Abstract
A total of 264 nonduplicate strains of imipenem (IPM)-nonsusceptible Pseudomonas aeruginosa were isolated from hospitals in 16 different regions throughout China. These 264 IPM-nonsusceptible clinical isolates of P. aeruginosa were examined by PCR, a metallo-β-lactamase (MBL) Etest, a double-disk synergy test (DDST), and a test using combined IPM disks supplemented with various amounts of EDTA. A total of 24 strains positive for MBLs were confirmed by PCR and DNA sequence analysis: 10 strains positive for the blaVIM-2 gene, 13 strains positive for the blaIMP-9 gene, and 1 strain positive for the blaIMP-1 gene. Real-time reverse transcriptase PCR (RT-PCR) was used to verify whether the isolates harboring MBL genes produced the enzyme and was considered the standard for evaluation of the methodology in this study. Of these 24 MBL-positive stains, 21 were confirmed as MBL-producing strains by real time RT-PCR for MBL expression and the other 3 had no expression of MBLs. The sensitivities, specificities, and positive and negative predictive values for the MBL Etest, the DDST, and the combined disk (CD) assay were evaluated. The best method for screening for MBL production in P. aeruginosa strains from China was the CD assay (IMP-EDTA) using 750 μg of EDTA/disk with a breakpoint of ≥6 mm. In the CD assay (IPM-EDTA) with 290 μg and 750 μg EDTA, the zone diameter increases for VIM-2-producing P. aeruginosa isolates were greater than those for IMP-9-producing P. aeruginosa isolates (P = 0.00).
The worldwide spread of acquired metallo-β-lactamases (MBLs) in clinically important pathogens, such as Pseudomonas spp., Acinetobacter spp., and members of the Enterobacteriaceae family, has become a great concern (9, 12). Increased mortality rates have been documented for patients infected with MBL-producing Pseudomonas aeruginosa, and these rates are especially due to inadequate empirical therapy (27). Most of the MBL-encoding genes reside on class 1 integrons and plasmids that usually confer high mobility to these genetic elements (8, 17, 22, 24, 26). Therefore, early detection of MBL-producing organisms is of crucial importance for prevention of their inter- and intrahospital dissemination, not only in institutions with high prevalences of such isolates but also in those in which phenotypes of resistance have never been detected. Various criteria for screening for MBL production in P. aeruginosa have been suggested (15). Currently, the most widely accepted standardized MBL functional screen is the MBL Etest (AB BioDisk, Solna, Sweden). However, due to the high cost and unavailability of Etest strips, many clinical microbiology laboratories use alternative screening methods, such as the double-disk synergy test (DDST) and the combined disk (CD) assay. Although the DDST and the CD assay are simple to perform and cheaper than the MBL Etest, they have shown discordant results, depending on the employed methodology, β-lactam substrates, MBL inhibitors (IMBL), and bacterial genus tested (7, 11, 14, 15). Standardization of a phenotypic method for screening for MBL-producing isolates is of crucial importance. It is desirable that the selection of the appropriate MBL test be based upon studies providing sensitivity (SN) and specificity (SP) results for that specific pathogen.
Although the prevalence of MBL-producing P. aeruginosa was lower in China than elsewhere, the MBL-encoding genes are usually carried by mobile genetic structures with great ability to spread (26). The aim of this study was to evaluate the accuracy of phenotypic tests for screening for MBL-producing isolates among P. aeruginosa isolates in China.
MATERIALS AND METHODS
Bacterial isolates.
From July 2006 to July 2007, a total of 264 nonduplicate imipenem (IPM)-nonsusceptible P. aeruginosa isolates were collected from hospitals in 16 different regions throughout China. All of these isolates were isolated from different patients and were identified with the API 20NE system (bioMérieux, Marcy l'Etoile, France). P. aeruginosa ATCC 27853 was used as an MBL-negative control. Four P. aeruginosa strains producing VIM-2 and two P. aeruginosa strains producing IMP-1 in our laboratory were used as MBL-positive controls (26).
Susceptibility testing.
The MICs of IPM and meropenem (MEM) were determined by agar dilution. The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI; 2007). P. aeruginosa ATCC 27853 was used as a control strain for susceptibility testing.
Phenotypic detection of MBL.
All 264 IPM-nonsusceptible clinical isolates of P. aeruginosa and the 6 MBL-positive control strains were tested by the three tests for phenotypic detection of MBL (the Etest, the DDST, and the CD test).
