Abstract
The incidence of Vibrio cholerae non-O1, non-O139 strains from hospitalized patients with acute diarrhea constituted 27.4% (n = 54) of the total 197 V. cholerae strains isolated from patients in Kolkata, India, in 2003. Of 197 strains, 135 were identified as O1 serotype Ogawa and 2 were identified as O139. In the same time period, six O1 background rough strains that possessed all known virulence factors were identified. Serotype analysis of the non-O1, non-O139 strains placed 42 strains into 19 serogroups, while 12 remained O nontypeable (ONT); the existing serotyping scheme involved antisera to 206 serogroups. Detection of a good number of ONT strains suggested that additional serogroups have arisen that need to be added to the current serotyping scheme. The non-O1, non-O139 strains were nontoxigenic except for an O36 strain (SC124), which regulated expression of cholera toxin as O1 classical strains did. Additionally, strain SC124 carried alleles of tcpA and toxT that were different from those of the O1 counterpart, and these were also found in five clonally related strains belonging to different serogroups. Strains carrying tcpA exhibited higher colonization in an animal model compared to those lacking tcpA. PCR-based analyses revealed remarkable variations in the distribution of other virulence factors, including hlyA, rtxA, Vibrio seventh pandemic island I (VSP-I), VSP-II, and type III secretion system (TTSS). Most strains contained hlyA (87%) and rtxA (81.5%) and secreted cytotoxic factors when grown in vitro. Approximately one-third of the strains (31.5%) contained the TTSS gene cluster, and most of these strains were more motile and hemolytic against rabbit erythrocytes. Partial nucleotide sequence analysis of the TTSS-containing strains revealed silent nucleotide mutations within vcsN2 (type III secretion cytoplasmic ATPase), indicating functional conservation of the TTSS apparatus.
In the recorded history of cholera, humans have experienced seven pandemics; the seventh pandemic is still ongoing since its commencement in 1961 (29). Cholera is a waterborne disease that afflicts millions of people every year. The etiological agent of the disease is Vibrio cholerae belonging to serogroups O1 and O139 (4, 29). Members of the O1 serogroup have been further subdivided into Ogawa and Inaba serotypes and El Tor and classical biotypes. The seventh pandemic strains are of the O1 El Tor biotype, which replaced the O1 classical biotype strains that caused previous pandemics (29). The current serotyping scheme of V. cholerae includes 206 serogroups (57). Strains belonging to serogroups other than O1 and O139 are collectively known as non-O1, non-O139 V. cholerae strains and exist in abundance in the aquatic environment. However, recent studies have concluded that considerable incidence of human diarrheal episodes in many countries, including India, is due to V. cholerae non-O1, non-O139 (10, 11, 27, 39, 40, 48). Studies of diarrheal cases in Thailand between 1993 and 1995 revealed that the prevalence of non-O1, non-O139 V. cholerae was comparable to that of O1 V. cholerae (11) and furthermore that these strains mostly belonged to O6 and O14 serogroups. Diarrheal episodes caused by V. cholerae belonging to various serogroups in different countries have been reported as follows: serogroups O10 and O12 in Peru (10), serogroup O10 in India (49), serogroup O37 in Sudan and Czechoslovakia (1), and serogroup O141 in many regions (12). An alarming upsurge in the isolation of non-O1, non-O139 V. cholerae strains from diarrheal cases in Kolkata, India, during 1996 has also been reported (54).
A great amount of research has focused on the identification and regulation of virulence factors in O1 and O139 V. cholerae (16, 19, 29, 35). However, virulence factors of diarrheagenic non-O1, non-O139 V. cholerae are yet to be elucidated clearly. The pathogenic potential of O1 and O139 strains is largely due to cholera toxin (CT), encoded by the ctxAB genes that reside on the lysogenic filamentous bacteriophage CTXφ (56). Another major virulence factor of O1 and O139 strains is the toxin coregulated pilus (TCP), which plays a crucial role in the intestinal colonization process (22, 55). The genes required for TCP biogenesis are located within a gene cluster known as the Vibrio pathogenicity island (VPI) (31, 34). In addition to its role in intestinal colonization, TCP also acts as the receptor for CTXφ infection (56). Unlike O1 and O139 strains, non-O1, non-O139 V. cholerae strains are generally devoid of ctxAB and VPI genes.
Potential virulence factors of the non-O1, non-O139 V. cholerae strains include hemolysin, repeat toxin (RTX), heat-stable enterotoxin, and Shiga-like toxin (2, 12, 13, 21, 37, 40, 50, 51). Comparative genomics revealed that ∼6% of the O1 V. cholerae genome is missing in certain non-O1, non-O139 strains. The missing portion includes CTX, VPI, Vibrio seventh pandemic island I (VSP-I), VSP-II, and two clusters of the small chromosome of O1 El Tor strain N16961 (17, 18). The VSP-I and VSP-II are proposed to have a role in adaptation to nutrient deprivation or physical/chemical stresses, and studies have revealed that VSP-I and VSP-II are rarely found in non-O1 non-O139 V. cholerae strains (17, 47). A number of non-O1, non-O139 strains possess a gene cluster not found in O1 and O139 strains that shares homology with Vibrio parahaemolyticus type three secretion systems (TTSS) (18); the prevalence of the TTSS in non-O1, non-O139 strains varies, depending on geographic location (18).
In this study, we demonstrate the prevalence of non-O1, non-O139 V. cholerae strains among hospitalized diarrheal patients of Kolkata, India, in 2003 and characterize these strains for their virulence traits and clonality.
MATERIALS AND METHODS
Bacteriology.
The study period included a span of 12 months from January to December 2003. Stool specimens were collected from diarrheal patients admitted to Infectious Diseases and Beliaghata General Hospital and BC Roy Children Hospital, Kolkata, India, and were collected before initiation of antibiotic therapy at the hospitals. Stool specimens or rectal swabs in Cary-Blair medium were processed for bacteriological analysis to detect the presence of V. cholerae. The serogrouping of V. cholerae was based on its agglutinibility with monospecific antisera to O1 and O139 serogroups. V. cholerae strains that did not show agglutination to either O1 or O139 antisera were considered to belong to non-O1, non-O139 serogroups. The serotype analyses of the non-O1, non-O139 strains were performed following the typing scheme developed at the National Institute of Infectious Diseases, Tokyo, Japan (57).
Disk diffusion assay.
Antimicrobial susceptibility testing was carried out using disk diffusion assay (3). Escherichia coli ATCC 25922 was used as quality control strain. V. cholerae strains were tested for susceptibility to different antimicrobial agents using commercially available disks (HiMedia, Mumbai, India) of ampicillin (10 μg), chloramphenicol (30 μg), cotrimoxazole (25 μg), ciprofloxacin (5 μg), furazolidone (100 μg), norfloxacin (10 μg), nalidixic acid (30 μg), gentamicin (10 μg), neomycin (30 μg), streptomycin (10 μg), and tetracycline (30 μg). Interpretation of an isolate as resistant, intermediately resistant, or susceptible to a particular antimicrobial agent was based on the inhibition zone size that matched the published interpretative criteria (43).
PCR assay.
The simplex and multiplex PCR assays were used to detect the presence of virulence genes in V. cholerae non-O1, non-O139 strains. In the multiplex PCR-based assay, a 301-bp amplicon of ctxA (encodes subunit A of CT) and a biotype-specific tcpA allele (encodes TcpA) were detected simultaneously (15, 32). In another multiplex PCR assay, O1 wb and O139 wbf along with ctxA were detected (25). Simplex PCR assays were used for the detection of ompW, cep, ctxB, hlyA, rtxA, toxT, and IS1004. The vcsC2, vcsN2, vspD, and vcsV2 genes were used as target loci for PCR-based amplification for the detection of the TTSS cluster in V. cholerae non-O1, non-O139 strains. Detection of PCR amplicons specific to VC0178 and VC0185 were considered positivity for VSP-I. Similarly, amplicon specific to VC0516 was considered an index for the presence of VSP-II. In separate PCR assays, allele-specific rstR primers were used for the detection of allelic types of rstR in clinical strains (5). PCR assays were carried out in a volume of 25 μl with appropriately diluted 10× PCR buffer (500 mM KCl, 100 mM Tris HCl, 15 mM MgCl2 [pH 8.3]) containing 25 μM each of the deoxynucleoside triphosphates, 1 pmol/μl each of the primers, 1.25 U of Taq polymerase (Takara Shuzo, Otsu, Japan), and 5 μl of whole-cell lysates as a source of DNA template. The simplex and multiplex PCRs were performed in an automated thermocycler (Applied Biosystems, CA). The PCR primers, amplicon sizes, and the PCR annealing temperatures used in this study are shown in Table 1. Polymerization time was adjusted according to the amplicon size considering 1 min per kb size of the amplicon.
