Abstract
The implementation of real-time PCR for the diagnosis of malaria has been hampered by poor sensitivity for the detection of mixed infections. We have optimized a method that enhances the sensitivity of detection of minor species in mixed infections within a single multiplex reaction. Our assay uses species-specific forward primers in combination with a conserved reverse primer and largely overcomes primer competition for the minor species DNA. With a blind panel of clinical samples, we successfully identified the species in 13/16 mixed infections. This assay was further validated with 91 blood samples and demonstrated a specificity and sensitivity for single infections of 100% compared with nested PCR as the “gold standard.” This test has been implemented for routine confirmation of malaria species in Alberta, Canada. In comparison with species identification by microscopy, the real-time PCR test demonstrated greater sensitivity for the identification of species causing low-level and mixed infections and for the discrimination of Plasmodium species other than Plasmodium falciparum. Our experience supports a role for real-time PCR in the identification of malarial species in conjunction with microscopy.
Malaria is the most lethal parasitic disease worldwide and is responsible for more than 1 million deaths per year, mostly of pregnant women and young children (2). Malaria is endemic to many developing countries, which bear the greatest loss-of-life and economic burdens of the disease (1). Although the number of malaria cases in areas where the disease is not endemic pales in comparison, a significant rise in the number of cases of imported malaria has been observed in recent years (11, 13). This is due to increases in travel by tourists to areas where the disease is endemic, the movement of refugees to adoptive countries, and immigrants visiting their country of origin who fail to take appropriate chemoprophylaxis (8).
Malaria is caused by an infection with Plasmodium, an intracellular parasite that infects the liver cells and red blood cells of its human host. There are five Plasmodium species that can cause human disease, including Plasmodium knowlesi, which is gaining widespread recognition as a human pathogen (5). The ability to correctly identify the species upon the diagnosis of malaria is critical to ensure an appropriate course of therapy. P. falciparum can cause a lethal infection and often requires emergency intervention. Other species can establish a chronic liver stage infection and require tissue schizonticides to destroy this stage and prevent relapses.
The erythrocytic cycle (blood stage) of infection is present in all cases and is responsible for symptoms and signs of infection. Thus, current diagnostic methods target the parasites in peripheral blood. The traditional and least expensive method of diagnosis is microscopic examination of thick and thin blood smears. The sensitivity and specificity of this method are contingent on the techniques used and the skill of the microscopist. In areas where malaria is not endemic, it can be quite challenging to maintain technical expertise in malaria identification (10, 16). Quality assurance programs have been implemented in various countries to maintain proficiency in malaria parasite identification to the species level. However, with low specimen numbers these standards are difficult to sustain. Rapid diagnostic tests (RDTs) based on immunochromatographic antigen detection systems have been implemented in many diagnostic laboratories as an adjunct to microscopy. RDTs are rapid and easy to use and may be cost-effective, providing many advantages over traditional microscopic methods. RDTs are useful for identifying P. falciparum and P. vivax infections but cannot be used to identify P. malariae and P. ovale infections (8).
In recent years, the development of molecular tests for malaria has introduced new diagnostic methods for the detection and identification of malaria parasites to the species level. In particular, real-time PCR has been promoted as an automated, quantitative, and closed system that reduces the risk of cross-contamination inherent in conventional PCR. Several real-time PCR methods for malaria have been described and validated within a research setting with very good sensitivity and specificity (8). However, specific limitations of each assay have largely precluded full implementation of these tests within diagnostic laboratories. Several assays have been developed that can identify P. falciparum but fail to distinguish among P. ovale, P. vivax, and P. malariae (7, 9). One method uses primer sets specific to P. falciparum, P. vivax, and P. ovale but does not include primers to detect P. malariae (18). This method also fails to capture DNA sequence variations in circulating strains of P. ovale (3, 4).
