Abstract
Many laboratories are experiencing growing shortages of trained microbiology technologists and technicians. Consequently, there is considerable interest in new automation that could potentially lessen labor demands for specimen processing. In this study, we present the first published evaluation of a new microbiology instrument, the Walk Away Specimen Processor (WASP), manufactured by Copan, Inc., in which we evaluated cross-contamination, the accuracy of plating, and the quality of the results. The absence of cross-contamination was demonstrated by plating a total of 200 alternating inoculated and sterile specimen tubes. The ability of the WASP to subculture enrichment broths was evaluated with 106 Lim broth specimens, with the results being identical to those obtained by testing by routine methods. Plating of urine specimens with the WASP was compared to plating with the Dynacon Inoculab instrument. Three hundred specimens were plated in duplicate on both instruments with 1-μl loops, and 293 specimens were plated in duplicate on both instruments with 10-μl loops. The results of duplicate plating with the same instrument (replicate plating) and of the consensus agreement between the two instruments were compared. The replicate plating results were comparable for both instruments, while the WASP had more specimens with significant results than the Inoculab with the 1-μl loop only. Lastly, for the plating of 113 specimens in ESwab tubes, the manual method and WASP plating each yielded 90 potential pathogens. In summary, we report the first evaluation of a new microbiology specimen-plating instrument, the WASP, which offers opportunities for the automated plating of microbiology specimens to an extent that has not been possible to date.
Clinical microbiology laboratories have largely been bypassed by the advances in automation that have benefitted other areas of the clinical laboratory in recent years. Continuously monitoring blood culture systems as well as automated microbial identification and susceptibility testing systems are widely utilized. However, specimen processing and culture workup specifically remain manual tasks, and few changes to the methods used to perform these tasks have been made in the recent past. While some larger laboratories utilize urine-plating instrumentation, most microbiology laboratories have no automation in their processing areas. In this report, we present the results of a preliminary evaluation of a new microbiology plating instrument that offers the potential to automate the plating of a variety of liquid-based microbiology specimens.
MATERIALS AND METHODS
Overview of WASP.
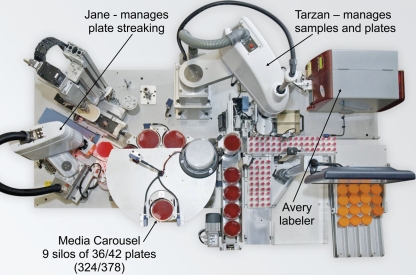
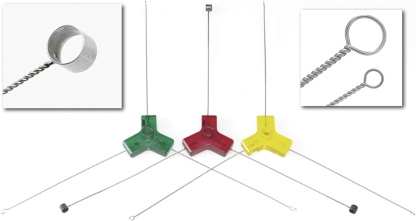
The Walk Away Specimen Processor (WASP) is a new instrument manufactured in Italy for Copan Diagnostics (Murrieta, CA) and is designed to plate liquid specimens from a variety of different transport devices (Fig. 1 and 2). The WASP utilizes two Toshiba selective compliant assembly robot arm (SCARA) robots. The first robot moves specimens and plated media and takes specimens to the decapping device, while the second robot does the actual inoculation and plate streaking. Barcode readers on the WASP scan the specimen tube, and a printer prints specimen information and a bar code on a label that is placed on the plated media. A nine-silo carousel holds 342 standard BD plates (Becton Dickinson Microbiology Systems, Cockeysville, MD) or 378 standard Remel (Lenexa, KS) plates. Only transport devices with the specimen in a liquid phase can be processed on the WASP. With the exception of large urine cups (120-ml cups in our laboratory), all specimens are loaded on the WASP by using special Teflon pallets that contain holes that are sized for specific containers. For example, one type of pallet holds 12 Vacutainer urine transport tubes, a second type of pallet holds 12 ESwab tubes, and a third type of pallet holds 6 enteric transport medium tubes. Up to six pallets can be loaded onto the instrument at one time, resulting in a maximum load of 72 Vacutainer tubes or ESwab tubes at one time. The WASP also has a vortex apparatus and a spinner/centrifuge that can be used to prepare specimens for plating. Actual plating is done by three metal loops incorporated in a triquetra (three-cornered) loop inoculation tool (Fig. 3). The loops are available in three sizes: 1 μl, 10 μl, and 30 μl. The WASP contains multiple sensors to verify proper operation, including one sensor that is connected to a camera that verifies that the loop contains the specimen after it is dipped into the specimen. A touch-screen computer facilitates the selection of inoculation protocols, media, and streaking patterns, as well as other functions, and guides the operation of the WASP.