TABLE 1.
Details of PCR primers and appropriate combination of factors to generate gene-specific amplicons
| Gene targeta | Primer sequence (5′-3′)
|
Annealing temp (°C) | Amplicon size (bp) | Reference | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| ctxA | CTCAGACGGGATTTGTTAGGCACG | TCTATCTCTGTAGCCCCTATTACG | 60 | 301 | 32 |
| cep | GCTACATGTTTAGCTCACTG | TTTAGCCTTACGAATTAAGCC | 60 | 251 | 5 |
| rstRCl | CTTCTCATCAGCAAAGCCTCCATC | TCGAGTTGTAATTCATCAAGAGTG | 55 | 474 | 5 |
| rstREl | GCACCATGATTTAAGATGCTC | TCGAGTTGTAATTCATCAAGAGTG | 55 | 501 | 5 |
| rstRCal | CTGTAAATCTCTTCAATCCTAGG | TCGAGTTGTAATTCATCAAGAGTG | 55 | 313 | 5 |
| rstREnv | GTTAACGCTTCAAGCCTG | TCGAGTTGTAATTCATCAAGAGTG | 55 | 400 | 44 |
| tcpACl | CACGATAAGAAAACCGGTCAAGAG | ACCAAATGCAACGCCGAATGGAGC | 60 | 618 | 32 |
| tcpAET | GAAGAAGTTTGTAAAAGAAGAACAC | GAAAGGACCTTCTTTCACGTTG | 60 | 472 | 32 |
| rtxA | GCGATTCTCAAAGAGATGC | CACTCATTCCGATAACCAC | 53.8 | 1,366 | 37 |
| hlyA | AGATCAACTACGATCAAGCC | AGAGGTTGCTATGCTTTCTAC | 54.2 | 1,677 | 47 |
| toxT | ACTGTCGACGCAAAGCATATTCAGAGA | CGCGGATCCATACAATCGAAAATAGGA | 65 | 1,101 | 7 |
| ctxB | GGTTGCTTCTCATCATCGAACCAC | GATACACATAATAGAATTAAGGAT | 55 | 460 | 46 |
| O1 wb | GTTTCACTGAACAGATGGG | GGTCATCTGTAAGTACAAC | 55 | 192 | 25 |
| O139 wbf | AGCCTCTTTATTACGGGTGG | GTCAAACCCGATCGTAAAGG | 55 | 449 | 25 |
| ompW | CTCAGACGGGATTTGTTAGGCACG | TCTATCTCTGTAGCCCCTATTACG | 64 | 588 | 41 |
| NAG-ST | CCTATTCATTAGCATAATG | CCAAAGCAAGCTGGATTGC | 55 | 215 | 45 |
| VSP-I (VC0178) | AGGAGGCGTGTAAGTCATAGC | AGACCACGAATACCTGCTCC | 55 | 1,110 | 47 |
| VSP-I (VC0185) | AGAGGCTTGTTTACTATCAG | ATCGGTACTGTCAGGGCT | 50 | 2,053 | 47 |
| VSP-II | GTTTTCTGCGTTGTTCGAG | TCCTGATGTCTCTCTTGCCG | 52 | 965 | 47 |
| TTSS (vcsC2) | GGAAAGATCTATGCGTCGACGTTACCGATGCTATGGGT | CATATGGAATTCCCGGGATCCATGCTCTAGAAGTCGGTTGTTTCGGTAA | 60 | 535 | This study |
| TTSS (vcsN2) | GGATCCCGGGAATTCCATATGCGTCGACAGTTGAGCCAATTCCATT | CGGGGTACCATGCTCTAGACGACCAAACGAGATAATG | 55 | 484 | This study |
| TTSS (vspD) | ATCGTCTAGAACTCGAAGAGCAGAAAAAAGC | ATCGGTCGACCTTCCCGCTTTTGATGAAATG | 55 | 422 | 18 |
| TTSS (vcsV2) | ATGCAGATCTTTTGGCTCACTTGATGGG | ATGCGTCGACGCCACATCATTGCTTGCT | 55 | 742 | 18 |
| IS1004 | ATTGTCATCCCTAAACCACC | AGGCGGTTTTAATATAAGCC | 60 | 603 | This study |
rstRCl and tcpACl, rstR gene and tcpA gene of the classical type; rstREl, rstR gene of El Tor biotype; rstRCal, rstR gene of Calcutta biotype; rstREnv, rstR gene of environmental strain VCE232; tcpAET, tcpA gene of El Tor biotype; NAG-ST, nonagglutinable V. cholerae heat-stable enterotoxin.
Southern hybridization and dot blot analysis.
The 7.5-kb BamHI fragment of the pKK3535 plasmid containing 16S and 23S rRNA genes of E. coli was used as the rRNA gene probe (6). The ctxA probe consisted of a 540-bp XbaI-ClaI fragment of ctxA cloned in pKTN901 using EcoRI linkers (28), and the 267-bp cep probe was prepared by EcoRI digestion of pSC01 (9). The IS1004 probe consisted of 603-bp EcoRI fragments of pNM506. The pNM506 plasmid construct was generated by cloning the IS1004 PCR amplicon in pGEM-T Easy vector (Promega). The identity of the cloned fragment as IS1004 was confirmed by nucleotide sequence analysis. Gel-purified PCR amplicons of 535 bp and 484 bp for vcsC2 and vcsN2, respectively, were generated and used as specific DNA probes for the respective genes.
In the dot blot assay, 2 to 3 μl of a DNA solution containing 1 μg of denatured genomic DNA obtained from V. cholerae cells was spotted onto Hybond N+ membrane (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). The samples were air dried, immobilized by UV cross-linking, and allowed to hybridize separately with specific DNA probes. For ctxA typing, genomic DNA was digested separately with AvaI, BglII, PstI, and MluI (Takara). The digested DNA materials were electrophoretically separated on agarose gels and transferred to Hybond N+ membrane (Amersham), immobilized by UV cross-linking, and allowed to hybridize with DNA probes specific to ctxA (ECL nucleic acid detection system; Amersham). The membranes were washed and exposed to Kodak film (Kodak India Limited, Mumbai, India), and the film was developed to detect the hybridization signals. Hybridization signals to cep were also generated in similar way. The ribotyping and IS1004 typing analyses were carried out with BglI-digested genomic DNA and with respective DNA probes. In a similar way, restriction fragment length polymorphism (RFLP) patterns of vcsC2 and vcsN2 were generated with BglI-digested genomic DNA.
Nucleotide sequencing and analysis.
PCR amplicons specific to target genes were amplified using high-fidelity Taq polymerase (Ex Taq; Takara). The amplicons were purified (PCR purification kit; Qiagen, Hilden, Germany) and used directly for nucleotide sequencing. Both strands were sequenced using Big-Dye terminator cycle sequencing kit (Applied Biosystems) through an automated nucleotide sequencer (ABI Prism 310; Applied Biosystems).
Suckling mouse colonization assay.
The potential of the V. cholerae strains to colonize the intestine was evaluated using the suckling mouse model (53). Suckling mice (3 to 4 days old) were orally challenged with 100 μl of bacterial suspension. The animals were sacrificed 18 h after challenge, and the intestines were removed, washed, and homogenized in normal saline. Challenge dose and intestinal homogenates were serially diluted and plated on Luria agar (LA) plates to determine the number of CFU. The colonization potential was determined as the number of intestinal CFU divided by the number of CFU in the challenge dose.
Hemolytic assay.
