Abstract
A study was designed to evaluate the ability of the DiversiLab fingerprinting kit, a type of repetitive element PCR (rep-PCR), to identify Escherichia coli clone ST131 producing β-lactamase CTX-M-15. A set of 53 nonduplicate isolates of extended-spectrum β-lactamase-producing E. coli underwent rep-PCR, pulsed-field gel electrophoresis, and multilocus sequence typing. The DiversiLab system successfully identified E. coli clone ST131 producing CTX-M-15 and provides a simple standardized typing protocol for monitoring the spread of this clone.
Multidrug-resistant, β-lactamase CTX-M-15-producing Escherichia coli is emerging worldwide in a simultaneous fashion as an important pathogen causing community-onset and hospital-acquired infections (9). An identical clone named ST131 has been identified using multilocus sequencing typing (MLST) among CTX-M-15-producing E. coli isolated during 2000 to 2006 from several countries, including Spain, France, Canada, Portugal, Switzerland, Lebanon, India, Kuwait, and Korea (2, 7). This clone belongs to the highly virulent phylogenetic group B2 and harbors multidrug-resistant IncFII plasmids. This suggests that the ST131 clone has emerged independently in different parts of the world either due to contaminated food/water sources or has been imported into different countries via foreign traveling. Clone ST131 has also recently been reported in the United Kingdom (5), Italy (1), and Turkey (12). Therefore, there is a need for a simple, standardized, and cost-effective typing protocol for monitoring the spread of clone ST131 throughout the world.
DiversiLab fingerprinting kits (previously called bacterial bar codes), a type of repetitive-element PCR (rep-PCR), is a rapid, semiautomated, PCR-based, commercial typing system that can be easily performed by bench technologists with some molecular experience (3). This technique has the potential to track and monitor resistant bacteria in a real-time basis but has not yet been validated for typing extended-spectrum β-lactamase (ESBL)-producing E. coli isolates.
The goal of this study was to assess the DiversiLab system as a potential rapid and reliable method to distinguish E. coli clone ST131 producing CTX-M-15 from E. coli isolates producing other ESBLs.
A set of 53 nonduplicate isolates of ESBL-producing E. coli collected at Calgary Laboratory Services during January 2000 to December 2006 were included in this study. These isolates were previously reported in a molecular epidemiology study and produced the following ESBLs (8): VEB-1 (n = 1), TEM-52 (n = 1), SHV-2 (n = 1), CTX-M-3 (n = 2), CTX-M-14 (n = 13), CTX-M-15 (n = 29), CTX-M-24 (n = 3), and CTX-M-27 (n = 3).
The CTX-M-15-producing E. coli isolates were typed with pulsed-field gel electrophoresis (PFGE) following the extraction of genomic DNA and digestion with XbaI using the standardized E. coli (O157:H7) protocol established by the Centers for Disease and Prevention, Atlanta, GA (4). The subsequent PFGE analyses were performed on a contour-clamped homogeneous electric field mapper apparatus (Bio-Rad Laboratories, Hercules, CA). DNA relatedness was calculated on the basis of the Dice coefficient, and isolates were considered to be genetically related if the Dice coefficient correlation was 80% or greater, which corresponds to the “possibly related (4- to 6-band difference)” criteria of Tenover et al. (11).
MLST was performed on the 29 CTX-M-15-producing isolates using the seven conserved housekeeping genes (aspC, clpX, fadD, icdA, lysP, mdh, and uidA). A detailed protocol of the MLST procedure, including allelic type and sequence type assignment methods, available at the EcMLST website (http://www.shigatox.net/mlst) was used in this study.
All DNA samples were amplified using the DiversiLab E. coli kit for DNA fingerprinting (bioMerieux, Inc., St. Laurent, Quebec, Canada) following the manufacturer's instructions and GeneAmp 9700 ThermoCycler instrument (Applied Biosystems, Norwalk, CT). Detection of rep-PCR products was implemented using the Agilent 2100 bioanalyzer (Agilent Technologies Inc., Santa Clara, CA), which employs microfluidics chip-based DNA fragment separation, rather than the gel electrophoresis traditionally employed for rep-PCR analyses. Thirteen samples can be analyzed simultaneously on a microfluidics chip, and internal DNA standards of known sizes are added to each well to allow for normalization and efficient chip-to-chip comparisons.