FIG. 1.
Front view of the WASP. The dimensions of the instrument are 75 in. wide, 75 in. high, and 43 in. deep.
FIG. 2.
Top view of the WASP. The two SCARA robots are referred to as Tarzan and Jane.
FIG. 3.
Triquetra loops: from left to right, 10-μl, 30-μl, and 1-μl loops.
The evaluation described here consisted of a preliminary evaluation of the first production version of a WASP in a clinical microbiology laboratory. At the time that this evaluation was performed, the WASP contained software that was able to process only urine specimens in Vacutainer, UriSwab, and ESwab tubes. Consequently, this evaluation was limited to those types of specimen transport devices.
Cross-contamination studies.
To determine whether cross-contamination occurs when sequential specimens are streaked by the WASP instrument, studies were performed with both Vacutainer Urine C&S Preservative Plus plastic tubes (BD) and ESwab tubes (Copan). A fresh subculture of Escherichia coli (ATCC 35218) was used to prepare a suspension equivalent to a 0.5 McFarland standard in sterile saline. Dilutions were performed to obtain organism suspensions of ∼105 CFU/ml and ∼106 CFU/ml.
Four milliliters of sterile saline was added to each of 50 Vacutainer tubes, and 4 ml of the 105-CFU/ml E. coli suspension was added to each of an additional 50 Vacutainer tubes. The tubes were then placed into the appropriate WASP pallet by alternating a tube filled with the E. coli suspension and a tube filled with sterile saline. The pallets were placed on the WASP instrument, and a standard urine-streaking pattern that included a blood agar plate (BAP) and MacConkey (MAC) agar plate was selected for each of the 100 Vacutainer tubes. The contents of all Vacutainer tubes were plated first by using a 1-μl loop and were then retested by using a 10-μl loop. This yielded a total of 200 sets of plates: 100 sets inoculated with sterile saline and 100 sets inoculated with the E. coli suspension. The plates were incubated at 37°C and were examined at 24 h for growth.
For testing of ESwab tubes, 0.1 ml of the 106-CFU/ml E. coli suspension was added to each ESwab tube (ESwab tubes contain 1 ml of liquid Amies transport medium). The tubes were placed into the appropriate WASP pallet by alternating a tube inoculated with the E. coli suspension and a tube filled only with the transport medium. The pallets were placed on the WASP instrument, and a three-quadrant streaking pattern that included a BAP and a MAC agar plate for each of the 100 ESwab tubes was selected. The contents of all ESwab tubes were processed first by using a 10-μl loop and were then retested by using a 30-μl loop. This yielded a total of 200 sets of plates: 100 sets inoculated with sterile saline and 100 sets inoculated with the E. coli suspension. The plates were incubated at 37°C and were examined at 24 h for growth.
Enrichment broth subculture.
To verify the accuracy of the WASP for the subculture of enrichment broths, testing was performed with Lim broths (Remel). Routine Lim broths inoculated for prenatal group B streptococcus screens from the Geisinger Microbiology Laboratory were used for this testing. Vaginal and rectal swab specimens were incubated in Lim broth for 18 to 24 h before they were subcultured for routine testing. After the routine testing was completed, the Lim broth tubes were vortexed and 1 ml was transferred from each Lim broth tube to an empty, sterile ESwab tube. The ESwab tubes with Lim broth were subsequently subcultured to neomycin-nalidixic acid agar plates (BD) and BAPs on the WASP by using the 10-μl loop. These cultures were read and worked up at 24 h and 48 h, and the results were compared with the routine culture results.
Urine cultures.