Hemolytic activity was assayed in cell-free culture supernatants against rabbit erythrocytes (24). V. cholerae strains were grown in brain heart infusion (BHI) broth at 37°C for 6 h and adjusted to unit opacity at 540 nm with BHI broth. Bacterial cells were then centrifuged at 12,000 × g for 15 min at 4°C, and the supernatant was serially diluted in a microtiter plate with 100 μl of 0.1 M phosphate-buffered saline, pH 7.2. One hundred microliters of a solution of 1% washed rabbit erythrocytes was added to each well and mixed by gentle rocking. Following an incubation of 2 h at 37°C, unlysed erythrocytes were removed by centrifugation, the optical density at 540 nm (OD540) was measured for the supernatants, and the OD value was compared to the reading that was obtained for 100% lysis. The hemolytic titer was considered the reciprocal of the highest dilution that was sufficient to cause at least 50% lysis of rabbit erythrocytes.
Biofilm assay.
For the biofilm assay (36), cells were grown in LB at 30°C under static condition in borosilicate glass tubes. Following 22 h of incubation, the cultures were removed, and the tubes were washed gently with water to remove loosely bound cells from the surface. Adherent cells were then stained with 0.01% crystal violet, washed thoroughly with water, and treated with dimethyl sulfoxide. Biofilm formation was measured by OD570, using V. cholerae O139 strain MO10, known to form a robust biofilm, as a positive control (26).
Motility assay.
V. cholerae strains were grown overnight at 30°C on LA plates, then inoculated into motility agar (LA with 0.3% agar), and incubated at 30°C for 18 h. The diameter of the swarm size was measured and compared to that of nonmotile variant KKV176 (33).
HeLa cell cytotoxicity.
Cytotoxic activity against HeLa cells was assayed by culturing HeLa cells in Eagle's modified minimum essential medium supplemented with 10% fetal bovine serum, 100 U/ml of penicillin G, and 100 μg/ml of streptomycin sulfate in 96-well flat-bottomed tissue culture plates at 37°C and 5% CO2. At 60 to 70% confluence, dilutions of filter-sterilized culture supernatants were added, and cells were incubated for 24 h at 37°C and 5% CO2. Alterations of morphology were observed using an optical microscope, and the cytotoxic titer was expressed as the reciprocal of the highest dilution that was sufficient to induce morphological changes in at least 50% of the HeLa cells.
CT production.
CT expression by the V. cholerae strains in vitro was assayed by the GM1 enzyme-linked immunosorbent assay method described previously (23) and expressed as nanogram/milliliter/opacity unit measured at 540 nm (42).
RESULTS
Prevalence of non-O1, non-O139 V. cholerae strains among diarrheal cases.
Stool specimens collected from hospitalized diarrheal patients were analyzed for V. cholerae. Microbiological analysis detected V. cholerae strains in 197 stool specimens; of these specimens, 135 specimens were determined to contain V. cholerae strains belonging to serogroup O1, and 2 specimens contained strains belonging to serogroup O139. The remaining 60 strains were nonagglutinable with either O1 or O139 antisera and were considered to belong to non-O1, non-O139 serogroups. PCR-based screening revealed that 6 of the 60 strains gave the 192-bp PCR amplicon-specific O1 wb region, which is known to be responsible for O1 antigen biosynthesis in V. cholerae. These data indicated that genetically these six strains were of the O1 background, but they lacked O1 antigen on the cell surface, and were considered rough variants. Therefore, 54 non-O1, non-O139 strains that were associated with hospitalized diarrheal cases in Kolkata, India, during 2003 were identified. It is worth mentioning here that 31 pediatric cases (≤12 years old) versus 23 adults (>12 years old) were identified for these 54 patients. In these cases, however, infection due to other enteric pathogens could not be ruled out.
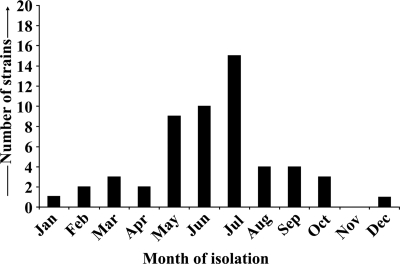
Serogrouping of 54 non-O1, non-O139 V. cholerae strains categorized 42 strains into 19 different serogroups with no apparent clustering of serogroups, with the exception of 6 and 5 strains belonging to O34 and O37 serogroups, respectively. The remaining 12 strains remained nonagglutinable with antisera against all known (currently 206) non-O1, non-O139 serogroups. Monthly isolation profiles of these non-O1, non-O139 strains are presented in Fig. 1. Analysis of the antimicrobial resistance profiles of these non-O1, non-O139 strains revealed no apparent clustering of particular profile type (data not shown). Most of the strains exhibited resistance to ampicillin (86.5%), furazolidone (78.8%), cotrimoxazole (50%), and nalidixic acid (48.1%). Most of the strains were also sensitive to neomycin (55.8%), norfloxacin (92.3%), and tetracycline (90.4%). Interestingly, the presence of strains with increased resistance to neomycin (42.3%) and tetracycline (7.7%) was noted.
FIG. 1.
Monthly isolation profile of non-O1, non-O139 V. cholerae strains isolated from diarrheal patients admitted to Infectious Diseases and Beliaghata General Hospital and BC Roy Children Hospital in Kolkata, India, in 2003.
Rough variants of V. cholerae O1.
Six rough variant V. cholerae O1 strains isolated during this study were screened by PCR for tcpA and ctxA (Table 2). All six strains possessed the El Tor tcpA allele. These strains were also positive by PCR for a panel of known virulence factors (ctxA, cep, tcpA, toxT, hlyA, rtxA, VSP-I, VSP-II, and the El Tor allele of rstR) of V. cholerae O1 El Tor strains. All six strains were also positive for the 192-bp PCR amplicon specific to the O1 (wb) biosynthetic gene cluster. The lipopolysaccharide (LPS) of these strains was isolated and analyzed by gel electrophoresis (Fig. 2A), which revealed the lack of O antigen on their LPS (lanes 3 to 8), unlike the O1 classical strain O395 (lane 2). These six rough variants were further subjected to BglI ribotyping analysis (Fig. 3A). All these rough variants displayed ribotype profiles that were identical to each other (lanes 1 to 6) and to the profile (RIII) of the O1 strains isolated in 2003 in Kolkata, India (9), represented by strain CO840 (lane 7).
TABLE 2.
Distribution of virulence genes among clinical non-O1, non-O139 V. cholerae strains isolated from patients with acute diarrhea in 2003
| No. of strains | PCR-based detectiona of virulence gene:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ctxA | cep | tcpA | toxT | hlyA | rtxA | TTSSb | VSP-Ic | VSP-IId | O1 wb | |
| 6e | + | + | + (El)f | + | + | + | − | + | + | + |
| 1 | + | + | + (Env)g | − (+)h | + | + | + | − | − | − |
| 5 | − | − | + (Env)g | − (+)h | + | + | + | − | − | − |
| 10 | − | − | − | − | + | + | + | − | − | − |
| 27 | − | − | − | − | + | + | − | − | − | − |
| 1 | − | − | − | − | + | + | − | + | + | − |
| 1 | − | − | − | − | + | − | + | − | − | − |
| 2 | − | − | − | − | + | − | − | − | − | − |
| 7 | − | − | − | − | − | − | − | − | − | − |
Symbols: +, PCR positive by generating the expected-size amplicon; −, no amplicon detected by the PCR assay.
PCR-based detection of the TTSS gene cluster was based on the detection of vcsV2, vcsC2, vcsN2, and vspD.
PCR-based detection of VSP-I was based on the detection of both VC0178 and VC0185.
PCR-based detection of VSP-II was based on the detection of VC0516.
All six strains were rough V. cholerae of O1 background.
+ (El), El Tor allele of tcpA.
These strains gave a tcpA amplicon identical to the classical allele. However, nucleotide sequences were identical to tcpAEnv.
These strains were PCR negative for toxT but positive by dot blot assay for toxT.
FIG. 2.

Lipopolysaccharide profiles of representative V. cholerae strains. The O1 background rough (A) and smooth non-O1, non-O139 (B) strains were isolated in 2003. (A) V. cholerae rough strains (lanes 3 to 8) included SC6, SC117, SC122, SC135, SC145, and SC156, respectively. (B) Smooth V. cholerae non-O1, non-O139 strains (lanes 3 to 8) included SC191 (O48), SC132 (O48), SC124 (O36), SC107a (O128), SC103a (O48), and SC72 (ONT), respectively (B). In panels A and B, lanes 1 and 2 show LPS profiles obtained with E. coli ATCC 25922 and V. cholerae O1 classical strain O395, respectively.