Analysis was performed with the DiversiLab software version 3.3. The resulting DNA fingerprint patterns were viewed as electropherograms, and reports included a dendrogram constructed from a similarity matrix and a virtual gel image of the fingerprint for each DNA sample. A correlation of 95% as a cutoff as recommended by the manufacturer was used. All library entries were typed in duplicate, and the observed reproducibility was consistent.
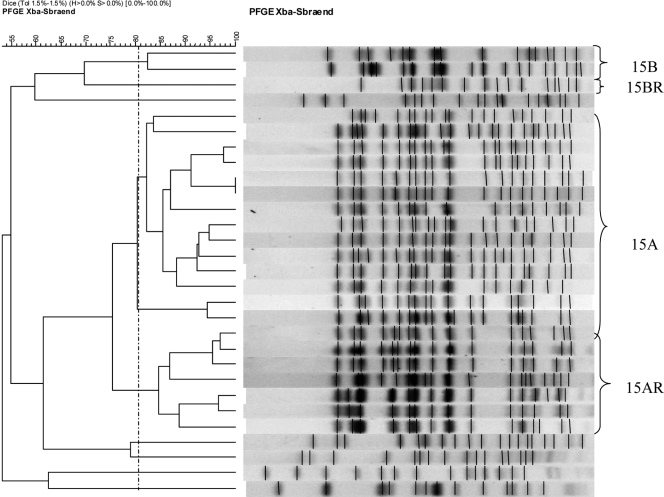
PFGE identified three closely related groups of E. coli isolates producing β-lactamase CTX-M-15 (designated 15A [n = 14] and 15AR [n = 7] [i.e., related to A]), as well as a separate clone designated 15B (n = 2) and a related strain called 15BR (n = 1). The 15A, 15AR, 15BR, and 15B isolates formed separate clones with >80% similar PFGE profiles. The 15AR isolates exhibited >60% similarity of profiles to 15A, which suggests that 15AR is related to 15A, while 15BR isolates exhibited >60% similarity of profiles to 15R, which suggests that 15BR is related to 15B. The remaining five CTX-M-15-producing isolates were not related to clones 15A, 15AR, 15B, or to each other and were designated 15NR (i.e., not related to 15A, 15AR, or 15B). These clones were previously reported in a molecular epidemiology study (8) (Fig. 1).
FIG. 1.
PFGE patterns and dendrogram of CTX-M-15-producing E. coli isolates. The 15A, 15AR, and 15B isolates formed separate clones with >80% similar PFGE profiles. The 15AR isolates exhibited >60% similarity of profiles to 15A, which suggests that 15AR is related to 15A, while 15BR isolates exhibited >60% similarity of profiles to 15R, which suggests that 15BR is related to 15B.
MLST performed on the CTX-M-15-producing isolates identified PFGE clones 15A and 15AR as ST131.
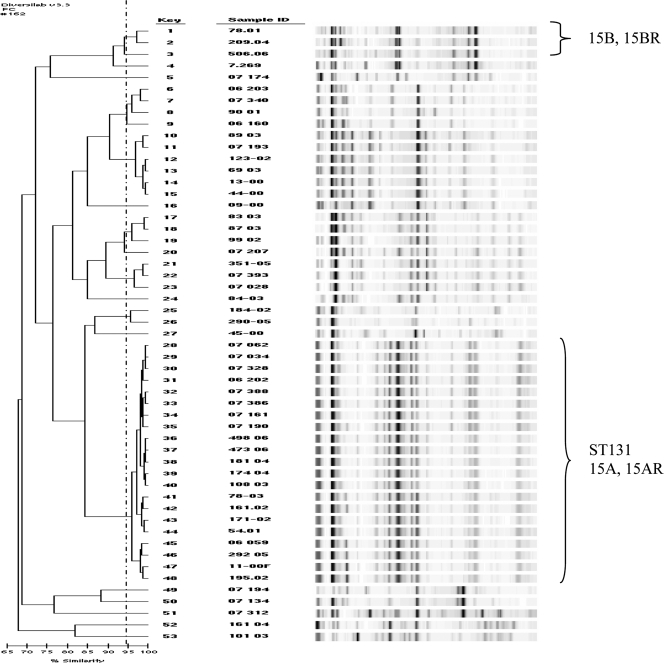
DiversiLab Rep-PCR using the >95% similarity index identified MLST clones ST131 (Fig. 2). This showed 100% concordance with MLST in identifying ST131 isolates. Rep-PCR performed very well in identifying known ST131 isolates; without MLST data on 24/53 isolates, it is uncertain if this method would have detected them. Some results were observed with Rep-PCR with other ESBL-producing E. coli isolates; CTX-M-14-producing isolates 89 03, 07 193, 123 02, 69 03, 13 00, and 44-00 were closely related to each other, while isolates 83 03, 87 03, and 99 02 were also closely related. Moreover, isolates 06 203 (CTX-M-3), 07 340 (CTX-M-14), 90 01 (CTX-M-15), and 06 160 (CTX-M-14) were closely related to each other and isolates 184 02 (CTX-M-24) and 290 05 (CTX-M-15) were related to each other, while isolates 351-05 (CTX-M-15), 07 393 (CTX-M-27), and 07 028 (CTX-M-24) were also closely related to each other (Fig. 2). These clones exhibited >60% similarity PGFE profiles (data not shown).