Another major limitation is the ability of these PCR assays to detect mixed Plasmodium infections. For assays that use conserved primers, this is attributed to competition of the primers for multiple templates in the sample (3, 12, 14, 19). Mixed species infections account for 3 to 5% of the malaria infections observed in areas where the disease is not endemic (15, 20). They are particularly challenging for microscopists and represent one aspect of malaria diagnosis where molecular methods can clearly support conventional microscopy. Failure to detect the minor species upon diagnosis can significantly impact patient care.
One of the most sensitive and specific real-time PCR assays for the detection of single-species infections was described by Rougemont et al. and incorporates conserved primers and species-specific probes (19). This is one of the few assays that have been carefully validated with the four major Plasmodium species but is limited by low sensitivity for mixed infections. We have refined this method and developed a real-time PCR assay that could fully support microscopists with species confirmation and identification of mixed infections. We have introduced species-specific forward primers in combination with a conserved reverse primer and species-specific probes multiplexed in a single reaction. Here we describe the validation of this modified assay for the detection of single- and mixed-species infections. We also discuss the implementation of this test for routine confirmatory diagnosis of malaria in Alberta.
MATERIALS AND METHODS
Blood samples.
Patient samples from Alberta were collected from regional laboratories in 2007 and 2008. Additional samples, including the panel of mixed infections, were selected from the malaria repository at the National Reference Centre for Parasitology/Tropical Diseases Centre (McGill University, Canada). Negative specimens were collected from healthy volunteers with no recent history of travel to areas where malaria is endemic. This study was approved by the Human Ethics Research Board at the University of Alberta.
DNA extraction.
Blood from patient samples (40 μl) was diluted in 160 μl of deionized water. DNA extraction was performed with the PSS GC12 instrument (Precision System Science Co. Ltd.) and the DNA 200 extraction protocol and kits (E2003). DNA was eluted into a 100-μl volume.
Plasmids.
The 18S gene for each Plasmodium species cloned into the pGEM-T vector (Promega) was kindly provided by Katia Jaton-Ogay and verified by DNA sequencing. In plasmid-mixing experiments, 103 copies of one plasmid were mixed with 103, 104, and 105 copies of the second plasmid and used as the template for the real-time PCR. Cycle threshold (CT) values were based on duplicate samples. Plasmid copy number quantification was performed by spectrophotometric analysis.
Real-time PCR.
Real-time PCR was performed under universal conditions (95°C for 15 s, 60°C for 1 min) with the ABI TaqMan 7500. The reaction was performed with a final volume of 25 μl containing 5 μl of DNA, 12.5 μl of TaqMan universal master mix (Applied Biosystems), primers, and probes. The primers and probes used in this study are listed in Table 1 with the respective concentrations for each reaction. The Plasprobe, Ovaprobe, and Malaprobe were synthesized by Applied Biosystems, and the Vivprobe, Falcprobe, and β2 M probe were synthesized by Biosearch Technologies. All probes were purified by high-performance liquid chromatography. Samples were confirmed Plasmodium positive with genus-specific primers Plasmo1 and Plasmo2 and the 6-carboxyfluorescein (FAM)-labeled Plasprobe to detect a region of the Plasmodium 18S gene that is conserved across all five species (19). The Plasmodium species present in the sample were determined with species-specific forward primers Plasmo2 and species-specific probes. The reaction was performed in a single tube with distinct fluorophores for each probe. A cutoff of 40 cycles was used to define positive samples. The testing algorithm also included a number of controls: extraction of TE (10 mM Tris-HCl [pH 8], 1 mM EDTA) as a negative control, β2-macroglobulin (β2 M) target amplification (CT, <40) as a positive extraction control for the specimen (23), a positive reference control to detect any variation between runs, and no-template controls for each of the master mixes.
TABLE 1.