MBL Etests.
We performed MBL Etests (IPM-EDTA; AB Biodisk), following the manufacturer's recommendations.
DDST.
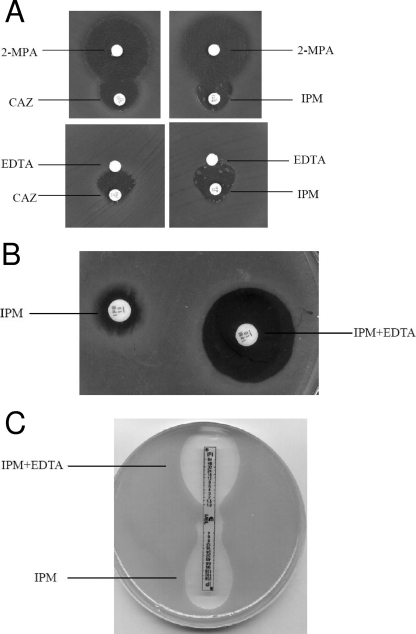
The phenotypic tests were performed, following the CLSI recommendations for the disk diffusion method. A 0.5 McFarland bacterial suspension was inoculated on a Mueller-Hinton (MH) agar plate (Oxoid, Basingstoke, England). IPM (10 μg) and ceftazidime (CAZ; 30 μg) disks were aligned around blank filter disks containing 3 μl mercaptopropionic acid (MPA; Sigma) or 10 μl 0.1 M EDTA (Sigma, Germany), added directly on the disk already placed on the MH agar plate. The following distances between the inhibitor and the substrates were tested: 1.5, 2.0, and 2.5 cm (from center to center). Enhancement of the zone of inhibition in the area between the antimicrobial agents and the inhibitor disk was considered to indicate positivity for MBL (Fig. 1).
FIG. 1.
Phenotypic tests for detection of MBL production. (A) Results for DDSTs using CAZ-MPA and IPM-EDTA for P. aeruginosa, with inhibition zone distortion toward the MPA and EDTA disks. (B) CD assays performed with an MBL producer P. aeruginosa strain. (C) Positive results for MBL Etests (IPM and IPM-EDTA).
CD.
For the CD assay, IPM disks (10 μg) were initially placed on the MH plates inoculated with the 0.5 McFarland bacterial suspension. The IMBL solutions (EDTA) added to the disks were 290 μg, 750 μg, and 930 μg, respectively (2, 6, 16, 25). After a 24-h incubation period at 35°C, the increase of the inhibition zone obtained with the CD was compared to that obtained with the antimicrobial disk alone. The positive criteria for classifying an isolate as an MBL producer are described below.
MBL gene PCR amplification and sequencing.
PCR assays were performed to amplify the sequences of the blaIMP, blaVIM, blaGIM, blaSPM, and blaSIM genes, as previously described (3, 10, 18, 19). The PCR products were purified by using a 3S spin PCR product purification kit (Shenergy Biocolor, China) and then sequenced.
Analysis of MBL gene expression by real-time RT-PCR.
For gene expression studies, total RNA was prepared using the TRIzol Max method (Invitrogen, Carlsbad, CA). Real-time reverse transcriptase PCR (RT-PCR) was performed using 250 ng of DNase-treated RNA, a PrimeScript RT reagent Kit (Takara, Japan), and specific internal IMP and VIM primer pairs. The primers for PCR amplification of cDNA were designed using the Primer3 program (available at http://frodo.wi.mit.edu/) and are shown in Table 1. Expression of the endogenous control gene rpsL was used to normalize data (5). Primer efficiency studies were carried out before the RNA expression levels of the isolates were compared. The primers showed 100% SN and 100% SP for detecting VIM and IMP MBLs. All of the MBL genes were sequenced. Searches for similarities between the sequences and those in sequence databases were performed with the BLAST program at the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/). All the genes were 100% identical. The efficiencies for amplification were 80% to 100%. Real-time RT-PCRs were carried out using an Opticon 2 real-time PCR detector, and the results were analyzed with the Opticon 2 real-time PCR detection software program. Relative quantification was determined by the 2−ΔΔCT method, where CT is the cycle threshold (13). The P. aeruginosa strain producing VIM-2, H22 (one of the MBL-positive control strains), was used as the reference strain.