The Inoculab instrument (Dynacon Inc., Mississauga, Ontario, Canada) is used for the routine plating of urine specimens submitted in Vacutainer tubes to the Geisinger Microbiology Laboratory. Plates on the Inoculab instrument can be inoculated with either 1-μl or 10-μl loops. For this validation, routine urine specimens received in BD Vacutainer tubes were first plated with the Inoculab instrument on BAPs and MAC agar plates by using a 1-μl loop for the routine microbiology laboratory culture, and these cultures were worked up and reported by using standard Geisinger Microbiology Laboratory protocols. All specimens were then plated a second time with the Inoculab instrument, and those plates were labeled for study purposes. The same specimens were then subsequently plated by using a 1-μl loop by the WASP on BAPs and MAC agar plates in duplicate and were identified for the study. This resulted in four total platings (two times on the Inoculab instrument and two times on the WASP). Additional specimens were then plated twice each on the Inoculab instrument and the WASP by using 10-μl loops.
All plates were placed in a non-CO2 incubator for a minimum of 16 h. One set of the plates from the Inoculab instrument inoculated with a 1-μl loop was read and reported by a Geisinger Microbiology Laboratory technologist, and the other sets of plates were read by one of us (B.L.S.). Standard Geisinger Microbiology Laboratory procedures were used for the workup of all cultures. A senior technologist or one of us (P.P.B.) reviewed all discrepant results.
ESwab tubes.
Routine cultures from a variety of sources submitted to the laboratory in ESwab tubes were plated by using the WASP. After routine manual plating was completed, the specimens were loaded onto the WASP and inoculated with a three-quadrant streaking pattern on BAPs, MAC agar plates, and chocolate agar plates. The WASP 30-μl loop was used to inoculate all plates.
All cultures plated by the WASP were worked up independently of the manually plated cultures. The results were compared after all testing with the manually plated and the WASP-plated specimens was completed.
RESULTS
Cross-contamination studies.
A total of 50 inoculated and 50 sterile Vacutainer tubes were alternately loaded on the WASP. They were plated with both 1-μl and 10-μl loops for a total of 200 cultures. No colonies were observed from the sterile tubes, and consistent streaking patterns were noted from the inoculated tubes. Similar results were observed with ESwab tubes for both the 10-μl and the 30-μl loops, with no growth occurring with samples from any of the sterile tubes and with consistent streaking patterns being obtained with samples from the inoculated tubes.
Enrichment broth subculture.
A total of 106 Lim broth specimens were subcultured with the WASP. Preliminary studies (data not shown) indicated that the use of the 30-μl loop did not result in a satisfactory number of isolated colonies. The use of the 10-μl loop, however, produced consistent numbers of isolated colonies. By use of the 10-μl loop, there was a 100% concordance of the results with those of the routine culture method, with each loop detecting 20 positive and 86 negative test results.
Urine cultures.
A total of 300 specimens were processed in duplicate on the WASP and Inoculab instruments by using 1-μl loops, while 293 specimens were processed in duplicate on both instruments by using 10-μl loops. The same specimens were not necessarily used for the 1-μl loops and the 10-μl loops, so the results were analyzed separately by loop size. Culture results were divided into those considered likely to be significant and those considered likely not to be significant.
For specimens plated with 1-μl loops, single or multiple isolates with <10,000 CFU/ml and cultures with three or more organisms each with >10,000 CFU/ml (multiple flora) were considered not significant, as were isolates of Micrococcus spp. and Lactobacillus spp. in any quantity. For the purposes of this study, coagulase-negative staphylococci and viridans group streptococci were considered likely to be significant if they were present at a concentration of ≥10,000 CFU/ml and were present as a single pathogen or with no more than one other organism. For tabulation purposes, if a culture contained ≥10,000 CFU of a potential pathogen (e.g., E. coli) and <10,000 CFU/ml of one or two other organisms, the E. coli isolate was included with the significant isolates and the other organism(s) was included with the not significant isolates. For specimens plated with 10-μl loops, the results obtained were interpreted in a similar manner but with the 10-fold difference in the dilution factor being accounted for. The results obtained with the Inoculab instrument and the WASP were evaluated in two ways. First, the replicate results were compared for each instrument. For example, the result obtained for specimen 1 plated with the WASP by using the 1-μl loop was compared with the results of the second plating of specimen 1 on the WASP by using the same size loop. Second, the significant isolates that the two instruments recovered were compared.
