quarter of all Middle Eastern patients were infected with East African/Indian isolates.
While the level of ongoing TB transmission in Montreal is quite low in comparison to other major urban centers (when identical RFLP matching criteria are used, transmission is estimated at 4 to 12% [18, 24, 35]), 100% of the 44 high-IS6110-copy-number clusters identified using identical matching criteria were found to be comprised of isolates belonging to the same LSP-defined lineage. Similarly, each of the 11 low-copy-number clusters identified through a combination of RFLP and spoligotyping contained isolates within the same strain lineage. However, in the case of the low-copy-number isolates, the analysis of IS6110 fingerprints or spoligotypes alone is not sufficient to correctly identify isolates implicated in chains of TB transmission (27, 33). Although not directly tested as part of this study, it is likely that the poor correlation of the spoligotyping technique is not a unique property of the low-IS6110-copy-number isolates. Hence, while obviously not a substitute for RFLP in contact or transmission investigations, we believe that use of LSP typing may serve as a supplement for ensuring that clustered or matched isolates have been correctly identified, particularly when dealing with low-IS6110-copy-number isolates or where spoligotyping is used in the absence of IS6110 RFLP. In support of this, Gutacker et al. have also recently observed that M. tuberculosis isolates bearing <6 IS6110 copies are able to be represented across very different SNP-defined phylogenetic lineages (17).
In Montreal, the proportion of foreign-born patients displaying extrapulmonary sites of TB disease is significantly higher than that of the Canadian-born cohort (31% versus 17.5%; χ2 = 7.16, P = 0.0075). Although these observations may, in part, reflect the greater strain diversity that exists among immigrant patients, the fact that we observed no significant association of any of the major lineages with extrapulmonary disease would tend to support the idea that one or more sociological, host, or clinical/diagnostic factors are involved. Human immunodeficiency virus infection is not expected to play a significant role here as the overall rate of TB-human immunodeficiency virus coinfection in Montreal is estimated to be below 10% (4, 34).
Gagneux et al. have recently proposed that the major M. tuberculosis lineages have evolved so as to become adapted to specific host genetic backgrounds and are much more likely to transmit and cause disease among patients of the same ethnicity (14). Although our data regarding the lack of assimilation of the predominant Euro-American/African lineage into immigrant communities in Montreal tend to support this interesting hypothesis (Fig. 3B), we also feel that at this stage we cannot discount the possibility that a lack of social mixing among different ethnic groups may also have contributed to this phenomenon.
Within the unique setting of Montreal, where we have a low overall incidence of TB and low rates of transmission, there is little evidence to support the suggestion that the East Asian lineage (RD105) is more highly virulent, more highly transmissible, or more likely to cause extrapulmonary disease as has been previously suggested (7, 10, 12, 19, 20, 23). However, our findings clearly do not preclude the possibility that this lineage may be associated with these phenotypes in other epidemiological settings where ongoing transmission of these strains is taking place and where clinical and public health interventions are less efficient than those in Montreal. Similarly, while it may be tempting to speculate that the East African/Indian lineage (RD750) is less transmissible (due to the fact that it does not appear among any of the clustered cases) and yet more likely to result in active disease (Fig. 3A), it would appear equally likely that this effect is attributable to any number of sociological factors associated with the Indian subcontinent communities that almost exclusively harbor this strain.
In conclusion, our data strongly support the global population structure of M. tuberculosis as originally proposed by Gagneux and colleagues. While the ranges of countries represented in the two independent studies are quite distinct, the geographical associations of the major LSP-defined M. tuberculosis lineages remain very clear. Finally, the need for a consensus among researchers as to how to define and describe families of genetically related isolates that presumably share common phenotypes or disease-related traits has never been more urgent for the types of strain-linked association studies that are becoming commonplace these days (6, 7, 10, 20, 23, 29). At present, the LSP/RD framework would appear to be the most robust and universally applicable model that will hopefully become the standard among epidemiological researchers.