FIG. 2.
Dendrogram analysis and virtual gel images of DiversiLab rep-PCR fingerprint analysis of ESBL-producing E. coli isolates. For the purpose of predicting different clones, the top match feature at >95% similarity was used (broken line). Clones 15A, 15AR, 15B, and 15BR were identified using PFGE, while ST131 was identified using MLST. MLST was performed on the CTX-M-15-producing isolates. Sample ID, sample identification.
Comparative calculations of costs (considering reagents, equipment, and labor) for different typing methods per isolate in Canadian dollars were $25 for PFGE, $75 for MLST, and $55 for DiversiLab Rep-PCR.
PFGE is considered the gold standard for typing of medically important bacteria and identifying different clusters or clones (10). Clones consist of bacteria that originate from the same parent, and PFGE is excellent for identifying different clones responsible for recent or ongoing outbreaks. Unfortunately, PFGE is labor-intensive and time-consuming and delivers the best results only when performed by a technologist with extensive technical experience with this method. This makes interlaboratory comparison of typing results extremely difficult.
MLST is a portable, universal, and definitive sequence-based typing method that examines the nucleotide sequences of multiple loci of housekeeping genes (6). This makes it the most suitable typing method for comparing data generated independently from different laboratories and ideal for tracking antimicrobial-resistant bacteria, such as clone ST131, on a worldwide basis. Unfortunately, MLST is expensive and time-consuming and not really suitable for tracking resistant clones in a rapid real-time fashion.
Rep-PCR is a rapid method for strain typing that relies on amplification of the sequences between various repetitive elements interspersed throughout the genome (3). Standardized, commercially available kits containing primers and PCR master mix reagents are marketed as DiversiLab systems. Gel electrophoresis has been replaced by separation in a microfluidics chamber, and digitized gel images are generated and compared by using proprietary computer software. However, digitized PFGE gel images obtained under standardized conditions can also be used for comparisons. Although the DiversiLab system rep-PCR method is limited by the requirement of significant expenditure for reagents and equipment, it has two major advantages. First, results are generated more rapidly than for either PFGE or MLST. A batch of 13 samples can be performed within a single day. Second, standardized, digitized gel images are stored for comparison between runs and between laboratories.
Our results show that DiversiLab system rep-PCR successfully identified E. coli clone ST131 producing CTX-M-15 and was able to distinguish it from other E. coli isolates producing CTX-M-15, CTX-M-3, CTX-M-14, CTX-M-24, and CTX-M-27. The typing results obtained with DiversiLab fingerprinting kits had 100% concordance with MLST in identifying ST131 isolates, while PFGE of clone ST131 divided them into two related but separate clones, 15A and 15AR (Fig. 1). Rep-PCR performed very well in identifying known ST131 isolates; without MLST data on 24/53 isolates, it is uncertain if this method would have detected them. However, it is unlikely that ST131 were present among the other ESBL-producing isolates, since the PFGE patterns of these isolates were different than those of ST131 (<60% similarity [data not shown]). PFGE patterns of known ST131 isolates from different parts of the world have similar PFGE patterns (>60% similarity) (2, 7). The results obtained with the DiversiLab system rep-PCR on the CTX-M-14 producing isolates correlated closely with PFGE patterns (data not shown). However, the clones obtained with the DiversiLab system rep-PCR among isolates producing different CTX-Ms were difficult to explain, since the patients infected with these bacteria were separated via time and space.