The results of the replicate results for specimens with significant isolates are summarized in Table 1. Both instruments yielded similar results in replicate plating for significant isolates for loops of both sizes. Overall, for quantitation (the same CFU/ml range, e.g., both replicates with >105 CFU/ml), there were no significant differences between the results for the two instruments.
TABLE 1.
Summary of replicate plating results for significant isolates
| Instrument | 1-μl inoculum
|
10-μl inoculum
|
||
|---|---|---|---|---|
| Totala | Same resultb (same CFU rangec) | Total | Same result (same CFU range) | |
| Inoculab | 80 | 80 (75) | 71 | 71 (70) |
| WASP | 81 | 80 (79) | 70 | 70 (70) |
Total number of significant isolates for that instrument.
Same result, number of organisms that were the same in replicate cultures with ≥104 CFU/ml for the 1-μl inoculum or ≥103 CFU/ml for the 10-μl inoculum.
Same CFU range, the number of organisms (indicated in parentheses) with similar results in both replicates (103 to 104 CFU/ml, 104 or 105 CFU/ml, or >105 CFU/ml).
For cultures with nonsignificant results, there was agreement for the Inoculab instrument between paired plates for the 1-μl loop for 184/223 results and for the 10-μl loop for 219/220 results. For the WASP, for specimens with nonsignificant results, there was agreement for the 1-μl loop for 192/220 results and for the 10-μl loop for 218/220 results. Most of the discrepant results involved specimens with no growth on one set of plates and <10 actual colonies on the paired culture plates.
The results obtained with the WASP and the Inoculab instrument for specimens with significant results not in agreement are summarized in Table 2. Among 74 specimens with significant results that were plated with 1-μl loops, there were 11 specimens with discrepant results. For five of the specimens, one Inoculab instrument result was not in agreement with the three other results, while for one specimen, one WASP result was not in agreement with the three other results. Four significant isolates were detected only by the two WASP cultures. Lastly, for one specimen, there was a difference in the CFU/ml counts between the two WASP cultures and the two Inoculab instrument cultures.
TABLE 2.
Results from WASP and Inoculab instrument for specimens with significant results not in agreementa
| Loop size | Organism(s) detected with:
|
|||
|---|---|---|---|---|
| Inoculab instrument
|
WASP
|
|||
| Replicate 1 | Replicate 2 | Replicate 1 | Replicate 2 | |
| 1 μl | Multiple flora | <104 mixed flora | 104-105Enterococcus, <104 two other organisms | 104-105Enterococcus, <104 2 other orgs |
| <104 mixed flora | <104 mixed flora | 104-105 beta-hemolytic streptococci | 104-105 beta-hemolytic streptococci | |
| <104 one type | <104 one type | 10-4105Enterococcus | 104-105Enterococcus | |
| <104 one type | No growth | 104-105K. pneumoniae, <104 one type | 104-105K. pneumoniae, <104 one type | |
| >105E. coli | 104-105E. coli | >105E. coli | >105E. coli | |
| >105E. coli | 104-105E. coli | >105E. coli | >105E. coli | |
| >105 beta-hemolytic streptococci | 104-105 beta-hemolytic streptococci | >105 beta-hemolytic streptococci | > 105 beta-hemolytic streptococci | |
| 104-105S. aureus | 104-105S. aureus | >105S. aureus | 104-105S. aureus | |
| 104-105Proteus mirabilis | 104-105P. mirabilis | >105P. mirabilis | >105P. mirabilis | |
| 104-105 NLF | >105 NLF | > 105 NLF | > 105 NLF | |
| 104-105 CoNS | >105 CoNS | > 105 CoNS | > 105 CoNS | |
| <104 one OT | <104 one OT | <104 one OT | <104 one OT | |
| 10 μl | Multiple flora | Multiple flora | 103 to 104 CoNS, <103 one OT | Multiple flora |
| <103 two organsims | <103 two organisms | 103-104Enterococcus, <103 one OT | 103-104Enterococcus, <103 one OT | |
| <102 Mixed Flora | 103-104 CoNS, <102 one OT | 103-104 CoNS, <103 one OT | 103-104 CoNS, <103 one OT | |
| 103-104E. coli | 103-104E. coli | >104E. coli | >104E. coli | |
| 103-104 GBS | 103-104 GBS | >104 GBS | >104 GBS | |
| 103-104Enterococcus | 103-104Enterococcus | >104Enterococcus | >104Enterococcus | |
| 103-104 viridans group streptococci | 103-104 viridans group streptococci | >104 viridans group streptococci | > 104 viridans group streptococci | |
| <102 one OT | <102 one OT | <102 two OTs | <102 one OT | |
OT, other type; GBS, group B streptococci; CoNS, coagulase-negative staphylococci; NLF, unidentified non-lactose-fermenting gram-negative bacilli.