Supplementary Material
Acknowledgments
This work was funded by Canadian Institutes of Health Research grants MOP82931 (M.B.R.), RRD153877 (D.M.), and MOP53184 (K.S.). A.M. was supported by a studentship from the Montreal Chest Institute Research Centre. M.B.R. is supported by a New Investigator Salary Award from the Canadian Institutes of Health Research. K.S. is supported by a Chercheur-Boursier Clinicien Career Award from the Fonds de la Recherche en Santé du Québec (FRSQ). M.A.B. is supported by a Chercheur-Boursier Senior Award from the FRSQ.
APPENDIX
Sublineage classification of the RD105 and RD9 isolates.
In order to more precisely define the nature of the atypical MTC samples in our collection, we first examined each of them for the RD711 and RD702 LSPs that are used to classify the type I (or subtype 1b) and type 2 (or subtype 1a) subbranches of M. africanum, respectively. M. africanum is endemic to western Africa, where it is responsible for up to 50% of smear-positive TB cases (11). As the disease caused by these organisms is perhaps indistinguishable from that of M. tuberculosis, Gagneux et al. have referred to these subbranches as M. tuberculosis West African-1 and West African-2 (14). Of the five RD9-deleted samples associated with a known country of origin, three contained the RD711 LSP and all three of these isolates were from West African patients (from Ghana, Mali, and Togo). None of the five samples were found to be deleted for RD702. One isolate from a Romanian patient was deleted for RD7, RD10, and RD13 but intact for RD4 (unique to “classical” M. bovis and its derivatives) and was therefore designated as M. caprae—a species that predominantly infects both wild and domesticated animals in several European countries (5). The remaining sample was isolated from a Central African patient (Democratic Republic of Congo) and was intact for each of the aforementioned deletions. Interestingly, it was also intact for the RD713 deletion that has been used previously to classify a small number of the type 1 M. africanum strains (M. tuberculosis West African-1) (22, 28). Although beyond the scope of the current study, it is likely that this isolate represents an intermediary step along the pathway leading to the differentiation of type 1 (RD9-deleted) and type 2 (RD7-, RD8-, RD9-, and RD10-deleted) M. africanum lineages.
We also decided to compare the sublineage distributions for one of the major strain families between the San Francisco and Montreal TB patient cohorts. For this purpose we chose the East Asian (W/Beijing) lineage, which has five well-defined sublineages based on the presence of specific LSPs (14, 39). Again, the overall distribution of the five sublineages was largely conserved between the two patient cohorts, with group 3 strains (RD105, RD207, and RD181 deleted) accounting for the vast majority of all East Asian isolates. This particular sublineage comprises 67% and 82% of East Asian isolates within the San Francisco (39) and Montreal databases, respectively. The next most common sublineage was the group 4 sublineage (RD105, RD207, RD181, and RD150 deleted), which accounts for 20.5% (San Francisco) and 11.3% (Montreal) of East Asian strains. Group 2 strains (RD105 and RD207 deleted) make up 8% (San Francisco) and 5% (Montreal) of isolates. Finally, the remainder of Montreal isolates fell within the group 1 subcategory (1.6%; RD105 deleted only), while the remainder of San Francisco isolates belong in group 5 (4.5%; RD105, RD207, RD181, and RD142 deleted). Whether these shared trends reflect the actual sublineage distribution extant within the East Asian and Southeast Asian regions or are reflective of certain patient characteristics common to both the San Francisco and Montreal immigrant populations is unknown at present. There were no obvious associations between particular countries of origin and any of the individual sublineages (data not shown).
For our set of isolates, the sequence of the RD207 deletion was determined not to be exactly as reported previously (14). A putative IS6110-related transposase sequence included in the original description of RD207 was found to be present within the RD207-deleted strains, albeit in an inverted orientation with respect to H37Rv. Immediately upstream of this sequence, we also identified an additional five unique spacer regions that form part of the direct repeat region of the genome (data not shown).
Primers used in LSP and DNA sequence analysis are shown in Table A1.