TABLE 1.
Primers used for real-time RT-PCR
| Primer | Nucleotide sequence | Length (bp) |
|---|---|---|
| IMP | 5′-GGAATAGAGTGGCTTAATTC-3′ | 277 |
| 5′-GCCAAGCTTCTATATTTGCG-3′ | ||
| VIM | 5′-GTGTTTGGTCGCATATCGC-3′ | 380 |
| 5′-CGCAGCACCAGGATAGAAG-3′ | ||
| rspL | 5′-GCAAGCGCATGGTCGACAAGA-3′ | 235 |
| 5′-CGCTGTGCTCTTGCAGGTTGTGA-3′ |
Statistical analysis.
SN, SP, positive predictive value (PPV), and negative predictive value (NPV) were calculated for the MBL Etest (IPM-EDTA), the DDST for each β-lactam-IMBL combination, and the CD assay for different increases of the inhibition zone. Since not all of the strains harboring MBL genes confirmed by PCR produce MBLs, real-time RT-PCR was used to detect the expression levels of MBL genes. Real-time RT-PCR results for MBL expression were used to verify whether the isolates harboring MBL genes produced the enzyme and were considered the standard for evaluation of the methodology.
The results from the CD phenotypic method were characterized by receiver operating characteristic (ROC) curves to determine the best cutoff values for indicating MBL production. For various amounts of EDTA, SN and SP were calculated successively according to the variation of inhibition zones of MBL-producing and -nonproducing isolates. CD results stratified into groups according to which MBL type was produced were also analyzed by ROC curve analysis and the Student t test.
RESULTS
Prevalence of MBL-producing isolates among IPM-nonsusceptible P. aeruginosa isolates.
Among the 264 IPM-nonsusceptible P. aeruginosa isolates collected throughout China, 24 were confirmed to be positive for MBLs by PCR and DNA sequence analysis (10 strains positive for the blaVIM-2 gene, 13 strains positive for the blaIMP-9 gene, and 1 strain positive for the blaIMP-1 gene), and no other MBLs were detected. These 24 strains were divided into nine pulsotypes by pulsed-field gel electrophoresis (PFGE): six pulsotypes positive for VIM-2, two pulsotypes positive for IMP-9, and one pulsotype positive for IMP-1 (see Table 4). The results for PFGE were interpreted according to the criteria of Tenover et al. (20).
TABLE 4.
MBL gene expression levels, MBL Etest results, CD results, and IPM and MEM MICs for the 24 MBL-positive strains in this study
| MBL gene | Strain | PFGE result | Relative expression level | MBL Etest resultb | Increase of diam in CD assayc (mm) | MIC (μg/ml)
|
|
|---|---|---|---|---|---|---|---|
| IPM | MEM | ||||||
| blaVIM-2 | H22a | 1 | + | 15 | 64 | 16 | |
| blaVIM-2 | Gf28 | E | 15.53 | + | 20 | 256 | 64 |
| blaVIM-2 | Hb12 | B | 0.87 | + | 13.5 | 128 | 8 |
| blaVIM-2 | Hb21 | B | 1.25 | + | 9.5 | 16 | 8 |
| blaVIM-2 | Hb28 | B | 1.37 | + | 10 | 8 | 8 |
| blaVIM-2 | Hb30 | B | 0.57 | + | 15 | 32 | 32 |
| blaVIM-2 | Tj4 | C | 1.27 | + | 13.5 | 8 | 8 |
| blaVIM-2 | Tj22 | C | 0.00 | − | 2 | 32 | 8 |
| blaVIM-2 | Ze5 | G | 14.32 | + | 19 | 64 | 64 |
| blaVIM-2 | Wt5 | I | 0.77 | + | 9 | 256 | 64 |
| blaVIM-2 | Ss37 | H | 0.00 | − | 2 | 16 | 4 |
| blaIMP-9 | Gf1 | A1 | 1.47 | + | 8 | 64 | 64 |
| blaIMP-9 | Gf2 | A1 | 1.13 | + | 7.5 | 128 | 64 |
| blaIMP-9 | Gf9 | A1 | 0.66 | + | 7 | 64 | 64 |
| blaIMP-9 | Gf10 | A1 | 0.5 | + | 7 | 64 | 16 |
| blaIMP-9 | Gf27 | A1 | 3.09 | + | 8 | 128 | 128 |
| blaIMP-9 | Ga2 | A2 | 2.09 | + | 8 | 32 | 64 |
| blaIMP-9 | Ga6 | A2 | 1.56 | + | 6 | 32 | 32 |
| blaIMP-9 | Ga7 | A2 | 1.34 | + | 8 | 64 | 64 |
| blaIMP-9 | Ga8 | A2 | 1.03 | − | 7 | 64 | 64 |
| blaIMP-9 | Ga9 | A1 | 1.89 | − | 7 | 32 | 64 |
| blaIMP-9 | Ga15 | A1 | 4.46 | + | 7 | 128 | 64 |
| blaIMP-9 | Ga16 | D | 0.15 | − | 6 | 64 | 256 |
| blaIMP-9 | Ga20 | A1 | 0.84 | + | 8 | 128 | 64 |
| blaIMP-1 | Zy8 | F | 0.00 | − | 4 | 32 | 64 |
The P. aeruginosa strain producing VIM-2, H22 (one of the MBL-positive control strains), was used as the reference strain for analyzing the relative expression levels of the MBL genes.