Among 65 specimens with significant results that were plated with 10-μl loops, there were 7 specimens with discrepant results. The result for one each of the WASP and Inoculab instrument specimens was not in agreement with the three other results. For five specimens, there was a difference in the CFU/ml counts between the two WASP cultures and the two Inoculab instrument cultures.
ESwab cultures.
A total of 113 specimens that were collected in ESwab tubes were plated on the WASP. These included 13 vaginal swab specimens and 100 specimens for routine culture of mixed types (wound, drainage, fluid, nares, skin, upper respiratory, and other specimens). All plates were inoculated with a 30-μl loop, and the results were read at 24 and 48 h of incubation. Plating both by the manual method and with the WASP yielded 90 potential pathogens. The yields of normal flora (skin, vaginal, and respiratory flora) was also the same both by the manual method and with the WASP. One of the WASP cultures grew one colony of mold that was not detected on the manually plated culture.
DISCUSSION
Many laboratories are experiencing growing shortages of trained microbiology technologists and technicians. This has been exacerbated not only by the growth in the rate of routine testing but also by the demand for testing performed for epidemiological purposes, such as for methicillin-resistant Staphylococcus aureus (3). Consequently, there is considerable interest in new automation that could potentially lessen labor demands for specimen processing (7).
The current instrumentation available for microbiology processing includes streaking and plating instruments. Three instruments that can perform plate inoculation and streaking are currently available: the Dynacon Inoculab instrument (models LQ and LQH), the bioMérieux MicroStreak instrument, and the WASP.
Inoculab instruments are designed to plate urine specimens from one type of container. The type of container is selected at the time of instrument purchase. The Dynacon LQH model that we have in our laboratory holds a total of 38 uninoculated Remel plates, giving it a capacity of 19 specimens without reloading if two plates are used for each culture or 38 specimens without reloading if one plate is used for each culture. Since the LQH model has only one silo for uninoculated plates, if more than one type of plate is used for a culture, the plates must be intercalated in the stack. Inoculab instruments can also be used with a streak-only function.
The MicroStreak instrument was released in 2008 (2). The MicroStreak instrument requires a plugged disposable pipette tip for each specimen and one disposable plastic applicator for each plate. It can plate either single plates or biplates, and the applicator spreads the inoculum over the entire area of a standard 100-mm plate or a biplate. The smallest pipette that the MicroStreak instrument can use is 10 μl, and currently, the specimen top must be removed at the time that the specimen is placed on the instrument. If the residual specimen is to be saved, the specimen must be recapped after plate inoculation (2).
This study was designed to be a preliminary evaluation of a new instrument named the WASP. The evaluation was restricted to two container types: Vacutainer tubes and ESwab tubes. Software that was not available when this study was performed permits the sampling of other types of specimen containers. We compared the inoculation of urine specimens in Vacutainer tubes by the WASP and the Inoculab LQH instrument. We did not perform manual plating for this study, as our in-house validation of the Inoculab instrument indicated that the results obtained with that instrument were more reproducible than those obtained by our manual plating method. Lue et al. have also shown in their laboratory that the Inoculab instrument is more accurate than manual plating with a 1-μl loop (5). Moreover, variability in the volume of urine specimens transferred manually has been demonstrated (1). We also evaluated the accuracy of the WASP compared to that of subculture using Lim broths. Lastly, we compared manual plating of routine specimens collected in ESwab tubes with the plating performed by the WASP.