Primers and probes used for screening and identification of Plasmodium species
| Species | Primer or probe | Concn (nM) | Sequence (5′-3′)d |
|---|---|---|---|
| Plasmodium spp. | Plasmo1-F primera | 200 | GTT AAG GGA GTG AAG ACG ATC AGA |
| Plasmodium spp. | Plasmo2-R primera | 200 | AAC CCA AAG ACT TTG ATT TCT CAT AA |
| Plasmodium spp. | Plasprobea | 50 | FAM-ACC GTC GTA ATC TTA ACC ATA AAC TAT GCC GAC TAG-TAMRA |
| P. falciparum | Fal-F primer | 200 | CCG ACT AGG TGT TGG ATG AAA GTG TTA A |
| P. falciparum | Falcprobeb | 80 | Quasar 670-AGC AAT CTA AAA GTC ACC TCG AAA GAT GAC T-BHQ-2 |
| P. vivax | Viv-F primer | 50 | CCG ACT AGG CTT TGG ATG AAA GAT TTT A |
| P. vivax | Vivprobeb | 80 | TAMRA-AGC AAT CTA AGA ATA AAC TCC GAA GAG AAA ATT CT-BHQ-2 |
| P. ovale | Ova-F primer | 50 | CCG ACT AGG TTT TGG ATG AAA GAT TTT T |
| P. ovale | Ovaprobea | 80 | VIC-CGA AAG GAA TTT TCT TAT T-MGBNFQ |
| P. malariae | Mal-F primer | 50 | CCG ACT AGG TGT TGG ATG ATA GAG TAA A |
| P. malariae | Malaprobea | 80 | FAM-CTA TCT AAA AGA AAC ACT CAT-MGBNFQ |
| Human | β2 M-F primerc | 900 | TGA GTA TGC CTG CCG TGT GA |
| Human | β2 M-R primerc | 900 | ACT CAT ACA CAA CTT TCA GCA GCT TAC |
| Human | β2 M probec | 100 | FAM-CCA TGT GAC TTT GTC ACA GCC CAA GAT AGT T-TAMRA |
Primer and probe sequences are as previously published (19).
Probe sequence is as previously published (19), with modified fluorophores.
Primer and probe sequences are as previously published (23).
TAMRA, 6-carboxytetramethylrhodamine; MGBNFQ, minor groove binding nonfluorescent quencher; Quasar 670, cyanine; BHQ-2, black hole quencher.
Nested PCR.
Nested PCR was performed by using primers and reaction conditions described previously (16, 17).
Microscopy.
Results from analysis of thick and thin blood smears were provided by pathologists from regional laboratories throughout Alberta who submitted specimens to ProvLab for PCR testing.
DNA sequencing.
Sequencing of PCR products was performed with the primers used for amplification. DNA sequencing was carried out on a Beckman Coulter CEQ2000XL sequencer at the Department of Biochemistry DNA Core Services Lab of the University of Alberta.
PCR confirmation at ProvLab.
Our acceptance criteria included EDTA blood samples from patients that were smear positive, regardless of species. Specimens with suspected subpatent parasitemia or an inconclusive smear result were also tested. DNA extracted from whole blood was tested in both the Plasprobe and species identification reactions.
RESULTS
Validation of the Plasmodium species identification reaction for single infections.
The method described by Rougemont et al. (19) consists of a “screening” reaction that uses conserved primers with a probe (Plasprobe) that detects all five Plasmodium species and a second “species identification” reaction that uses the same conserved primers with species-specific probes. For the species identification reaction, we designed species-specific forward primers that are used with the conserved reverse primer and the species-specific probes. We first compared the sensitivity of our modified species identification reaction with that of the Plasprobe screening reaction. A panel of 91 samples from 86 patients was evaluated in a blind fashion under the two reaction conditions. Of the 56 samples positive by Plasprobe, all were detected by the species identification reaction. No false positives were observed, demonstrating 100% specificity for Plasmodium detection. These results were further confirmed by nested PCR.
Nested PCR also served as the “molecular gold standard” for species identification. We observed 100% concordance between the species identified by nested PCR and by our real-time PCR. This panel included two mixed infections for which the major and minor species were both correctly identified with the new species identification reaction.