+, positive result by the MBL Etest (IPM-EDTA); −, negative result by the MBL Etest (IPM-EDTA).
Using IPM-EDTA with 750 μg EDTA.
Expression of MBL genes by real-time RT-PCR.
Among these 24 MBL-positive strains, 3 had no expression of MBL: zy8 (IMP-1), tj22 (VIM-2), and ss37 (VIM-2). The expression levels of MBL genes among the 24 MBL-positive strains are shown in Table 4. The expression levels of MBL genes were also confirmed by isoelectric focusing according to established methods with the PhastSystem (Pharmacia, Uppsala, Sweden) (23).
Phenotypic MBL detection by the DDST.
Table 2 shows the SN, SP, PPV, and NPV results for the DDST. Among the 264 IPM-nonsusceptible P. aeruginosa isolates, EDTA provided the best results with CAZ used as a substrate. The SN, SP, PPV, and NPV results at 1.5 cm were 90.5, 97.5, 76.0, and 99.2%, respectively. Other DDST results with better SN and SP were obtained with either CAZ-MPA at 2.5 cm (95.2% and 91.4%) or IPM-MPA at 1.5 cm (85.7% and 93.8%). However, the PPVs for CAZ-MPA and IPM-MPA were lower (48.8% and 54.6%), overestimating the number of MBL-positive isolates.
TABLE 2.
MBL detection by DDST, with various distances between the inhibitor and the substratesa
| Distance (cm) | Level (%)
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ-MPA
|
CAZ-EDTA
|
IPM-MPA
|
IPM-EDTA
|
|||||||||||||
| SN | SP | PPV | NPV | SN | SP | PPV | NPV | SN | SP | PPV | NPV | SN | SP | PPV | NPV | |
| 1.5 | 95.2 | 90.1 | 45.5 | 99.6 | 90.5 | 97.5 | 76 | 99.2 | 85.7 | 93.8 | 54.6 | 98.7 | 76.2 | 97.9 | 76.2 | 97.9 |
| 2.0 | 95.2 | 90.5 | 46.5 | 99.5 | 28.6 | 99.6 | 85.7 | 94.2 | 66.7 | 95.1 | 53.9 | 97.1 | 33.3 | 98.4 | 63.6 | 94.5 |
| 2.5 | 95.2 | 91.4 | 48.8 | 99.6 | 28.6 | 99.6 | 85.7 | 94.2 | 66.7 | 95.1 | 53.9 | 97.1 | 33.3 | 99.2 | 77.8 | 94.5 |
PPV and NPV values were calculated as a/(a + b) and d/(c + d), respectively, where a is the number of isolates correctly identified as MBL producers, c is the number of MBL producers incorrectly identified as MBL nonproducers, d is the number of isolates correctly identified as MBL nonproducers, and b is the number of isolates incorrectly identified as MBL producers by the DDST and the CD assay.
Phenotypic MBL detection by the CD assay.