Overall, the results obtained with the Inoculab instrument and WASP for the plating of urine specimens were comparable for specimens plated with 1-μl and 10-μl loops. Interestingly, there were four specimens plated with the 1-μl loop that grew a significant pathogen at a concentration of 104 to 105 CFU/ml on both WASP cultures (two with Enterococcus spp. and one each with Klebsiella pneumoniae and a beta-hemolytic streptococcus), while the Inoculab instrument culture was mixed or had multiple flora. Similarly, there was also one specimen plated with the 10-μl loop that grew a Enterococcus sp. at a concentration of 103 to 104 CFU/ml on both WASP cultures and mixed flora on the Inoculab instrument culture.
There are several possible explanations for the different results between the WASP and the Inoculab instruments for these specimens: (i) random differences in the numbers of CFU near the breakpoints for reporting organisms, (ii) failure of the loop to pick up a specimen (loss of the meniscus), and (iii) a more homogeneous specimen preparation with one instrument than with the other. Differences between the operation of the Inoculab instrument and the WASP support the last two possibilities. The WASP has a camera that detects the presence of a droplet inside the inoculating loop after the specimen is sampled. If no specimen is detected, the loop returns to the tube a second time and, if necessary, a third time. If the camera fails to again detect a droplet, the tube is moved to a discard bin. The Inoculab instrument has no sensor to detect the presence of a specimen in the loop. A second difference between the instruments is the preparation of the specimen prior to sampling. The manual for the Inoculab instrument states that the specimen should be agitated before it is placed on the instrument but does not specify how that agitation should be accomplished. In contrast, the WASP has a vortex mixer that vigorously mixes the tube with urine before it is sampled. The exact reasons for the observed differences are clearly speculative on our part; nonetheless, the ability to verify the presence of actual specimen in the inoculating loop as well as the vigorous vortexing that takes place before the specimen is plated may offer more than a theoretical advantage for the WASP over the Inoculab instrument.
Clearly, there is a need for studies that will assess the speed, throughput, and labor savings of the WASP. At the time that this preliminary study was performed, the WASP required 27 min to plate specimens from 24 Vacutainer tubes onto two plates each, while the Inoculab instrument required 24 min to plate specimens from 24 Vacutainer tubes onto two plates. We anticipate the collection of more data once the Sunquest Laboratory Information System interface is fully validated and software upgrades have been installed.
There are also significant capacity differences between the Inoculab instrument and the WASP. The Dynacon Inoculab model LQH instrument, which was used in our laboratory, holds a total of 38 uninoculated Remel plates, whereas the WASP holds 378 Remel plates. Since the Dynacon Inoculab model LQH instrument has only one silo for uninoculated plates, if more than one type of plate is used for a culture, the plates must be intercalated in the stack. This would not be necessary if a single plate was used. The Dynacon Inoculab model LQ instrument has two silos for uninoculated media that permits the use of one silo for each of two types of media for each specimen. The WASP has nine silos, each of which holds 42 Remel plates, and one to nine types of media can be loaded at one time. More than one silo can be loaded with the same type of media. For example, if a laboratory uses a BAP for most specimen types, it might chose to load three silos with BAP and fewer silos with other types of media.
In our evaluation of the WASP for the subculture of broths to plated media, we encountered no problems with the WASP. By using a 10-μl loop, the WASP consistently produced isolated colonies. For this study, the Lim broths were transferred to empty ESwab tubes. We do not anticipate that laboratories would want to do this as a standard practice. The resolution to this issue could be the production of broths in smaller tubes by either Copan and or another manufacturer or changes to the WASP software to accommodate taller tubes.