The species identification reaction consists of primers and probes for the four major Plasmodium species. In light of the emergence of P. knowlesi as a simian parasite capable of causing malaria infections in humans (5), an additional probe would need to be designed to detect this species. We have successfully detected genomic DNA from P. knowlesi with the Plasprobe and verified that the species-specific primers and probes do not cross-react with this species. This implies that the test can be used as a screening tool for P. knowlesi, although validation with clinical infections of P. knowlesi is required. At present, if a specimen is positive with the Plasprobe but negative in the species identification reaction and the patient travel history is consistent with exposure to P. knowlesi, then further investigation is warranted.
Multiplex species identification can simultaneously detect all four Plasmodium species.
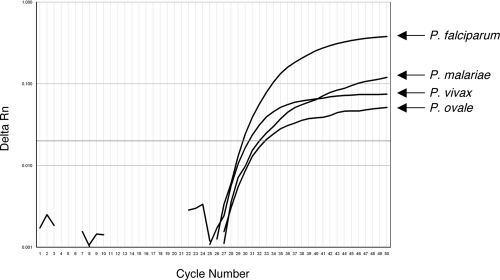
Primer competition is a technical concern with multiplex PCRs. As proof of the principle that the species-specific primers and probes are not limiting the detection of all four species, we tested a pool of four templates in the species identification reaction. Genomic DNA was extracted from four clinical samples containing each of the four Plasmodium species at similar concentrations, as judged by their CT values in the Plasprobe reaction. The DNA was pooled in equal volumes and served as the template in the real-time PCR. All four species were successfully amplified and detected in the species identification reaction (Fig. 1). The amplitude of the fluorescence curves varied, perhaps due to the different efficiency of each PCR. Since this pool was generated from clinical specimens, this could be a result of sequence variations within the probe or primer regions for the particular strain. This experiment, along with the validation of the blind panel described above, demonstrates the high specificity of these primers for their target species.
FIG. 1.
Simultaneous detection of four Plasmodium species. A mixture of genomic DNAs of four Plasmodium species served as the template in the species identification real-time PCR. Rn, normalized reporter.
Assay sensitivity for detection of mixed infections.
One of the major limitations of the Rougemont method is the ability to detect mixed infections (3, 4, 19). This is likely due to competition of the conserved primers for the different templates and favors amplification of the species with the highest level of infection. We showed that our species-specific primers can simultaneously detect all four species within a single reaction when the templates are present in nearly equal concentrations. With mixed infections, the most important clinical application of this assay is to detect lower levels of P. falciparum in the presence of other species. To determine the analytical sensitivity of the assay for P. falciparum as the minor species, we prepared artificial mixtures of plasmids containing the 18S genes from all four Plasmodium species with different ratios of the P. falciparum plasmid relative to the other three species (1:1, 1:10, and 1:100). We could reproducibly detect the P. falciparum plasmid at a 1:100 ratio relative to the other three plasmids (Table 2). We did observe greater competition for the conserved reverse primer at this higher ratio, resulting in higher CT values for the P. falciparum template, but this did not compromise the sensitivity within this range. We also tested P. vivax as the minor species against P. falciparum and were able to detect this plasmid at a 1:100 ratio (data not shown).
TABLE 2.
Detection of P. falciparum as the minor species in mixed plasmid templates
| Combination and ratio |
CT
|
|||
|---|---|---|---|---|
| P. falciparum | P. vivax | P. ovale | P. malariae | |
| P. falciparum-P. vivax | ||||
| 1:1 | 31.60 | 33.43 | ||
| 1:10 | 30.99 | 30.06 | ||
| 1:100 | 31.77 | 26.42 | ||
| P. falciparum-P. ovale | ||||
| 1:1 | 31.44 | 30.69 | ||
| 1:10 | 31.39 | 27.27 | ||
| 1:100 | 34.16 | 23.95 | ||
| P. falciparum-P. malariae | ||||
| 1:1 | 31.61 | 30.20 | ||
| 1:10 | 32.86 | 26.18 | ||
| 1:100 | 36.53 | 22.12 | ||
In clinical samples from mixed infections, the ratios between the two species can vary substantially. To test whether these primers would improve the detection of mixed infections, a blind panel of clinical samples consisting of 16 mixed infections was procured. Nested PCR was used as the molecular gold standard for identifying the mixed species. Our real-time PCR species identification reaction detected 13 of 16 mixed infections (Table 3). For three of the mixed infections, only one species was detected. In two of these, we did observe an amplification curve corresponding to the minor species but the curves crossed the threshold between cycles 40 and 45. In addition, we discovered that one of our mixed infections contained a third species. This species was detected by nested PCR but not by real-time PCR. It is likely that this third species represents a very low level of infection that is below the sensitivity of the real-time PCR assay.