FIG. 3.

The BglI ribotype patterns of V. cholerae O1 background rough (A) and smooth non-O1, non-O139 (B) strains isolated in 2003. (A) V. cholerae rough strains (lanes 1 to 6) included SC6, SC117, SC122, SC135, SC145, and SC156, respectively. The smooth variant of O1 strains with RIII ribotype pattern (9) (lane 7, CO840) was included as reference in panel A. (B) Smooth V. cholerae non-O1, non-O139 strains (lanes 1 to 9) included SC17 (O37), SC110 (O34), SC182 (O15), SC72 (ONT), SC103a (O48), SC107a (O128), SC132 (O48), SC191 (O48), and SC124 (O36), respectively. The positions of the HindIII-digested λ molecular size markers (in kilobases) are indicated at the sides of the gels in the figure.
Toxigenic traits and colonization potentials of clinical non-O1, non-O139 V. cholerae strains.
The 54 non-O1, non-O139 V. cholerae strains failed to produce O1 wb- and O139 wbf-specific amplicons (Table 2). The LPS profiles of these strains were distinct from those of the O1 and O139 strains and showed remarkable variations (Fig. 2B). All the non-O1, non-O139 strains were PCR negative for ctxA, with the exception of the O36 strain SC124 (Table 2). Strain SC124 was also PCR positive for another CTX prophage gene, cep, whereas the other non-O1, non-O139 strains were PCR negative for cep. Strain SC124 was further analyzed by Southern hybridization analysis (Fig. 4), using DNA probes specific for ctxA and cep. Hybridization with the ctxA probe revealed the presence of a single ∼7-kb hybridizing fragment with AvaI, PstI, and MluI digestions and a larger hybridizing fragment with BglII digestions (Fig. 4A). However, hybridization with the cep probe demonstrated the presence of four hybridizing fragments in the AvaI, PstI, and MluI digestions and three hybridizing fragments in the BglII digestion (Fig. 4B). Sequencing of the ctxB gene of strain SC124 revealed that is was identical to that of environmental strain VCE232 (GenBank accession no. AF414369) (5). Among the 53 ctxA-negative non-O1, non-O139 strains, 12 were PCR positive for the El Tor rstR allele, and interestingly only one, strain SC124 (ctxA positive) was PCR positive for the classical rstR allele as well as an environmental allele of rstR (47).
FIG. 4.
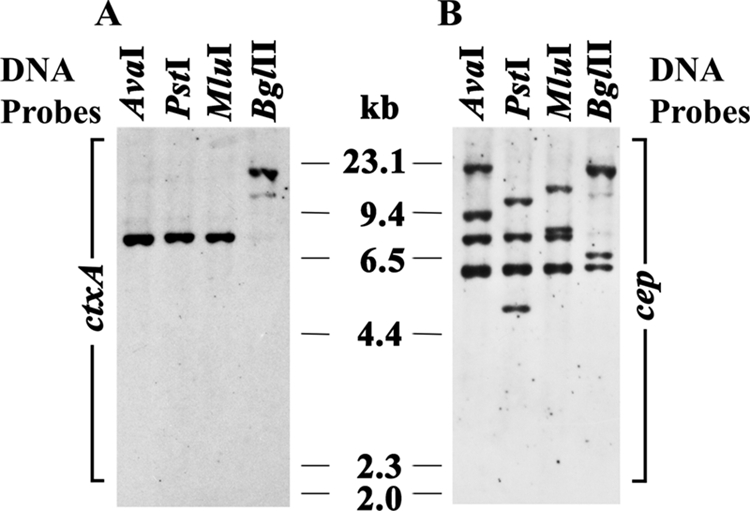
RFLP patterns of the CTX genetic elements of toxigenic non-O1, non-O139 V. cholerae strain SC124 (O36). Genomic DNA was digested with different restriction enzymes as indicated in the figure. DNA probes specific for ctxA (A) and cep (B) were used. The positions of the HindIII-digested λ molecular size markers (in kilobases) are indicated at the sides of the gels in the figure.
The five ctxA-negative strains (three O48 strains [SC103a, SC132, and SC191], an O128 strain [SC107a], and the O nontypeable [ONT] strain SC72) and the toxigenic strain SC124 were PCR positive for tcpA. The nucleotide sequences of the tcpA genes of these six strains were identical to the nucleotide sequence of the tcpA gene of environmental strain VCE22 (GenBank accession no. AF414371) (5) and are therefore referred to as “environmental” tcpA (tcpAEnv) hereafter. Interestingly, all six tcpA-positive strains failed to produce a PCR amplicon specific to toxT, the regulatory gene normally found within the tcpA-encoding VPI (35). These strains were further analyzed in a dot blot assay using DNA probes specific for toxT. The results demonstrated that all these strains reacted strongly with both tcpA and toxT probes. The ctxA-positive, tcpAEnv strain SC124 and the ctxA-negative, tcpAEnv strain SC132 showed comparable colonization potentials in the infant mouse intestine, displaying ∼39- and ∼42-fold increases in intestinal bacterial counts, respectively (Table 3). In contrast, the tcpA-negative strain SC182 showed only ∼2.5-fold increase in intestinal bacterial counts. The tcpACl (tcpA of the classical type) O1 strain O395 showed much higher (∼277-fold) increases in intestinal bacterial counts, approximately 10-fold higher than for the tcpAEnv non-O1, non-O139 strains.
TABLE 3.
Colonization potential of clinical non-O1, non-O139 V. cholerae strains assayed in a suckling mouse model
| V. cholerae strain | Serogroup/biotypea | tcpA geneb | Challenge dose (mean ± SEM) (CFU/ml)c | Recovery (mean ± SEM) (CFU/mouse)d | Fold increase in CFUe |
|---|---|---|---|---|---|
| SC132 | O48/− | + (Env) | (1.69 ± 0.38) × 105 | (7.08 ± 0.74) × 106 | 41.9 |
| SC124 | O36/− | + (Env) | (1.57 ± 2.50) × 105 | (6.11 ± 0.89) × 106 | 38.9 |
| SC182 | O15/− | − | (2.32 ± 0.64) × 105 | (5.80 ± 0.90) × 105 | 2.5 |
| O395 | O1/classical | + (Cl) | (2.32 ± 0.64) × 105 | (6.44 ± 2.03) × 107 | 277 |
The serogroup is shown before the slash, and the biotype is shown after the slash. −, no biotype.
The presence (+) or absence (−) of the tcpA gene and the type of tcpA gene are shown. V. cholerae O1 classical strain O395 possesses tcpA of the classical type (tcpACl), and strains SC132 and SC124 possessed environmental tcpA (tcpAEnv).
Each mouse was challenged with 100 μl of bacterial suspension mixed with 0.01% Evans blue. The number of viable cells present in 100 μl was determined by the serial dilution method.
After 18 h of challenge, all mice were sacrificed, and intestinal homogenates were plated separately for each of the groups from appropriately diluted suspensions. The number of viable cells was counted, and the number of cells recovered per intestine was estimated.
Increase of bacterial CFU due to colonization is expressed as the increase in CFU from the challenge dose to the number of CFU recovered per mouse.
Detection and distribution of potential virulence factor genes.
PCR analyses were performed on the non-O1, non-O139 V. cholerae strains to detect a number of virulence genetic elements, including hlyA, rtxA, TTSS, VSP-I, and VSP-II. Most of the 54 non-O1, non-O139 strains possessed hlyA (87%) and rtxA (81.5%) (Table 2). The TTSS gene cluster was detected by PCR analysis targeting four different TTSS genes (vcsC2, vcsN2, vspD, and vcsV2). Of the non-O1, non-O139 strains, 31.5% possessed the TTSS, since these strains were positive for all four TTSS genes. Only one non-O1, non-O139 strain (strain SC75, O11) was PCR positive for both VSP-I and VSP-II genes, while the rest of the non-O1, non-O139 strains were negative for both VSP-I and VSP-II (Table 2). None of the non-O1, non-O139 strains were PCR positive for nonagglutinable V. cholerae heat-stable enterotoxin (data not shown).
Segregation of different clones of non-O1, non-O139 V. cholerae strains.