Footnotes
Published ahead of print on 11 February 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Alland, D., D. W. Lacher, M. H. Hazbon, A. S. Motiwala, W. Qi, R. D. Fleischmann, and T. S. Whittam. 2007. Role of large sequence polymorphisms (LSPs) in generating genomic diversity among clinical isolates of Mycobacterium tuberculosis and the utility of LSPs in phylogenetic analysis. J. Clin. Microbiol. 4539-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2002. Short protocols in molecular biology, 5th ed., vol. 1. John Wiley & Sons, Inc., Hoboken, NJ.
- 3.Barnes, P. F., and M. D. Cave. 2003. Molecular epidemiology of tuberculosis. N. Engl. J. Med. 3491149-1156. [DOI] [PubMed] [Google Scholar]
- 4.Brassard, P., and R. S. Remis. 1999. Incidence of tuberculosis among reported AIDS cases in Quebec from 1979 to 1996. CMAJ 1601838-1842. [PMC free article] [PubMed] [Google Scholar]
- 5.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 993684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caws, M., G. Thwaites, S. Dunstan, T. R. Hawn, N. T. N. Lan, N. T. T. Thuong, K. Stepniewska, M. N. T. Huyen, N. D. Bang, T. H. Loc, S. Gagneux, D. van Soolingen, K. Kremer, M. van der Sande, P. Small, P. T. H. Anh, N. T. Chinh, H. T. Quy, N. T. H. Duyen, D. Q. Tho, N. T. Hieu, E. Torok, T. T. Hien, N. H. Dung, N. T. Q. Nhu, P. M. Duy, N. V. V. Chau, and J. Farrar. 2008. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 4e1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caws, M., G. Thwaites, K. Stepniewska, T. N. Nguyen, T. H. Nguyen, T. P. Nguyen, N. T. Mai, M. D. Phan, H. L. Tran, T. H. Tran, D. van Soolingen, K. Kremer, V. V. Nguyen, T. C. Nguyen, and J. Farrar. 2006. Beijing genotype of Mycobacterium tuberculosis is significantly associated with human immunodeficiency virus infection and multidrug resistance in cases of tuberculous meningitis. J. Clin. Microbiol. 443934-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 9.Constant, P., E. Perez, W. Malaga, M. A. Laneelle, O. Saurel, M. Daffe, and C. Guilhot. 2002. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex. Evidence that all strains synthesize glycosylated p-hydroxybenzoic methyl esters and that strains devoid of phenolglycolipids harbor a frameshift mutation in the pks15/1 gene. J. Biol. Chem. 27738148-38158. [DOI] [PubMed] [Google Scholar]
- 10.de Jong, B. C., P. C. Hill, A. Aiken, T. Awine, M. Antonio, I. M. Adetifa, D. J. Jackson-Sillah, A. Fox, K. Deriemer, S. Gagneux, M. W. Borgdorff, K. P. McAdam, T. Corrah, P. M. Small, and R. A. Adegbola. 2008. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J. Infect. Dis. 1981037-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Jong, B. C., P. C. Hill, A. Aiken, D. J. Jeffries, A. Onipede, P. M. Small, R. A. Adegbola, and T. P. Corrah. 2007. Clinical presentation and outcome of tuberculosis patients infected by M. africanum versus M. tuberculosis. Int. J. Tuberc. Lung Dis. 11450-456. [PubMed] [Google Scholar]
- 12.European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis. 2006. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg. Infect. Dis. 12736-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gagneux, S., K. DeRiemer, T. Van, M. Kato-Maeda, B. C. de Jong, S. Narayanan, M. Nicol, S. Niemann, K. Kremer, M. C. Gutierrez, M. Hilty, P. C. Hopewell, and P. M. Small. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 1032869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gagneux, S., and P. M. Small. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7328-337. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, A. L., R. C. Huard, N. C. Gey van Pittius, L. C. Lazzarini, J. Driscoll, N. Kurepina, T. Zozio, C. Sola, S. M. Spindola, A. L. Kritski, D. Fitzgerald, K. Kremer, H. Mardassi, P. Chitale, J. Brinkworth, D. Garcia de Viedma, B. Gicquel, J. W. Pape, D. van Soolingen, B. N. Kreiswirth, R. M. Warren, P. D. van Helden, N. Rastogi, P. N. Suffys, J. Lapa e Silva, and J. L. Ho. 2008. Application of sensitive and specific molecular methods to uncover global dissemination of the major RDRio sublineage of the Latin American-Mediterranean Mycobacterium tuberculosis spoligotype family. J. Clin. Microbiol. 461259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutacker, M. M., B. Mathema, H. Soini, E. Shashkina, B. N. Kreiswirth, E. A. Graviss, and J. M. Musser. 2006. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J. Infect. Dis. 193121-128. [DOI] [PubMed] [Google Scholar]
- 18.Haase, I., S. Olson, M. A. Behr, I. Wanyeki, L. Thibert, A. Scott, A. Zwerling, N. Ross, P. Brassard, D. Menzies, and K. Schwartzman. 2007. Use of geographic and genotyping tools to characterise tuberculosis transmission in Montreal. Int. J. Tuberc. Lung Dis. 11632-638. [PubMed] [Google Scholar]
- 19.Hanekom, M., G. D. van der Spuy, N. C. Gey van Pittius, C. R. McEvoy, S. L. Ndabambi, T. C. Victor, E. G. Hoal, P. D. van Helden, and R. M. Warren. 2007. Evidence that the spread of Mycobacterium tuberculosis strains with the Beijing genotype is human population dependent. J. Clin. Microbiol. 452263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanekom, M., G. D. van der Spuy, E. Streicher, S. L. Ndabambi, C. R. McEvoy, M. Kidd, N. Beyers, T. C. Victor, P. D. van Helden, and R. M. Warren. 2007. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J. Clin. Microbiol. 451483-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsh, A. E., A. G. Tsolaki, K. DeRiemer, M. W. Feldman, and P. M. Small. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA 1014871-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huard, R. C., M. Fabre, P. de Haas, L. C. Lazzarini, D. van Soolingen, D. Cousins, and J. L. Ho. 2006. Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J. Bacteriol. 1884271-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong, Y., M. D. Cave, L. Zhang, B. Foxman, C. F. Marrs, J. H. Bates, and Z. H. Yang. 2007. Association between Mycobacterium tuberculosis Beijing/W lineage strain infection and extrathoracic tuberculosis: insights from epidemiologic and clinical characterization of the three principal genetic groups of M. tuberculosis clinical isolates. J. Clin. Microbiol. 45409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulaga, S., M. Behr, K. Musana, J. Brinkman, D. Menzies, P. Brassard, D. Kunimoto, T. N. Tannenbaum, L. Thibert, L. Joseph, J. F. Boivin, and K. Schwartzman. 2002. Molecular epidemiology of tuberculosis in Montreal. CMAJ 167353-354. [PMC free article] [PubMed] [Google Scholar]
- 25.Malik, A. N., and P. Godfrey-Faussett. 2005. Effects of genetic variability of Mycobacterium tuberculosis strains on the presentation of disease. Lancet Infect. Dis. 5174-183. [DOI] [PubMed] [Google Scholar]
- 26.Marmiesse, M., P. Brodin, C. Buchrieser, C. Gutierrez, N. Simoes, V. Vincent, P. Glaser, S. T. Cole, and R. Brosch. 2004. Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150483-496. [DOI] [PubMed] [Google Scholar]
- 27.Mathema, B., N. E. Kurepina, P. J. Bifani, and B. N. Kreiswirth. 2006. Molecular epidemiology of tuberculosis: current insights. Clin. Microbiol. Rev. 19658-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mostowy, S., A. Onipede, S. Gagneux, S. Niemann, K. Kremer, E. P. Desmond, M. Kato-Maeda, and M. Behr. 2004. Genomic analysis distinguishes Mycobacterium africanum. J. Clin. Microbiol. 