TABLE 3.
Detection of mixed infections by real-time PCR compared with nested PCR in a blind panel of clinical samples
| Species detected (no. of specimens)c by:
| |
|---|---|
| Real-time PCR | Nested PCRb |
| P. falciparum and P. malariae (4)a | P. falciparum and P. malariae (4) |
| P. falciparum and P. ovale (5); P. falciparum only (1) | P. falciparum and P. ovale (6) |
| P. falciparum and P. vivax (3); P. falciparum only (1) | P. falciparum and P. vivax (4) |
| P. ovale and P. vivax (1); P. ovale only (1) | P. ovale and P. vivax (2) |
One P. falciparum-P. malariae infection included a third species (P. ovale), which was detected by nested PCR but not by real-time PCR.
Gold standard.
A total of 13 mixed infections were detected by real-time PCR, and16 were detected by nested PCR.
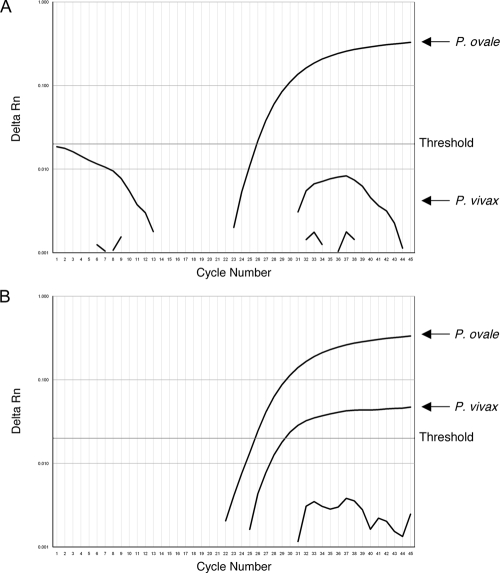
One important consideration for the species identification assay is the analysis of baseline values with the SDS software from Applied Biosystems. The sensitivity was greater when this parameter was set to the manual function and was calculated based on the CT values between cycles 3 and 15. With the default “Auto CT” baseline algorithm, the minor species in two of our mixed infections were undetected (Fig. 2A). Upon analysis under “Manual CT,” these were clearly detected in the reaction (Fig. 2B). Occasionally, we observed a nonspecific fluorescent signal for the P. malariae probe with this baseline setting (Fig. 2B, lower curving line). Although this line occasionally crossed the threshold and yielded a CT value close to the cutoff at cycle 40, examination of the shape of the amplification curve and the component profile clearly showed this signal to be nonspecific. Together, the modifications with species-specific forward primers and analysis of baseline parameters mark a significant improvement in the sensitivity of the assay for detecting mixed infections.
FIG. 2.
Manual baseline correction improves sensitivity for mixed infections. In this specimen, the minor species P. vivax was undetected with the Auto CT baseline parameter (A) but was clearly identified with the Manual CT baseline (B) (cycles 3 to 15). Rn, normalized reporter.
Test implementation for species confirmation in a diagnostic laboratory.
Following its superior performance during our validation, we implemented this assay as a confirmatory test at the Provincial Laboratory for Public Health (ProvLab) in Alberta. This test serves as a secondary confirmation of malaria species following morphological identification by a microscopist.