In conclusion, we found that a rapid standardized typing protocol could be established for monitoring the spread of E. coli clone ST131 producing CTX-M-15 throughout the world. Typing images can be stored and forwarded to a centralized center for comparison purposes to ensure the tracking of these important CTX-M-producing bacteria. Our results suggest that DiversiLab fingerprinting rep-PCR might provide such an option to clinical laboratories worldwide and will benefit infection control procedures with the rapid introduction of control measures and significantly improve patient care. However, the significant cost of this procedure (55 Canadian dollars per isolate) limits its usefulness.
Acknowledgments
This work was financed by a grant from the Calgary Laboratory Services (grant 73-4063). J.D.D.P., D.L.C., and D.B.G. have previously received funding from Merck (Canada) and Wyeth (Canada) for research projects.
Footnotes
Published ahead of print on 9 February 2009.
REFERENCES
- 1.Cagnacci, S., L. Gualco, E. Debbia, G. C. Schito, and A. Marchese. 2008. European emergence of ciprofloxacin-resistant Escherichia coli clonal groups O25:H4-ST 131 and O15:K52:H1 causing community-acquired uncomplicated cystitis. J. Clin. Microbiol. 462605-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coque, T. M., A. Novais, A. Carattoli, L. Poirel, J. Pitout, L. Peixe, F. Baquero, R. Canton, and P. Nordmann. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg. Infect. Dis. 14195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Healy, M., J. Huong, T. Bittner, M. Lising, S. Frye, S. Raza, R. Schrock, J. Manry, A. Renwick, R. Nieto, C. Woods, J. Versalovic, and J. R. Lupski. 2005. Microbial DNA typing by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 43199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter, S. B., P. Vauterin, M. A. Lambert-Fair, M. S. Van Duyne, K. Kubota, L. Graves, D. Wrigley, T. Barrett, and E. Ribot. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 431045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lau, S. H., M. E. Kaufmann, D. M. Livermore, N. Woodford, G. A. Willshaw, T. Cheasty, K. Stamper, S. Reddy, J. Cheesbrough, F. J. Bolton, A. J. Fox, and M. Upton. 2008. UK epidemic Escherichia coli strains A-E, with CTX-M-15 β-lactamase, all belong to the international O25:H4-ST131 clone. J. Antimicrob. Chemother. 621241-1244. [DOI] [PubMed] [Google Scholar]
- 6.Maiden, M. C. 2006. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60561-588. [DOI] [PubMed] [Google Scholar]
- 7.Nicolas-Chanoine, M. H., J. Blanco, V. Leflon-Guibout, R. Demarty, M. P. Alonso, M. M. Canica, Y. J. Park, J. P. Lavigne, J. Pitout, and J. R. Johnson. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61273-281. [DOI] [PubMed] [Google Scholar]
- 8.Pitout, J. D., D. L. Church, D. B. Gregson, B. L. Chow, M. McCracken, M. R. Mulvey, and K. B. Laupland. 2007. Molecular epidemiology of CTX-M-producing Escherichia coli in the Calgary Health Region: emergence of CTX-M-15-producing isolates. Antimicrob. Agents Chemother. 511281-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitout, J. D., and K. B. Laupland. 2008. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8159-166. [DOI] [PubMed] [Google Scholar]
- 10.Tenover, F. C. 2007. Rapid detection and identification of bacterial pathogens using novel molecular technologies: infection control and beyond. Clin. Infect. Dis. 44418-423. [DOI] [PubMed] [Google Scholar]
- 11.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yumuk, Z., G. Afacan, M. H. Nicolas-Chanoine, A. Sotto, and J. P. Lavigne. 2008. Turkey: a further country concerned by community-acquired Escherichia coli clone O25-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 62284-288. [DOI] [PubMed] [Google Scholar]