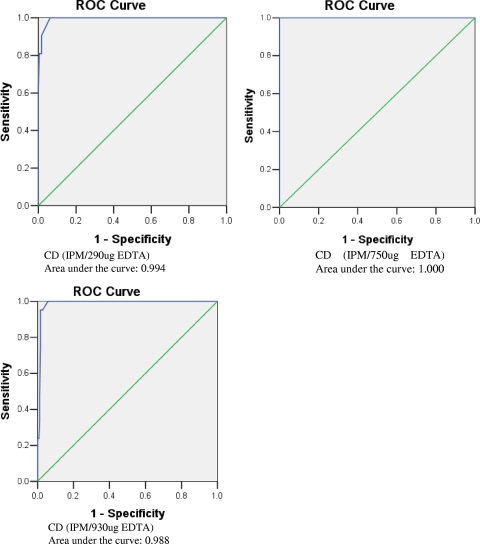
In the CD assay, the zone diameters were found to be similar and reproducible when the procedure was repeated. All the CD (IPM-EDTA) test results were applied in ROC curve analysis to establish the best breakpoint (increase in mm) for MBL detection (Fig. 2). The best breakpoint for the CD assay (IPM-EDTA) with 290 μg EDTA for indicating MBL production was 2 to 3 mm, which was difficult to discriminate (Fig. 2). The best breakpoint for the CD assay (IPM-EDTA) with 750 μg and 930 μg EDTA for indicating MBL producers was 5 to 7 mm (Fig. 2). So, the breakpoints of 4 mm, 6 mm, and 7 mm were selected to evaluate the values for the CD assays. Table 3 shows the CD results for various breakpoints. The best results for indicating MBL producers were obtained using 750 μg of EDTA/disk with a breakpoint of ≥6 mm. The SN, SP, PPV, and NPV for the CD assay (IMP-EDTA) with these criteria were all 100% (Table 3). The results for these criteria were better than those for the DDST and even the MBL Etest (Table 3). However, among several MBL-negative isolates, the zone diameters of IPM increased greatly (to 16 and 19 mm) when 930 μg EDTA was added to the CD assay. So, the EDTA amounts of 290 μg and 930 μg were not adapted to separate MBL producers.
FIG. 2.
ROC curves for different volumes of EDTA in combination with IPM for indicating MBL production in P. aeruginosa. The resulting SN values were plotted against the corresponding SP values, producing a ROC curve. The area under the ROC curve and its standard error were calculated, and statistical significance was then evaluated by the nonparametric method. Differences between the areas under the curve for the variables were evaluated through a comparison of the 95% confidence intervals for the corresponding areas.
TABLE 3.
Results of phenotypic MBL detection
| Method, agent(s), and distance | No. of MBL-positive control strains | SN (%) | SP (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|
| DDST | |||||
| CAZ-MPA (2.5 cm) | 6 | 95.2 | 91.4 | 48.8 | 99.6 |
| CAZ-EDTA (1.5 cm) | 5 | 90.5 | 97.5 | 76 | 99.2 |
| IPM-MPA (1.5 cm) | 5 | 85.7 | 93.8 | 54.6 | 98.7 |
| IPM-EDTA (1.5 cm) | 1 | 76.2 | 97.9 | 76.2 | 97.9 |
| CD (IPM-EDTA) | |||||
| 290 μg EDTA | |||||
| ≥4 mm | 4 | 81.0 | 98.0 | 77.3 | 98.4 |
| ≥6 mm | 3 | 61.9 | 100.0 | 100.0 | 96.8 |
| ≥7 mm | 1 | 33.3 | 100 | 100 | 94.6 |
| 750 μg EDTA | |||||
| ≥4 mm | 6 | 100.0 | 84.8 | 36.2 | 100.0 |
| ≥6 mm | 6 | 100.0 | 100.0 | 100.0 | 100.0 |
| ≥7 mm | 4 | 90.5 | 100.0 | 100.0 | 99.2 |
| 930 μg EDTA | |||||
| ≥4 mm | 6 | 100.0 | 68.3 | 21.4 | 100.0 |
| ≥6 mm | 6 | 100.0 | 94.2 | 60.0 | 100.0 |
| ≥7 mm | 6 | 95.2 | 98.4 | 83.3 | 99.6 |
| Etest | 5 | 85.7 | 100 | 100 | 98.8 |
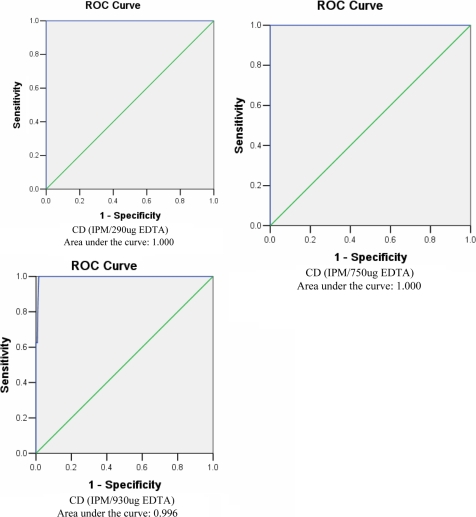
In addition, it is interesting that distinct results for the zone diameter increase were observed between the IMP-9- and VIM-2-producing P. aeruginosa isolates. The best breakpoints in the CD assay with 290 μg and 750 μg EDTA added to separate these two MBL types (VIM-2 and IMP-9) were 6.25 and 8.5 mm, respectively, by ROC curve analysis (Fig. 3). The zone diameter increases for VIM-2-type producers (11.75 ± 3.41 mm and 13.69 ± 4.19 mm) were greater than those for IMP-9-type producers (4.85 ± 1.20 mm and 7.27 ± 0.73 mm) in the CD assay with 290 μg and 750 μg EDTA. The difference in diameter increase between the IMP-9-producing isolate group and the VIM-2-producing isolate group was substantial (P = 0.00).