All of the specimens submitted to ProvLab for species confirmation are accompanied by the smear result with the species identification. A comparison of the species interpretation by microscopy with the real-time species identification assay was carried out with specimens from 30 patients. The comparison was performed with the microscopy data provided with the first specimen submitted for each patient (Table 4). Nearly half of the specimens were identified as P. falciparum, and these correlated precisely between microscopy and PCR. However, there were a number of discrepancies in the identification of Plasmodium species other than P. falciparum. Two P. ovale infections were identified by microscopy as P. vivax, and one P. falciparum infection was identified as P. ovale. Given that species identification by microscopy was performed by readers in different laboratories with varied levels of expertise, nested PCR was considered the gold standard test for all discrepant results (Table 4). These results were further confirmed by DNA sequencing.
TABLE 4.
Plasmodium species identification by microscopy versus PCR during routine testing of patient samples
| Microscopy | Real-time PCRa | Nested PCRa,d |
|---|---|---|
| P. falciparum (15)e | P. falciparum (15) | P. falciparum (15) |
| P. vivax (5) | P. vivax (3), P. ovale (2) | P. vivax (3), P. ovale (2) |
| P. ovale (1) | P. falciparum (1) | P. falciparum (1) |
| Plasmodium species other than P. falciparum (6) | P. vivax (4), P. ovale (1), P. malariae-P. falciparum (1) | P. vivax (4), P. ovale (1), P. malariae-P. falciparum (1) |
| Suspected mixed infectionb (1) | P. falciparum (1) | P. falciparum (1) |
| Unidentified speciesc (2) | P. falciparum (2) | P. falciparum (2) |
PCR results discordant with microscopy results are in bold.
Major species identified as P. falciparum with suspected P. vivax coinfection.
Parasitemia level too low for species identification (<0.1%).
Gold standard.
The values in parentheses are the numbers of specimens tested.
For six samples, results were reported as Plasmodium spp. or Plasmodium species other than P. falciparum but differentiation among P. ovale, P. vivax, and P. malariae was not possible. Real-time PCR was able to identify the species in all of these samples and, in fact, identified one mixed infection with P. falciparum as the minor species. Two additional specimens could not be identified to the species level by microscopy because of low parasitemia, and both of these were identified as P. falciparum infections by real-time PCR. One suspected mixed infection on the basis of microscopy was confirmed as a single infection by both real-time and nested PCRs. These results strengthen the validity of the real-time PCR test as a confirmation of the morphological identification of malarial species.
DISCUSSION
Over the past decade, a number of real-time PCR tests have been described and evaluated for the detection and species identification of malaria parasites. Four methods can detect and identify the four major Plasmodium species (6, 12, 19, 22). Two of these methods provide good detection of mixed infections but require four separate species identification reactions, increasing the cost per test (6, 22). The original Rougemont method condensed the species identification into two reactions, one with probes for P. falciparum and P. vivax and another with probes for P. ovale and P. malariae. In one report comparing two real-time PCR methods and nested PCR as the gold standard (3), the Rougemont method only detected 4/14 mixed infections. In this study, we designed sequence-specific forward primers and multiplexed the reaction in a single tube. This method reduced the cost per test, did not compromise sensitivity for the detection of single infections, and improved the detection of mixed infections to 13/16 in a blind panel.
With these modifications, we implemented this assay as a confirmatory test for malaria species identification in a routine diagnostic laboratory. In comparison with traditional microscopy, the real-time PCR assay was useful for the identification of Plasmodium species other than P. falciparum and identified specimens where the morphological species identification was discordant with the PCR result. Of important clinical significance, four patients with P. falciparum were undiagnosed or misdiagnosed by microscopy. One specimen was incorrectly identified as containing P. ovale. A second specimen was reported as a Plasmodium species other than P. falciparum and in fact contained a mixed infection of P. malariae/P. falciparum. The two other specimens had a low level of parasitemia (<0.1%), which precluded species identification. In one of these cases, a returning traveler presented with a low-level infection of parasites that could not be identified to the species level by microscopy. The next day, the parasitemia had climbed to 5% and the species was identified by microscopy as P. falciparum. However, real-time PCR was able to identify the species as P. falciparum on the initial blood sample and thus could have provided an accurate diagnosis at an early stage of infection. Our current service provides results within 1 week of specimen submission, although a result with this assay can be obtained within 3 h. This shorter turnaround time could rapidly identify undiagnosed or misdiagnosed cases, with an early, direct impact on patient management.