423594-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newton, S. M., R. J. Smith, K. A. Wilkinson, M. P. Nicol, N. J. Garton, K. J. Staples, G. R. Stewart, J. R. Wain, A. R. Martineau, S. Fandrich, T. Smallie, B. Foxwell, A. Al-Obaidi, J. Shafi, K. Rajakumar, B. Kampmann, P. W. Andrew, L. Ziegler-Heitbrock, M. R. Barer, and R. J. Wilkinson. 2006. A deletion defining a common Asian lineage of Mycobacterium tuberculosis associates with immune subversion. Proc. Natl. Acad. Sci. USA 10315594-15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen, D., P. Brassard, D. Menzies, L. Thibert, R. Warren, S. Mostowy, and M. Behr. 2004. Genomic characterization of an endemic Mycobacterium tuberculosis strain: evolutionary and epidemiologic implications. J. Clin. Microbiol. 422573-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed, M. B., P. Domenech, C. Manca, H. Su, A. K. Barczak, B. N. Kreiswirth, G. Kaplan, and C. E. Barry III. 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 43184-87. [DOI] [PubMed] [Google Scholar]
- 32.Reed, M. B., S. Gagneux, K. Deriemer, P. M. Small, and C. E. Barry III. 2007. The W-Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and has the DosR dormancy regulon constitutively upregulated. J. Bacteriol. 1892583-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhee, J. T., M. M. Tanaka, M. A. Behr, C. B. Agasino, E. A. Paz, P. C. Hopewell, and P. M. Small. 2000. Use of multiple markers in population-based molecular epidemiologic studies of tuberculosis. Int. J. Tuberc. Lung Dis. 41111-1119. [PubMed] [Google Scholar]
- 34.Rivest, P., T. Tannenbaum, and L. Bedard. 1998. Epidemiology of tuberculosis in Montreal. CMAJ 158605-609. [PMC free article] [PubMed] [Google Scholar]
- 35.Scott, A. N., D. Menzies, T. N. Tannenbaum, L. Thibert, R. Kozak, L. Joseph, K. Schwartzman, and M. A. Behr. 2005. Sensitivities and specificities of spoligotyping and mycobacterial interspersed repetitive unit-variable-number tandem repeat typing methods for studying molecular epidemiology of tuberculosis. J. Clin. Microbiol. 4389-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Somerville, W., L. Thibert, K. Schwartzman, and M. A. Behr. 2005. Extraction of Mycobacterium tuberculosis DNA: a question of containment. J. Clin. Microbiol. 432996-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 949869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thwaites, G., M. Caws, T. T. H. Chau, A. D'Sa, N. T. N. Lan, M. N. T. Huyen, S. Gagneux, P. T. H. Anh, D. Q. Tho, E. Torok, N. T. Q. Nhu, N. T. H. Duyen, P. M. Duy, J. Richenberg, C. Simmons, T. T. Hien, and J. Farrar. 2008. Relationship between Mycobacterium tuberculosis genotype and the clinical phenotype of pulmonary and meningeal tuberculosis. J. Clin. Microbiol. 461363-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsolaki, A. G., S. Gagneux, A. S. Pym, Y. O. Goguet de la Salmoniere, B. N. Kreiswirth, D. Van Soolingen, and P. M. Small. 2005. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J. Clin. Microbiol. 433185-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. O. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 1014865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Soolingen, D., P. E. de Haas, P. W. Hermans, P. M. Groenen, and J. D. van Embden. 1993. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol. 311987-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wirth, T., F. Hildebrand, C. Allix-Beguec, F. Wolbeling, T. Kubica, K. Kremer, D. van Soolingen, S. Rusch-Gerdes, C. Locht, S. Brisse, A. Meyer, P. Supply, and S. Niemann. 2008. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4e1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.World Health Organization. 2006. Global tuberculosis control—surveillance, planning, financing. World Health Organization report 2006. World Health Organization, Geneva, Switzerland.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.