FIG. 3.
ROC curves for different volumes of EDTA in combination with IPM for separating levels of VIM-2 and IMP-9 production in P. aeruginosa.
MBL Etest.
In our study, the overall performance of the MBL Etest (IPM-EDTA) revealed SN, SP, PPV, and NPV results of 85.7%, 100%, 100%, and 98.8%, respectively (Table 3). No false-positive results were found by the MBL Etest (IPM-EDTA) among the 240 isolates of P. aeruginosa negative for MBL genes. There were six MBL-positive strains with negative results for the MBL Etest, with three of these strains (Tj22, Ss37, and Zy8) having no expression of MBL genes (the VIM-2 and IMP-1 genes) and three (Ga8, Ga9, and Ga16) negative for the IMP-9 gene (Table 4).
DISCUSSION
The prevalence of MBL-positive strains among IPM-nonsusceptible P. aeruginosa isolates from hospitals in China was estimated at 9.1% (24/264), lower than those for some developed countries, such as Japan. The MICs of IPM and MEM have no correlation with the expression levels of MBL genes. Also, most of the non-MBL producers in our study had high-level resistance to IPM and MEM. Decreased permeability and increased efflux are the most prevalent carbapenem resistance mechanisms in Chinese clinical isolates of P. aeruginosa (data not shown). PFGE of SpeI-digested genomic DNA showed that organisms from different regions are grouped into various PFGE types. These findings suggest that transmission of the blaVIM-2 and blaIMP-9 genes among clinical strains with different genetic backgrounds may be associated with mobile genetic elements, such as transposons and transferable plasmids, instead of a clonal expansion of an MBL-carrying strain throughout China. Since most of the MBL-encoding genes reside on class 1 integrons and/or plasmids that usually confer high mobility to these genetic elements, early detection of MBL-producing isolates is important for avoiding dissemination of such strains.
Various criteria for screening for MBL production in P. aeruginosa have been suggested. However, there are no standard guidelines provided by the CLSI for detection of these enzymes in various bacterial species. It is desirable that the selection of the appropriate MBL test be based upon studies providing SN and SP results for that specific pathogen. The MBL Etest has been evaluated in several studies and found to be a sensitive method for detection of MBL production in P. aeruginosa (21). However, in this study, several MBL-producing strains could not be detected by the MBL Etest (IMP-EDTA). Also, the MBL Etest, PCR, and even real-time RT-PCR were expensive and not adaptable for extensive use in clinical microbiology laboratories. The DDST and the CD assay have been reported to be simple, inexpensive phenotypic resources for detection of MBL that could be easily incorporated into the routines of clinical laboratories.
In the CD assay, the best separation between MBL-positive and -negative isolates was obtained using 750 μg of EDTA/disk with a breakpoint of ≥6 mm. It is known that EDTA may increase bacterial cell wall permeability and that zinc (chelated by EDTA) accelerates IPM decomposition and decreases OprD expression of P. aeruginosa. (4). These nonspecific effects might cause false-positive MBL results in the CD assay with 930 μg EDTA added but not in that with 290 μg and 750 μg EDTA added. Interestingly, the zone diameter increases for VIM-2-producing P. aeruginosa isolates were found to be greater than those for IMP-9-producing P. aeruginosa isolates in the CD assay (IPM-EDTA) with 290 μg and 750 μg EDTA. The phenotypic difference may be associated with the difference in inhibition ability of EDTA between the VIM- and IMP-type MBLs. This presumption needs to be confirmed by more MBL producers. However, no isolates containing other MBLs are available in this study. It is a conceded possibility that these results may not apply in general to other MBLs not evaluated in these experiments.