Six tcpAEnv-positive, non-O1, non-O139 V. cholerae strains and three arbitrarily selected tcpA-negative, non-O1, non-O139 V. cholerae strains were analyzed by Southern blotting with the rRNA gene probe to determine their ribotype pattern (Fig. 3B). Interestingly, five nontoxigenic tcpAEnv-positive, non-O1, non-O139 strains displayed identical ribotype profiles (Fig. 3B, lanes 4 to 8). The ctxA-positive tcpAEnv strain SC124 displayed a different ribotype profile from that of the other tcpAEnv-positive strains (Fig. 3B, lane 9). The ribotype profiles of the other three nontoxigenic, tcpA-negative, non-O1, non-O139 strains analyzed (Fig. 3B, lanes 1 to 3) showed patterns different from those of the tcpA-positive strains. The five nontoxigenic, tcpAEnv-positive, non-O1, non-O139 strains were also subjected to Southern blot analysis using IS1004 and TTSS (vcsN2) probes (Fig. 5). The five nontoxigenic, tcpAEnv-positive, non-O1, non-O139 strains reacted weakly only with the IS1004 probe (Fig. 5A, lanes 4 to 8) but showed identical single bands when hybridized with the vcsN2 probe (Fig. 5B, lanes 4 to 8). The toxigenic tcpAEnv-positive strain SC124 gave patterns with both the IS1004 and vcsN2 probes that were distinct from those of the other nontoxigenic tcpAEnv-positive strain (Fig. 5A and B, lanes 9). The profiles of two of the three nontoxigenic, tcpA-negative, non-O1, non-O139 strains analyzed (Fig. 5A, lanes 1 and 3) showed IS1004 patterns that were different from those of the tcpA-positive strains, whereas the third nontoxigenic, tcpA-negative, non-O1, non-O139 strain (lane 2) reacted weakly with this probe, similar to the nontoxigenic, tcpAEnv non-O1, non-O139 strains. All three nontoxigenic, tcpA-negative, non-O1, non-O139 strains gave patterns with the vcsN2 probe (Fig. 5B, lanes 1 to 3) that were distinct from those of the nontoxigenic, tcpAEnv non-O1, non-O139 strains.
FIG. 5.

DNA fingerprinting patterns generated with IS1004 (A) and vcsN2 (B) probes against BglI-digested genomic DNA obtained from non-O1, non-O139 V. cholerae strains. V. cholerae strains include SC17 (O37), SC110 (O34), SC182 (O15), SC72 (ONT), SC103a (O48), SC107a (O128), SC132 (O48), SC191 (O48), and SC124 (O36) in lanes 1 to 9, respectively. The positions of the HindIII-digested λ molecular size markers (in kilobases) are indicated at the sides of the gels in the figure.
Virulence factor production by non-O1, non-O139 V. cholerae strains.
The toxigenic non-O1, non-O139 strain SC124 (O36) was assayed for its ability to produce CT in vitro (Table 4). Growing strain SC124 in LB (pH 6.5) at 30°C induced CT expression to a level that was similar to that in an El Tor O1 strain grown under these conditions, but approximately 10-fold less than that expressed by a classical O1 strain. Growth of strain SC124 under AKI growth conditions induced only low-level CT expression that was again approximately 10-fold less than that expressed by a classical O1 strain grown under these conditions.
TABLE 4.
CT production ability of V. cholerae strains grown under different cultural conditions
| Strain (serogroup/biotype) | Amt of CTa produced in medium:
|
|
|---|---|---|
| LBb | AKIc | |
| SC124 (O36) | 2.9 | 0.45 |
| O395 (O1/classical) | 39.1 | 4.38 |
| VC20 (O1/El Tor) | 3.6 | 13.01 |
Assayed by the GM1 enzyme-linked immunosorbent assay method (23) and expressed as nanograms of toxin per milliliter of culture supernatant per unit of opacity of bacterial suspension measured at 540 nm.
LB medium, pH 6.5, incubation temperature of 30°C, 16 h of culture.
AKI medium, pH 7.4, incubation temperature of 30°C, 4 h of static culture with limited aeration, followed by 16 h of mild shaking culture with adequate aeration.
A panel of 13 clinical non-O1, non-O139 V. cholerae strains were assayed for the expression of other virulence phenotypes, including hemolytic activity against rabbit erythrocytes and secretion of cytotoxic factors, as well as motility and biofilm phenotypes. All strains were positive for all four phenotypes; however, variability in the phenotypes was evident (Table 5). The culture filtrate obtained from all 13 non-O1, non-O139 strains tested induced cell rounding in HeLa cells, and the effect was reversed by replacing the culture filtrate with fresh medium. Treating the culture filtrate with heat (10 min in a boiling water bath) reduced the cell rounding induction by 2- to 16-fold in most cases. Interestingly, the supernatants of three strains (SC103a, SC65, and SC108) were capable of inducing equivalent levels of cell rounding even after heat treatment (data not presented).
TABLE 5.
Assays of clinical non-O1, non-O139 V. cholerae strains to determine their ability to lyse rabbit erythrocytes, extent of motility, formation of biofilm, and cytotoxic effect
| Strain/serogroup | Presence or absence of potential virulence genesa
|
Phenotypes
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| TTSSb | tcpAc | ctxA | rtxA | hlyA | Hemolytic titerd | Motility (cm)e | Biofilm assayf | Cytotoxic titerg | |
| SC124/O36 | + | + | + | + | + | 64 | 2.9 | 0.62 | 128 |
| SC72/ONT | + | + | − | + | + | 16 | 4.1 | 0.31 | 256 |
| SC103(a)/O48 | + | + | − | + | + | 32 | 5.2 | 0.78 | 128 |
| SC113/O23 | + | − | − | + | + | 64 | 4.0 | 0.47 | 64 |
| SC132/O48 | + | + | − | + | + | 32 | 4.2 | 0.41 | 128 |
| SC182/O15 | + | − | − | + | + | 2 | 3.0 | 0.20 | 64 |
| SC191/O48 | + | + | − | + | + | 16 | 4.1 | 0.43 | 128 |
| SC130/O37 | − | − | − | + | + | 8 | 2.6 | 0.47 | 2048 |
| SC164/O59 | − | − | − | + | + | 16 | 2.4 | 0.73 | 512 |
| SC42/ONT | − | − | − | + | − | 4 | 2.6 | 0.64 | 32 |
| SC133/O6 | − | − | − | + | − | 8 | 3.0 | 0.58 | 128 |
| SC65/ONT | − | − | − | − | − | 2 | 2.3 | 0.25 | 32 |
| SC108/O113 | − | − | − | − | − | 2 | 2.1 | 0.21 | 64 |
The presence (+) or absence (−) of potential virulence genes is shown.
PCR-based detection of the TTSS cluster by the presence of vcsC2, vcsN2, vspD, and vcsV2.
The strains possessed the tcpAEnv allele.
The hemolytic titer was defined as the highest dilution of cell-free culture supernatants (BHI, 6 h, 37°C) sufficient to cause 50% lysis of rabbit erythrocytes in 2 h at 37°C. Photometric assay (OD540) was used to quantify the percent lysis.
The motility of V. cholerae strains was determined in centimeters and represents the diameter of the culture zone formed on the surface of the plate incubated at 30°C for 15 h.
Retention of crystal violet dye by V. cholerae cells that formed the biofilm on a glass surface was estimated at 570 nm.
Inducing altered morphology in at least 50% HeLa cells was considered the endpoint.
DISCUSSION
This study characterized a number of non-O1, non-O139 V. cholerae clinical strains that were isolated from hospitalized diarrheal patients admitted to the Infectious Diseases and Beliaghata General Hospital and BC Roy Children Hospital in Kolkata, India. Bacteriological analysis detected 60 non-O1, non-O139 V. cholerae strains in the stool specimens collected from patients in Kolkata, India, in 2003. V. cholerae non-O1, non-O139 strains isolated from diarrheal patients comprised 30.4% of the total cases (n = 197) that were microbiologically positive for V. cholerae; this is consistent with an earlier report on the prevalence of non-O1, non-O139 strains isolated from acute diarrheal cases in Kolkata, India (54). All 60 non-O1, non-O139 V. cholerae strains were positive for the 588-bp PCR amplicon specific to V. cholerae ompW (41). Six of these strains were determined to be rough O1 strains (i.e., lacking the O1 antigen but containing the O1-specific gene cluster) by a PCR-based assay (Table 2); these strains were initially classified as non-O1, non-O139 due to their lack of agglutination with either O1- or O139-specific antisera. Therefore, 54 non-O1, non-O139 strains were identified; these strains were associated with hospitalized diarrheal cases in Kolkata, India, in 2003 (Table 2). Although no apparent serogroup clustering was observed, six and five strains belonged to O34 and O37 serogroups, respectively. In fact, 42 strains were classified into 19 different serogroups and 12 strains remained O nontypeable. These results demonstrate the potential for strains of diverse serogroups within an region where cholera is endemic, like Kolkata, India, to cause acute diarrhea. Moreover, the detection of a large number of ONT strains also suggests that additional serogroups have arisen that need to be added to the current serotyping scheme.