Although our evaluation of the WASP for the plating of routine cultures was limited to 113 specimens, the results obtained were comparable to those for the manually plated cultures. We developed a three-quadrant streaking pattern for use with the 30-μl loop that was used for all of these specimens. The WASP has the potential to process containers of different sizes. The jaws on the robot have three settings to accommodate three sizes of containers. There are three docking stations (the docking station holds the bottom of the container during the uncapping process) that also accommodate containers of three sizes. The docking station can easily be swapped out. There are two decapping devices on the WASP. One is fixed and is designed for Vacutainer urine tubes. A variety of other decappers can be placed in the second decapping station, including an ESwab tube decapper. For this study, we used the decapper and docking station designed to hold ESwab tubes. During this evaluation, we encountered no problems with the opening or closing of either Vacutainer or ESwab tubes. Subsequent to the performance of this study, an interface was established between the WASP and the Sunquest Laboratory Information System. While it is now operational, it has yet to be fully validated. Unlike the MicroStreak instrument and similar to the Inoculab instrument, the WASP requires no disposable products for plating. The option of plating urine specimens with 1-μl or 10-μl loops provides laboratories with the flexibility to address clinical needs (4, 6).
The only mechanical (hardware) problem that we encountered during this study was a burnt out incinerator, which we replaced. There were several software problems that were systematically addressed by Copan as they appeared. None of those problems persisted.
This study was designed to be a preliminary evaluation of a new instrument, the WASP. As we expand the use of the WASP to other specimen types, we are working to optimize the work-flow pattern for each specimen type. In our routine use of ESwab tubes, specimens are manually plated with a disposable pipette that is manufactured to deliver approximately 28 μl/drop. Each piece of medium receives 1 drop of the inoculum, and for specimens requiring a Gram stain, 1 drop is placed on a slide. We envision a similar work flow with the WASP, with the preparation of any necessary slides occurring before the containers are loaded onto the WASP for plating. It is important to understand that the only type of swab specimen that can be plated by the WASP is one that transfers the specimen to a liquid phase. ESwab is currently the only swab with that capability. ESwabs are more expensive than traditional wound swabs, and for laboratories that do not routinely use ESwabs, this increase in cost should be included in the operational budget for the WASP.
We anticipate that the relative amount of specimens that a laboratory can plate on a WASP will be strongly influenced by the specimen volume for a particular test and the medium required for that particular specimen and test. For example, if each day a laboratory plants an average of five specimens of a particular specimen type for a particular test and that test requires media not used for other tests, the removal of media from the silos and the addition of the required media to the silos may not be an efficient use of time. Importantly, the current operation of the WASP is designed for batch testing and not random-access testing.
In summary, we report on a first evaluation of a new microbiology specimen-processing instrument, the WASP. We were satisfied with the performance of the WASP and believe that it offers opportunities to automate the processing of microbiology specimens to an extent that has not been possible to date. Additional studies by this and other laboratories are merited to test the full potential of the instrument.
Acknowledgments
This investigation was supported by a grant from Copan, Inc.
Footnotes
Published ahead of print on 21 January 2009.
REFERENCES
- 1.Albers, A. C., and R. D. Fletcher. 1983. Accuracy of calibrated-loop transfer. J. Clin. Microbiol. 1840-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glasson, J. H., L. H. Guthrie, D. J. Nielsen, and F. A. Bethell. 2008. Evaluation of an automated instrument for inoculating and spreading samples onto agar plates. J. Clin. Microbiol. 461281-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones, G. A., and R. B. Matthews. 2008. Abstr. 108th Gen. Meet. Am. Soc. Microbiol., abstr. 169.
- 4.Kass, E. 1956. Asymptomatic infections of the urinary tract. Trans. Assoc. Am. Phys. 6956-63. [PubMed] [Google Scholar]
- 5.Lue, Y. A., C. J. Krause, and A. B. John. 2002. Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. 81.
- 6.Stamm, W., G. Counts, K. Running, S. Fihn, M. Turck, and K. Holmes. 1982. Diagnosis of coliform infection in acutely dysuric women. N. Engl. J. Med. 307463-468. [DOI] [PubMed] [Google Scholar]
- 7.Tilton, R. C., and R. W. Ryan. 1978. Evaluation of an automated agar plate streaker. J. Clin. Microbiol. 7298-304. [DOI] [PMC free article] [PubMed] [Google Scholar]