The SN, SP, PPV, and NPV for the CD assay (IPM-EDTA) using 750 μg of EDTA/disk with a breakpoint of ≥6 mm were better than those for the DDSTs and even the MBL Etest (IPM-EDTA). Additionally, interpretation of the CD assay results is more objective than that of the DDST results because the DDST depends upon the technician's expertise in discriminating true synergism from intersection of inhibition zones. Our results were in accordance with those obtained by Berges et al. but not with those obtained by Picão et al. (2, 15). Most previous studies evaluating MBL phenotypic detection were performed under distinct experimental conditions, jeopardizing comparison of their results (6, 11, 14). The sizes of inhibition zones produced by β-lactam-IMBL combinations may differ according to the way that IMBL is incorporated into the β-lactam disks (1). In the current study, we added the IMBL solutions directly on β-lactam disks already placed on the agar plate, as described by Picão et al., whereas some authors first prepare and freeze IMBL-β-lactam disks; thus, the results of our CD assay may be comparable to those of studies using the same methodology (1, 15, 25). It has been suggested that the selection of the optimal MBL screening method be based not only on bacterial species but also on the strains collected and the local prevalence of MBL producers (11, 15).
In conclusion, in our study, the best method for screening for MBL production in P. aeruginosa strains from China was the CD assay (IMP-EDTA) using 750 μg of EDTA/disk with a breakpoint of ≥6 mm. This method also provides a simple, inexpensive, and reproducible functional screen for MBL-producing P. aeruginosa strains in China.
Acknowledgments
This work was supported by a research grant from the National Basic Research Program (the 973 Program) of China (no. 2005CB523101) and by a research grant from the National Natural Science Foundation of China (no. NSFC30670930).
Footnotes
Published ahead of print on 11 February 2009.
REFERENCES
- 1.Andrade, S. S., R. C. Picao, E. H. Campana, A. G. Nicoletti, A. C. Pignatari, and A. C. Gales. 2007. Influence of disk preparation on detection of metallo-β-lactamase-producing isolates by the combined disk assay. J. Clin. Microbiol. 452058-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berges, L., H. Rodriguez-Villalobos, A. Deplano, and M. J. Struelens. 2007. Prospective evaluation of imipenem/EDTA combined disc and Etest for detection of metallo-beta-lactamase-producing Pseudomonas aeruginosa. J. Antimicrob. Chemother. 59812-813. [DOI] [PubMed] [Google Scholar]
- 3.Castanheira, M., M. A. Toleman, R. N. Jones, F. J. Schmidt, and T. R. Walsh. 2004. Molecular characterization of a beta-lactamase gene, blaGIM-1, encoding a new subclass of metallo-β-lactamase. Antimicrob. Agents Chemother. 484654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conejo, M. C., I. Garcia, L. Martinez-Martinez, L. Picabea, and A. Pascual. 2003. Zinc eluted from siliconized latex urinary catheters decreases OprD expression, causing carbapenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 472313-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumas, J. L., C. van Delden, K. Perron, and T. Kohler. 2006. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol. Lett. 254217-225. [DOI] [PubMed] [Google Scholar]
- 6.Franklin, C., L. Liolios, and A. Y. Peleg. 2006. Phenotypic detection of carbapenem-susceptible metallo-beta-lactamase-producing gram-negative bacilli in the clinical laboratory. J. Clin. Microbiol. 443139-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galani, I., P. D. Rekatsina, D. Hatzaki, D. Plachouras, M. Souli, and H. Giamarellou. 2008. Evaluation of different laboratory tests for the detection of metallo-beta-lactamase production in Enterobacteriaceae. J. Antimicrob. Chemother. 61548-553. [DOI] [PubMed] [Google Scholar]
- 8.Giakkoupi, P., G. Petrikkos, L. S. Tzouvelekis, S. Tsonas, N. J. Legakis, and A. C. Vatopoulos. 2003. Spread of integron-associated VIM-type metallo-beta-lactamase genes among imipenem-nonsusceptible Pseudomonas aeruginosa strains in Greek hospitals. J. Clin. Microbiol. 41822-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta, V. 2008. Metallo beta lactamases in Pseudomonas aeruginosa and Acinetobacter species. Expert Opin. Investig. Drugs 17131-143. [DOI] [PubMed] [Google Scholar]