In addition to species confirmation of acute malaria, this test can be applied to a number of clinical contexts that require greater assay sensitivity. For example, it may be relevant for screening refugees arriving from areas where malaria is endemic. These individuals are semi-immune and likely asymptomatic but may harbor low numbers of parasites that could result in a malaria attack within weeks or months of arrival in their destination country (16, 21). This technology may also prove useful in the context of clinical trials for antimalarials, such as the determination of species-specific effects of a given drug combination, quantifying the parasite response to treatment, and identifying resistance that can be correlated with the genotype of the isolate. At the moment, this assay holds great value in support of conventional approaches to malaria diagnosis in correctly identifying malarial species and mixed infections.
Acknowledgments
We thank Katia Jaton-Ogay from the University Hospital of Lausanne for providing plasmids and technical advice during the validation of this assay. We also appreciate the cooperation of the hematopathologists and pathologists from DynaLife Dx and the University of Alberta Hospital who forwarded samples for assay validation. MRA-456G genomic DNA of the P. knowlesi H strain was obtained through the MR4 Malaria Research and Reference Reagent Resource Center. P. falciparum 3D7 malaria parasites were contributed by D. J. Carucci.
We thank Dick MacLean from the McGill University Tropical Diseases Centre and PHAC/National Microbiology Laboratory for financial support of the NRCP. This work was supported by Alberta Health and Wellness.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Amexo, M., R. Tolhurst, G. Barnish, and I. Bates. 2004. Malaria misdiagnosis: effects on the poor and vulnerable. Lancet 3641896-1898. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 2008. World malaria report 2008. World Health Organization, Geneva, Switzerland. http://www.who.int/malaria/wmr2008/malaria2008.pdf.
- 3.Bialasiewicz, S., D. M. Whiley, M. D. Nissen, and T. P. Sloots. 2007. Impact of competitive inhibition and sequence variation upon the sensitivity of malaria PCR. J. Clin. Microbiol. 451621-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calderaro, A., G. Piccolo, F. Perandin, C. Gorrini, S. Peruzzi, C. Zuelli, L. Ricci, N. Manca, G. Dettori, C. Chezzi, and G. Snounou. 2007. Genetic polymorphisms influence Plasmodium ovale PCR detection accuracy. J. Clin. Microbiol. 451624-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox-Singh, J., T. M. Davis, K. S. Lee, S. S. Shamsul, A. Matusop, S. Ratnam, H. A. Rahman, D. J. Conway, and B. Singh. 2008. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin. Infect. Dis. 46165-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Monbrison, F., C. Angei, A. Staal, K. Kaiser, and S. Picot. 2003. Simultaneous identification of the four human Plasmodium species and quantification of Plasmodium DNA load in human blood by real-time polymerase chain reaction. Trans. R. Soc. Trop. Med. Hyg. 97387-390. [DOI] [PubMed] [Google Scholar]
- 7.Elsayed, S., K. Plewes, D. Church, B. Chow, and K. Zhang. 2006. Use of molecular beacon probes for real-time PCR detection of Plasmodium falciparum and other Plasmodium species in peripheral blood specimens. J. Clin. Microbiol. 44622-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erdman, L. K., and K. C. Kain. 2008. Molecular diagnostic and surveillance tools for global malaria control. Travel Med. Infect. Dis. 682-99. [DOI] [PubMed] [Google Scholar]