Through this study, six rough variant O1 strains were identified. These O1 background rough variants were PCR positive for a panel of known virulence factors (ctxA, cep, tcpA, toxT, hlyA, rtxA, VSP-I, VSP-II, and the El Tor allele of rstR) of V. cholerae O1 El Tor strains and displayed a ribotype profile (RIII) of the O1 strains isolated in 2003 in Kolkata, India (9). All of these data confirmed that these rough variants were derived from O1 El Tor. The rough variant O1 V. cholerae isolated from clinical cases has been reported previously (14, 38). The significance of the high incidence of such strains isolated in the present study (10% of the non-O1, non-O139 strains) is not clear, but the lack of O antigen may facilitate phage resistance against O1-specific phages (20) and may also facilitate horizontal gene transfer (38), and thus, these strains may represent a previously unrecognized important role in cholera epidemiology.
PCR-based screening revealed detection of only one toxigenic strain (SC124 [serogroup O36]) of the 54 non-O1, non-O139 V. cholerae strains. These data were further extended through Southern hybridization analysis, which indicated the existence of a complete CTX prophage in tandem with additional copies of truncated CTX prophages in the SC124 genome. Sequencing of the ctxB gene of strain SC124 revealed it to be identical to that of the environmental strain VCE232 (5), and this result is in line with the concept of non-O1, non-O139 CTX prophages having a lineage different from the lineage of their O1 counterpart. Strain SC124 (O36) produced very low levels of CT in vitro when assayed in both the conditions known to be optimal for O1 classical and El Tor strains. Previous reports have also noted the low level of CT expression by toxigenic non-O1, non-O139 strains grown under these conditions (52, 58), which may be due to suboptimal in vitro inducing conditions derived for clinical O1 strains.
Apart from toxigenic traits, these strains were also tested for the presence of the tcpA gene that encodes the pilus colonization factor of V. cholerae O1. The five ctxA-negative strains (three O48 strains [SC103a, SC132, and SC191], an O128 strain [SC107a], and the ONT strain SC72) and the toxigenic strain SC124 were PCR positive for tcpA. The tcpA genes of these non-O1, non-O139 strains were identical to that of an environmental strain VCE22 (5) and thus referred to as “environmental” tcpA (tcpAEnv). Identification of the existence of tcpAEnv type among the non-O1, non-O139 strains prompted us to check the presence of toxT, the regulatory gene normally found within the tcpA-encoding VPI (34). These strains were negative for toxT PCR but reacted strongly in the dot blot assay. The apparent discrepancy between toxT negativity in the PCR-based assay to the dot blot assay result can be explained by considering the presence of toxT alleles different from canonical ones. The data suggest that these six non-O1, non-O139 strains possess an altered VPI with variant tcpA and toxT alleles.
The clonal relationships among these tcpA-positive strains were determined. The RFLP profiles generated with the rRNA gene probe and the IS1004- and TTSS (vcsN2)-specific DNA probes revealed that five nontoxigenic, tcpAEnv-positive, non-O1, non-O139 V. cholerae strains were clonally related, even though these strains belong to three different serogroups (one strain belonged to serogroup O128, one strain was ONT, and three strains belonged to serogroup O48). The profiles of the ctxA-positive tcpAEnv strain SC124 were different from those of the other tcpAEnv-positive strains. The five nontoxigenic tcpAEnv-positive strains were isolated on different dates spanning five months (May to September), suggesting that these strains were not isolated from a single cluster of cases. The other non-O1, non-O139 strains (SC17, SC110, and SC182) that were included in the RFLP analysis exhibited patterns different from those of the tcpA-positive strains. Collectively, these results indicate a close relationship between the five nontoxigenic, tcpAEnv non-O1, non-O139 strains and the lack of a close relationship among and between all other non-O1, non-O139 strains.
The function of tcpAEnv was evaluated by measuring the colonization potential of several non-O1, non-O139 V. cholerae strains with tcpAEnv, as well as that of one of the non-O1, non-O139 strains lacking tcpA. The ctxA-positive, tcpAEnv strain SC124 and the ctxA-negative, tcpAEnv strain SC132 showed comparable colonization potentials in the infant mouse intestine (Table 3). In contrast, the tcpA-negative strain SC182 showed only ∼2.5-fold increase in intestinal bacterial counts. The tcpACl O1 strain O395 showed much higher (∼277-fold) increases in intestinal bacterial counts. Further studies are required to determine whether the alternative tcpA and toxT alleles or other aspects of the strain background are responsible for lowering the colonization potential of the non-O1, non-O139 strains compared to that of the O1 classical strain O395.
The presence of major virulence genes (ctxA and tcpAEnv) and their regulators (alleles of toxT) in some of the non-O1, non-O139 V. cholerae strains prompted us to screen the strains for the occurrence of other potential virulence factors. PCR-based analysis targeting various virulence gene loci showed that the hlyA (87%) and rtxA (81.5%) genes (Table 2) were prevalent in these strains, which is consistent with previous reports including strains that were isolated in Kolkata, India (8, 47, 54). The prevalence of the RTX cluster in most of the non-O1, non-O139 strains isolated is consistent with the hypothesis that RTX contributes to virulence in nontoxigenic V. cholerae (37, 47). The incidence of TTSS among the non-O1, non-O139 strains was 31.5%, which is similar to that reported in Bangladesh (38.9%) (17). The nucleotide sequence of the ATPase homologue vcsN2 from the non-O1, non-O139 strains revealed variations at the nucleotide level (GenBank accession no. EU689098 to EU689102), but the predicted amino acid sequence remained identical to the amino acid sequence of the reference vcsN2 sequence AM19226 (GenBank accession no. DQ124262). Detection of silent nucleotide mutations within vcsN2 indicated functional conservation of the TTSS apparatus. Only one non-O1, non-O139 strain (SC75 [serogroup O11]) was PCR positive for both VSP-I and VSP-II genes, while the rest of the non-O1, non-O139 strains were negative for both VSP-I and VSP-II (Table 2), consistent with previous published reports (47).
Functional expression of virulence phenotypes was assayed by using a panel of 13 clinical non-O1, non-O139 V. cholerae strains. These strains caused hemolysis of rabbit erythrocytes, secreted cytotoxic factors in the culture-free supernatant, produced biofilm, and were motile; however, interesting patterns could be detected. For example, a tendency to have a higher hemolytic titer and higher cytotoxic activity was evident in strains that carried the TTSS gene cluster (Table 5). The strains that possess the TTSS cluster also appeared to be more motile than those lacking the TTSS, consistent with previous observations regarding the TTSS (18). No clear correlation could be obtained between the presence of specific virulence genes and biofilm formation. The culture filtrate obtained from these strains induced cell rounding in HeLa cells, which can be reversed by replacing the culture filtrate with fresh medium. Interestingly, the strains that lacked ctxA, hlyA, and TTSS (SC42, SC133, SC65, and SC108) and strains that lacked rtxA (SC65 and SC108) were capable of inducing cell rounding (Table 5).