- 10.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 431584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, K., D. Yong, J. H. Yum, Y. S. Lim, A. Bolmstrom, A. Qwarnstrom, A. Karlsson, and Y. Chong. 2005. Evaluation of Etest MBL for detection of blaIMP-1 and blaVIM-2 allele-positive clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 43942-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lincopan, N., J. A. McCulloch, C. Reinert, V. C. Cassettari, A. C. Gales, and E. M. Mamizuka. 2005. First isolation of metallo-β-lactamase-producing multiresistant Klebsiella pneumoniae from a patient in Brazil. J. Clin. Microbiol. 43516-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 14.Marchiaro, P., M. A. Mussi, V. Ballerini, F. Pasteran, A. M. Viale, A. J. Vila, and A. S. Limansky. 2005. Sensitive EDTA-based microbiological assays for detection of metallo-β-lactamases in nonfermentative gram-negative bacteria. J. Clin. Microbiol. 435648-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picão, R. C., S. S. Andrade, A. G. Nicoletti, E. H. Campana, G. C. Moraes, R. E. Mendes, and A. C. Gales. 2008. Metallo-β-lactamase detection: comparative evaluation of double-disk synergy versus combined disk tests for IMP-, GIM-, SIM-, SPM-, or VIM-producing isolates. J. Clin. Microbiol. 462028-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pitout, J. D., D. B. Gregson, L. Poirel, J. A. McClure, P. Le, and D. L. Church. 2005. Detection of Pseudomonas aeruginosa producing metallo-β-lactamases in a large centralized laboratory. J. Clin. Microbiol. 433129-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel, L., T. Lambert, S. Turkoglu, E. Ronco, J. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senda, K., Y. Arakawa, S. Ichiyama, K. Nakashima, H. Ito, S. Ohsuka, K. Shimokata, N. Kato, and M. Ohta. 1996. PCR detection of metallo-beta-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum beta-lactams. J. Clin. Microbiol. 342909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh, T. R., A. Bolmstrom, A. Qwarnstrom, and A. Gales. 2002. Evaluation of a new Etest for detecting metallo-β-lactamases in routine clinical testing. J. Clin. Microbiol. 402755-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong, J., M. F. Hynes, H. Ye, H. Chen, Y. Yang, F. M'Zali, and P. M. Hawkey. 2006. blaIMP-9 and its association with large plasmids carried by Pseudomonas aeruginosa isolates from the People's Republic of China. Antimicrob. Agents Chemother. 50355-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan, J. J., P. R. Hsueh, W. C. Ko, K. T. Luh, S. H. Tsai, H. M. Wu, and J. J. Wu. 2001. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 452224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yatsuyanagi, J., S. Saito, S. Harata, N. Suzuki, Y. Ito, K. Amano, and K. Enomoto. 2004. Class 1 integron containing metallo-β-lactamase gene blaVIM-2 in Pseudomonas aeruginosa clinical strains isolated in Japan. Antimicrob. Agents Chemother. 48626-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yong, D., K. Lee, J. H. Yum, H. B. Shin, G. M. Rossolini, and Y. Chong. 2002. Imipenem-EDTA disk method for differentiation of metallo-β-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 403798-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu, Y. S., T. T. Qu, J. Y. Zhou, J. Wang, H. Y. Li, and T. R. Walsh. 2006. Integrons containing the VIM-2 metallo-β-lactamase gene among imipenem-resistant Pseudomonas aeruginosa strains from different Chinese hospitals. J. Clin. Microbiol. 444242-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zavascki, A. P., A. L. Barth, A. L. Goncalves, A. L. Moro, J. F. Fernandes, A. F. Martins, F. Ramos, and L. Z. Goldani. 2006. The influence of metallo-beta-lactamase production on mortality in nosocomial Pseudomonas aeruginosa infections. J. Antimicrob. Chemother. 58387-392. [DOI] [PubMed] [Google Scholar]