During the last couple of years there has been a delightful increase in interest in genetic typing of human rhinoviruses. This is to a large extent due to the discovery of a proposed novel clade, human rhinovirus C (HRV-C). As a consequence, new methods have been reported aiming at unequivocal distinction of traditional HRV prototype strains as well as the newly found uncultivable HRV-C strains. We read with interest the article by Kiang and coworkers (2) describing reverse transcription-PCR-sequencing applications for genetic typing of human rhinoviruses targeting the 5′noncoding region (5′ NCR). A similar approach with largely similar results was published earlier (6). Relatively conserved areas within this region enable broad-spectrum primer design for sensitive methods. However, there are several issues to consider when using the 5′ NCR for genetic typing of HRV.
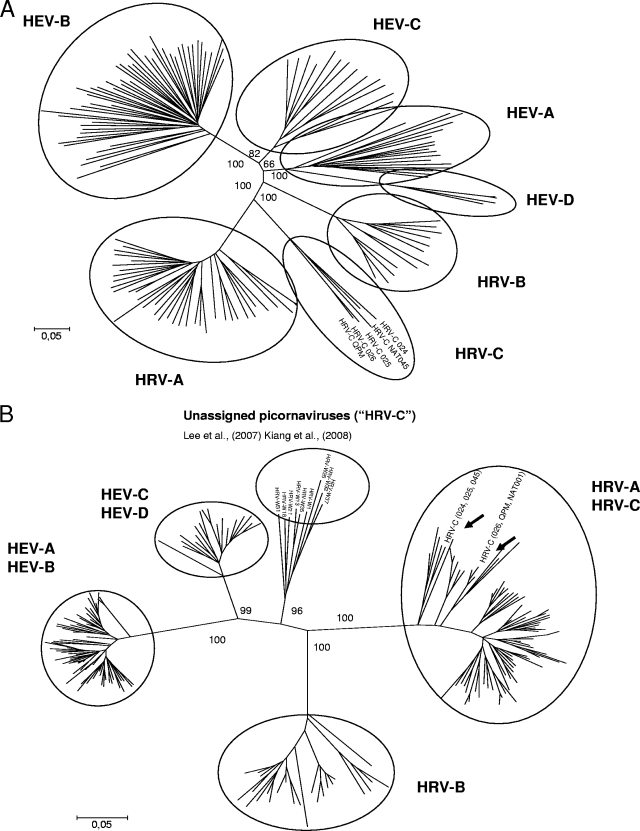
Current taxonomy and classification of picornaviruses are based on capsid region coding sequences. On the basis of this region, a group of previously characterized novel HRV strains form one distinct clade (5, 7) (Fig. 1A), a fact that has also been the basis of the proposal of the Picornavirus Study Group to form a new species, HRV-C, within the Enterovirus genus (4). However, in the article by Kiang et al., on the basis of the partial 5′ NCR sequences, the designated HRV-C strains clustered within the HRV-A clade (2). In contrast, the strains labeled HRV-C in this article formed a clade of their own. As a consequence, because 5′ NCR sequences do not segregate the designated HRV-C from HRV-A (Fig. 1B), they should not be used for typing of new strains. Nevertheless, this region is quite suitable for selected topics of molecular epidemiology, such as analysis of short-term transmission routes (1, 8) or tentative prediction of genetic type as in human enteroviruses (HEV) (9). The sequences nominated as HRV-C by Kiang et al. (2) and by Lee et al. (6) form a new clade in the 5′ NCR. The exact taxonomic position of this clade should be determined according to the clustering of these strains in the capsid region. Clearly, it is divergent from all known HRV and HEV clades in the 5′ NCR, but the decision on whether the strains represent HRV-C or some other picornavirus group cannot be made on the basis of the 5′ NCR alone.
FIG. 1.
Phylogenetic trees in the VP4/VP2 capsid coding region (A) and in the 5′ NCR (B) of different species of the enterovirus genus showing different clustering of the species in the two regions. Trees were constructed with MEGA4 using the neighbor-joining method with Tamura-Nei model and 1,000 bootstrap replicates.
The area close to the beginning of the open reading frame in the 5′ NCR is known to be a recombination hot spot in HEV. Although frequent recombination has not yet been shown for HRV, the analysis of the complete genome sequence data of all HRV prototype strains has not yet been published. Furthermore, the number of completely sequenced genomes of circulating HRV strains has remained low and is too low to conclude that the evolution in the 5′ NCR is always congruent with that of the capsid. Therefore, we would see phylogenetic analysis of the 5′ NCR of HRV as a welcome addition to HRV research, but not a surrogate of capsid coding sequence-based typing.
REFERENCES
- 1.Jartti, T., W. M. Lee, T. Pappas, M. Evans, R. F. Lemanske, Jr., and J. E. Gern. 2008. Serial viral infections in infants with recurrent respiratory illnesses. Eur. Respir. J. 32:314-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiang, D., I. Kalra, S. Yagi, J. K. Louie, H. Boushey, J. Boothby, and D. P. Schnurr. 2008. Assay for 5′ noncoding region analysis of all human rhinovirus prototype strains. J. Clin. Microbiol. 46:3736-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Knowles, N. J. 2008. New species in the genus Enterovirus (Picornaviridae). http://talk.ictvonline.org/media/p/687.aspx.
- 5.Lau, S. K., C. C. Yip, H. W. Tsoi, R. A. Lee, L. Y. So, Y. L. Lau, K. H. Chan, P. C. Woo, and K. Y. Yuen. 2007. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J. Clin. Microbiol. 45:3655-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, W. M., C. Kiesner, T. Pappas, I. Lee, K. Grindle, T. Jartti, B. Jakiela, R. F. Lemanske, P. A. Shult, and J. E. Gern. 2007. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS ONE 2:e966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McErlean, P., L. A. Shackelton, E. Andrews, D. R. Webster, S. B. Lambert, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2008. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C). PLoS ONE 3:e1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peltola, V., M. Waris, R. Osterback, P. Susi, O. Ruuskanen, and T. Hyypia. 2008. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J. Infect. Dis. 197:382-389. [DOI] [PubMed] [Google Scholar]
- 9.Thoelen, I., E. Moes, P. Lemey, S. Mostmans, E. Wollants, A. M. Lindberg, A. M. Vandamme, and M. Van Ranst. 2004. Analysis of the serotype and genotype correlation of VP1 and the 5′ noncoding region in an epidemiological survey of the human enterovirus B species. J. Clin. Microbiol. 42:963-971. [DOI] [PMC free article] [PubMed] [Google Scholar]