Our study characterized a number of non-O1, non-O139 V. cholerae clinical strains that were isolated from hospitalized patients with diarrhea. Certain interesting characteristics were evident, including the identification of strains carrying ctxA, and alternate forms of tcpA and toxT (otherwise found almost exclusively in O1 strains). Interestingly, the tcpA- and toxT-positive strains appear to be closely related, yet they do not all express the same O antigen. We also found a number of strains carrying the TTSS, and in vitro, the presence of the TTSS correlated with increased hemolytic titers and increased motility, suggesting a role for this secretion system in human infections by non-O1, non-O139 strains. Most strains isolated also contained RTX and/or HlyA, but two of the strains identified in this study contained none of these defined factors, indicating that additional virulence factors associated with human disease remain to be identified. We also identified several rough O1 strains that had originally been characterized as non-O1, non-O139 due to lack of agglutination with O1 antisera, suggesting that rough strains resulting from immune pressure or phage predation may represent a previously unappreciated cause of disease among “non-O1, non-O139” cholera-like diarrhea cases.
Disease-causing strains need to survive in the environment for a substantial period of time between successive infections in order to maintain an efficient transmission cycle. Previous studies have suggested that toxigenic non-O1, non-O139 V. cholerae strains are distinct from toxigenic V. cholerae O1 and O139 strains and represent a separate reservoir of cholera enterotoxin genes (30). Our study showed that clinical V. cholerae non-O1, non-O139 strains possess additional virulence factors, some of which are commonly found in environmental strains. The continuous development of genetic diversity in non-O1, non-O139 strains within the environment and the association of V. cholerae non-O1, non-O139 strains of several serogroups with clinical diarrheal cases are likely to complicate the development of an effective cholera vaccine.
Acknowledgments
This work was supported in part by the Council of Scientific and Industrial Research (CSIR) (grant 37/1206/04/EMR II), Department of Biotechnology (DBT) (grant BT/PR6918/BRB/10/454/2005), the government of India, and Japan International Cooperation Agency (JICA/NICED project 054-1061-E-O). S.C. received an ASM travel grant in 2005 and worked in the Department of Biology, University of Texas at San Antonio. K.G. was the recipient of a research fellowship from the University Grants Commission (UGC) of the government of India.
Footnotes
Published ahead of print on 21 January 2009.
REFERENCES
- 1.Aldova, E., K. Laznickova, E. Stepankova, and J. Lietava. 1968. Isolation of nonagglutinable vibrios from an enteritis outbreak in Czechoslovakia. J. Infect. Dis. 11825-31. [DOI] [PubMed] [Google Scholar]
- 2.Basu, I., R. Mitra, P. K. Saha, A. N. Ghosh, J. Bhattacharya, M. K. Chakrabarti, Y. Takeda, and G. B. Nair. 1999. Morphological and cytoskeletal changes caused by non-membrane damaging cytotoxin of Vibrio cholerae on int 407 and HeLa cells. FEMS Microbiol. Lett. 179255-263. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, A. W., W. M. Kirby, J. C. Sherris, and M. Turck. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45493-496. [PubMed] [Google Scholar]
- 4.Bhattacharya, S. K., M. K. Bhattacharya, G. B. Nair, D. Dutta, A. Deb, T. Ramamurthy, S. Garg, P. K. Saha, P. Dutta, A. Moitra, B. K. Mandal, T. Shimada, Y. Takeda, and B. C. Deb. 1993. Clinical profile of acute diarrhoea cases infected with the new epidemic strain of Vibrio cholerae O139: designation of the disease as cholera. J. Infect. 2711-15. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya, T., S. Chatterjee, D. Maiti, R. K. Bhadra, Y. Takeda, G. B. Nair, and R. K. Nandy. 2006. Molecular analysis of the rstR and orfU genes of the CTX prophages integrated in the small chromosomes of environmental Vibrio cholerae non-O1, non-O139 strains. Environ. Microbiol. 8526-534. [DOI] [PubMed] [Google Scholar]
- 6.Brosius, J., A. Ullrich, M. A. Raker, A. Gray, T. J. Dull, R. R. Gutell, and H. F. Noller. 1981. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid 6112-118. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, P. A., K. T. Tashima, M. B. Rogers, V. J. DiRita, and S. B. Calderwood. 1997. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol. Microbiol. 251099-1111. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborty, S., P. Garg, T. Ramamurthy, M. Thungapathra, J. K. Gautam, C. Kumar, S. Maiti, S. Yamasaki, T. Shimada, Y. Takeda, A. Ghosh, and G. B. Nair. 2001. Comparison of antibiogram, virulence genes, ribotypes and DNA fingerprints of Vibrio cholerae of matching serogroups isolated from hospitalised diarrhoea cases and from the environment during 1997-1998 in Calcutta, India. J. Med. Microbiol. 50879-888. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee, S., K. Ghosh, A. Raychoudhuri, A. Pan, M. K. Bhattacharya, A. K. Mukhopadhyay, T. Ramamurthy, S. K. Bhattacharya, and R. K. Nandy. 2007. Phenotypic and genotypic traits and epidemiological implication of Vibrio cholerae O1 and O139 strains in India during 2003. J. Med. Microbiol. 56824-832. [DOI] [PubMed] [Google Scholar]
- 10.Dalsgaard, A., M. J. Albert, D. N. Taylor, T. Shimada, R. Meza, O. Serichantalergs, and P. Echeverria. 1995. Characterization of Vibrio cholerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J. Clin. Microbiol. 332715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalsgaard, A., A. Forslund, L. Bodhidatta, O. Serichantalergs, C. Pitarangsi, L. Pang, T. Shimada, and P. Echeverria. 1999. A high proportion of Vibrio cholerae strains isolated from children with diarrhoea in Bangkok, Thailand are multiple antibiotic resistant and belong to heterogenous non-O1, non-O139 O-serotypes. Epidemiol. Infect. 122217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalsgaard, A., O. Serichantalergs, A. Forslund, W. Lin, J. Mekalanos, E. Mintz, T. Shimada, and J. G. Wells. 2001. Clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the CTX phage and the genes encoding the toxin-coregulated pili. J. Clin. Microbiol. 394086-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalsgaard, A., O. Serichantalergs, T. Shimada, O. Sethabutr, and P. Echeverria. 1995. Prevalence of Vibrio cholerae with heat-stable enterotoxin (NAG-ST) and cholera toxin genes: restriction fragment length polymorphisms of NAG-ST genes among V. cholerae O serogroups from a major shrimp production area in Thailand. J. Med. Microbiol. 43216-220. [DOI] [PubMed] [Google Scholar]
- 14.De, K., T. Ramamurthy, S. M. Faruque, S. Yamasaki, Y. Takeda, G. B. Nair, and R. K. Nandy. 2004. Molecular characterisation of rough strains of Vibrio cholerae isolated from diarrhoeal cases in India and their comparison to smooth strains. FEMS Microbiol. Lett. 23223-30. [DOI] [PubMed] [Google Scholar]
- 15.De, K., T. Ramamurthy, A. C. Ghose, M. S. Islam, Y. Takeda, G. B. Nair, and R. K. Nandy. 2001. Modification of the multiplex PCR for unambiguous differentiation of the El Tor & classical biotypes of Vibrio cholerae O1. Indian J. Med. Res. 11477-82. [PubMed] [Google Scholar]
- 16.DiRita, V. J. 1992. Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae. Mol. Microbiol. 6451-458. [DOI] [PubMed] [Google Scholar]
- 17.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 991556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dziejman, M., D. Serruto, V. C. Tam, D. Sturtevant, P. Diraphat, S. M. Faruque, M. H. Rahman, J. F. Heidelberg, J. Decker, L. Li, K. T. Montgomery, G. Grills, R. Kucherlapati, and J. J. Mekalanos. 2005. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. USA 1023465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 621301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faruque, S. M., I. B. Naser, M. J. Islam, A. S. Faruque, A. N. Ghosh, G. B. Nair, D. A. Sack, and J. J. Mekalanos. 2005. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc. Natl. Acad. Sci. USA 1021702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guhathakurta, B., D. Sasmal, S. Pal, S. Chakraborty, G. B. Nair, and A. Datta. 1999. Comparative analysis of cytotoxin, hemolysin, hemagglutinin and exocellular enzymes among clinical and environmental isolates of Vibrio cholerae O139 and non-O1, non-O139. FEMS Microbiol. Lett. 179401-407. [DOI] [PubMed] [Google Scholar]
- 22.Herrington, D. A., R. H. Hall, G. Losonsky, J. J. Mekalanos, R. K. Taylor, and M. M. Levine. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 1681487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmgren, J. 1973. Comparison of the tissue receptors for Vibrio cholerae and Escherichia coli enterotoxins by means of gangliosides and natural cholera toxoid. Infect. Immun. 8851-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honda, T., and R. A. Finkelstein. 1979. Purification and characterization of a hemolysin produced by Vibrio cholerae biotype El Tor: another toxic substance produced by cholera vibrios. Infect. Immun. 261020-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoshino, K., S. Yamasaki, A. K. Mukhopadhyay, S. Chakraborty, A. Basu, S. K. Bhattacharya, G. B. Nair, T. Shimada, and Y. Takeda. 1998. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol. Med. Microbiol. 20201-207. [DOI] [PubMed] [Google Scholar]
- 26.Joelsson, A., Z. Liu, and J. Zhu. 2006. Genetic and phenotypic diversity of quorum-sensing systems in clinical and environmental isolates of Vibrio cholerae. Infect. Immun. 741141-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamble, T. K., S. R. More, S. S. Chavan, N. D. Kulkarni, N. S. Lodha, and A. S. Kamble. 2000. Clinical profile of non-O1 strain-O139 of Vibrio cholerae in the region of Ambajogai, Maharashtra. J. Assoc. Physicians India 48505-506. [PubMed] [Google Scholar]
- 28.Kaper, J. B., J. G. J. Morris, and M. Nishibuchi. 1988. DNA probes for pathogenic Vibrio species, p. 65-77. In F. C. Jenover (ed.), DNA probes for infectious diseases. CRC Press, Boca Raton, FL.