- 9.Farcas, G. A., K. J. Zhong, T. Mazzulli, and K. C. Kain. 2004. Evaluation of the RealArt Malaria LC real-time PCR assay for malaria diagnosis. J. Clin. Microbiol. 42636-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kain, K. C., M. A. Harrington, S. Tennyson, and J. S. Keystone. 1998. Imported malaria: prospective analysis of problems in diagnosis and management. Clin. Infect. Dis. 27142-149. [DOI] [PubMed] [Google Scholar]
- 11.Kain, K. C., D. W. MacPherson, T. Kelton, J. S. Keystone, J. Mendelson, and J. D. MacLean. 2001. Malaria deaths in visitors to Canada and in Canadian travellers: a case series. Can. Med. Assoc. J. 164654-659. [PMC free article] [PubMed] [Google Scholar]
- 12.Mangold, K. A., R. U. Manson, E. S. Koay, L. Stephens, M. Regner, R. B. Thomson, Jr., L. R. Peterson, and K. L. Kaul. 2005. Real-time PCR for detection and identification of Plasmodium spp. J. Clin. Microbiol. 432435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martens, P., and L. Hall. 2000. Malaria on the move: human population movement and malaria transmission. Emerg. Infect. Dis. 6103-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNabb, A., R. Chen, T. Lo, Q. Wong, and J. Isaac-Renton. 2008. Detection and speciation of Plasmodium species in blood by real-time PCR; is there a role in today's diagnostic laboratory? Can. J. Med. Lab. Sci. 7088-98. [Google Scholar]
- 15.Morassin, B., R. Fabre, A. Berry, and J. F. Magnaval. 2002. One year's experience with the polymerase chain reaction as a routine method for the diagnosis of imported malaria. Am. J. Trop. Med. Hyg. 66503-508. [DOI] [PubMed] [Google Scholar]
- 16.Ndao, M., E. Bandyayera, E. Kokoskin, D. Diemert, T. W. Gyorkos, J. D. MacLean, R. St. John, and B. J. Ward. 2005. Malaria “epidemic” in Quebec: diagnosis and response to imported malaria. Can. Med. Assoc. J. 17246-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ndao, M., E. Bandyayera, E. Kokoskin, T. W. Gyorkos, J. D. MacLean, and B. J. Ward. 2004. Comparison of blood smear, antigen detection, and nested-PCR methods for screening refugees from regions where malaria is endemic after a malaria outbreak in Quebec, Canada. J. Clin. Microbiol. 422694-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perandin, F., N. Manca, A. Calderaro, G. Piccolo, L. Galati, L. Ricci, M. C. Medici, M. C. Arcangeletti, G. Snounou, G. Dettori, and C. Chezzi. 2004. Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. J. Clin. Microbiol. 421214-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rougemont, M., S. M. Van, R. Sahli, H. P. Hinrikson, J. Bille, and K. Jaton. 2004. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J. Clin. Microbiol. 425636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubio, J. M., I. Buhigas, M. Subirats, M. Baquero, S. Puente, and A. Benito. 2001. Limited level of accuracy provided by available rapid diagnosis tests for malaria enhances the need for PCR-based reference laboratories. J. Clin. Microbiol. 392736-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stauffer, W. M., M. Weinberg, R. D. Newman, L. M. Causer, M. J. Hamel, L. Slutsker, and M. S. Cetron. 2008. Pre-departure and post-arrival management of P. falciparum malaria in refugees relocating from sub-Saharan Africa to the United States. Am. J. Trop. Med. Hyg. 79141-146. [PubMed] [Google Scholar]
- 22.Vo, T. K., P. Bigot, P. Gazin, V. Sinou, J. J. De Pina, D. C. Huynh, F. Fumoux, and D. Parzy. 2007. Evaluation of a real-time PCR assay for malaria diagnosis in patients from Vietnam and in returned travellers. Trans. R. Soc. Trop. Med. Hyg. 101422-428. [DOI] [PubMed] [Google Scholar]
- 23.Watzinger, F., M. Suda, S. Preuner, R. Baumgartinger, K. Ebner, L. Baskova, H. G. Niesters, A. Lawitschka, and T. Lion. 2004. Real-time quantitative PCR assays for detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. J. Clin. Microbiol. 425189-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]