- 29.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 848-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaper, J. B., J. P. Nataro, N. C. Roberts, R. J. Siebeling, and H. B. Bradford. 1986. Molecular epidemiology of non-O1 Vibrio cholerae and Vibrio mimicus in the U.S. Gulf Coast region. J. Clin. Microbiol. 23652-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karaolis, D. K., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 953134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keasler, S. P., and R. H. Hall. 1993. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet 3411661. [DOI] [PubMed] [Google Scholar]
- 33.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28501-520. [DOI] [PubMed] [Google Scholar]
- 34.Kovach, M. E., M. D. Shaffer, and K. M. Peterson. 1996. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology 1422165-2174. [DOI] [PubMed] [Google Scholar]
- 35.Krukonis, E. S., and V. J. DiRita. 2003. From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6186-190. [DOI] [PubMed] [Google Scholar]
- 36.Lauriano, C. M., C. Ghosh, N. E. Correa, and K. E. Klose. 2004. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 1864864-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, W., K. J. Fullner, R. Clayton, J. A. Sexton, M. B. Rogers, K. E. Calia, S. B. Calderwood, C. Fraser, and J. J. Mekalanos. 1999. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc. Natl. Acad. Sci. USA 961071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitra, R. K., R. K. Nandy, T. Ramamurthy, S. K. Bhattacharya, S. Yamasaki, T. Shimada, Y. Takeda, and G. B. Nair. 2001. Molecular characterisation of rough variants of Vibrio cholerae isolated from hospitalised patients with diarrhoea. J. Med. Microbiol. 50268-276. [DOI] [PubMed] [Google Scholar]
- 39.Morris, J. G., Jr. 1994. Non-O group 1 Vibrio cholerae strains not associated with epidemic disease, p. 103-115. In I. K. Wachsmuth, P. A. Blake, and O. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. ASM Press, Washington, DC.
- 40.Mukhopadhyay, A. K., P. K. Saha, S. Garg, S. K. Bhattacharya, T. Shimada, T. Takeda, Y. Takeda, and G. B. Nair. 1995. Distribution and virulence of Vibrio cholerae belonging to serogroups other than O1 and O139: a nationwide survey. Epidemiol. Infect. 11465-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nandi, B., R. K. Nandy, S. Mukhopadhyay, G. B. Nair, T. Shimada, and A. C. Ghose. 2000. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J. Clin. Microbiol. 384145-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nandy, R. K., T. K. Sengupta, S. Mukhopadhyay, and A. C. Ghose. 1995. A comparative study of the properties of Vibrio cholerae O139, O1 and other non-O1 strains. J. Med. Microbiol. 42251-257. [DOI] [PubMed] [Google Scholar]
- 43.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing, 12th informational supplement. Approved standard M100-S12. National Committee for Clinical Laboratory Standard, Wayne, PA.
- 44.Nusrin, S., G. Y. Khan, N. A. Bhuiyan, M. Ansaruzzaman, M. A. Hossain, A. Safa, R. Khan, S. M. Faruque, D. A. Sack, T. Hamabata, Y. Takeda, and G. B. Nair. 2004. Diverse CTX phages among toxigenic Vibrio cholerae O1 and O139 strains isolated between 1994 and 2002 in an area where cholera is endemic in Bangladesh. J. Clin. Microbiol. 425854-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogawa, A., J. Kato, H. Watanabe, K. Suzuki, S. Dohi, and T. Takeda. 1990. Cloning and nucleotide sequence determination of a heat-stable enterotoxin gene from Vibrio cholerae non-O1. Jpn. J. Med. Sci. Biol. 43255. [PubMed] [Google Scholar]
- 46.Olsvik, O., J. Wahlberg, B. Petterson, M. Uhlen, T. Popovic, I. K. Wachsmuth, and P. I. Fields. 1993. Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J. Clin. Microbiol. 3122-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Shea, Y. A., F. J. Reen, A. M. Quirke, and E. F. Boyd. 2004. Evolutionary genetic analysis of the emergence of epidemic Vibrio cholerae isolates on the basis of comparative nucleotide sequence analysis and multilocus virulence gene profiles. J. Clin. Microbiol. 424657-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramamurthy, T., P. K. Bag, A. Pal, S. K. Bhattacharya, M. K. Bhattacharya, T. Shimada, T. Takeda, T. Karasawa, H. Kurazono, Y. Takeda, et al. 1993. Virulence patterns of Vibrio cholerae non-O1 strains isolated from hospitalised patients with acute diarrhoea in Calcutta, India. J. Med. Microbiol. 39310-317. [DOI] [PubMed] [Google Scholar]
- 49.Rudra, S., R. Mahajan, M. Mathur, K. Kathuria, and V. Talwar. 1996. Cluster of cases of clinical cholera due to Vibrio cholerae O10 in east Delhi. Indian J. Med. Res. 10371-73. [PubMed] [Google Scholar]
- 50.Saha, P. K., H. Koley, and G. B. Nair. 1996. Purification and characterization of an extracellular secretogenic non-membrane-damaging cytotoxin produced by clinical strains of Vibrio cholerae non-O1. Infect. Immun. 643101-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saha, P. K., and G. B. Nair. 1997. Production of monoclonal antibodies to the non-membrane-damaging cytotoxin (NMDCY) purified from Vibrio cholerae O26 and distribution of NMDCY among strains of Vibrio cholerae and other enteric bacteria determined by monoclonal-polyclonal sandwich enzyme-linked immunosorbent assay. Infect. Immun. 65801-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarkar, A., R. K. Nandy, G. B. Nair, and A. C. Ghose. 2002. Vibrio pathogenicity island and cholera toxin genetic element-associated virulence genes and their expression in non-O1 non-O139 strains of Vibrio cholerae. Infect. Immun. 704735-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sengupta, D. K., T. K. Sengupta, and A. C. Ghose. 1992. Major outer membrane proteins of Vibrio cholerae and their role in induction of protective immunity through inhibition of intestinal colonization. Infect. Immun. 604848-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma, C., M. Thungapathra, A. Ghosh, A. K. Mukhopadhyay, A. Basu, R. Mitra, I. Basu, S. K. Bhattacharya, T. Shimada, T. Ramamurthy, T. Takeda, S. Yamasaki, Y. Takeda, and G. B. Nair. 1998. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J. Clin. Microbiol. 36756-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 842833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 2721910-1914. [DOI] [PubMed] [Google Scholar]
- 57.Yamai, S., T. Okitsu, T. Shimada, and Y. Katsube. 1997. Distribution of serogroups of Vibrio cholerae non-O1 non-O139 with specific reference to their ability to produce cholera toxin, and addition of novel serogroups. Kansenshogaku Zasshi 711037-1045. [DOI] [PubMed] [Google Scholar]
- 58.Yu, R. R., and V. J. DiRita. 2002. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 43119-134. [DOI] [PubMed] [Google Scholar]