Abstract
Thirty-three clinical isolates of the highly pathogenic Yersinia enterocolitica bioserotype 1B/O8 were collected from sporadic cases in Poland from January 2004 to July 2008. The isolates carried major virulence markers and were strongly clonal. This is the first report of the emergence and dissemination of highly related clones of Y. enterocolitica 1B/O8 in Europe.
Yersinia enterocolitica is known to be an important human enteric pathogen, causing a large variety of clinical and immunological manifestations (2). Infections in humans are mostly sporadic and related to contaminated food or water. Seven biotypes (1A, 1B, 2, 3, 4, 5, and 6) and more than 60 serotypes have been described for Y. enterocolitica. However, certain serotypes (O3; O5,27; O8; and O9) are prevalent among human isolates (2, 23). This species encompasses three grades of pathogenicity: mostly nonpathogenic strains (biotype 1A), weakly pathogenic strains of biotypes 2 to 6, and highly pathogenic strains (biotype 1B). The high pathogenicity is attributed to the yersiniabactin siderophore-mediated iron uptake system. Yersiniabactin is maintained by genes located in the Yersinia enterocolitica high-pathogenicity island (3). The geographical distribution of Y. enterocolitica 1B/O8 is generally restricted to North America (2, 4); however, this pathogen has been isolated sporadically in Japan (8, 9, 11, 15). Recently a single isolate was reported in Germany (17). Other reports of Y. enterocolitica O8 occurrence in Europe were published elsewhere (for a review, see reference 17); however, only strain 893/87 from Italy (16) and the German isolate were confirmed by molecular methods. In Poland, bioserotype 4/O3 is the predominant causative agent of human yersiniosis (5). To the best of our knowledge, bioserotype 1B/O8 was not isolated in Poland until 2004 (13). In the present study, we characterize 33 clinical isolates of Y. enterocolitica 1B/O8 collected from January 2004 to July 2008 from a variety of sporadic cases of human yersiniosis in Poland (Table 1).
TABLE 1.
Characteristics of tested isolates of Yersinia enterocolitica 1B/O8
| Strain | Epidemiological data
|
Presence of pYVa | PFGE result
|
|||||
|---|---|---|---|---|---|---|---|---|
| Yr of isolation | Region of origin | Age of patient | Main clinical manifestation | Cloneb | NotI | XbaI | ||
| WA-314 | NAc | NA (reference strain) | NA | NA | − | 4 | B | I |
| 27/04 | 2004 | Konin | 38 | Mesenteritis | + | 1 | A | II |
| 67/04 | 2004 | Poznań | NDd | ND | − | 1 | A | II |
| 152/05 | 2005 | Czştochowa | 1 | Diarrhea | − | 1 | A | II |
| 85/05 | 2005 | Czştochowa | 2 | Diarrhea | − | 1 | A | II |
| 82/06 | 2006 | Poznań | 2 | Diarrhea | − | 1 | A | II |
| 86/06 | 2006 | Kielce | 14 | Mesenteritis | + | 1 | A | II |
| 51/07 | 2007 | Kielce | 1 | ND | − | 1 | A | II |
| 52/07 | 2007 | Poznań | 2 | Diarrhea | + | 1 | A | II |
| 71/07 | 2007 | Poznań | 1 | Diarrhea | − | 1 | A | II |
| 72/07 | 2007 | Poznań | 3 | ND | − | 1 | A | II |
| 84/07 | 2007 | Poznań | 9 | Diarrhea | − | 2 | A1 | II |
| 85/07 | 2007 | Poznań | 27 | Cecal abscess | − | 1 | A | II |
| 89/07 | 2007 | Grodzisk Maz.e | 36 | Cecal abscess | + | 1 | A | II |
| 200/07 | 2007 | Zabrze | 1 | Diarrhea | − | 1 | A | II |
| 361/07 | 2007 | Kielce | 24 | Pyoperitoneum | − | 2 | A1 | II |
| 379/07 | 2007 | Poznań | 28 | Pyoperitoneum | − | 1 | A | II |
| 404/07 | 2007 | Katowice | 25 | Abdominal ulcer | − | 2 | A1 | II |
| 31/08 | 2008 | Warsaw | 3 | Diarrhea | + | 1 | A | II |
| 46/08 | 2008 | Łódź | ND | ND | + | 1 | A | II |
| 47/08 | 2008 | Zabrze | 2 | Diarrhea | − | 1 | A | II |
| 51/08 | 2008 | Kielce | 2 | Diarrhea | − | 1 | A | II |
| 61/08 | 2008 | Pułtusk | 78 | Bacteremia | − | 1 | A | II |
| 93/08 | 2008 | Warszawa | 10 | Diarrhea | − | 2 | A1 | II |
| 128/08 | 2008 | Katowice | 1 | ND | − | 1 | A | II |
| 146/08 | 2008 | Tarnów | 1 | Diarrhea | + | 1 | A | II |
| 160/08 | 2008 | Szczecin | 1 | Diarrhea | − | 1 | A | II |
| 161/08 | 2008 | Szczecin | 50 | Diarrhea | − | 1 | A | II |
| 162/08 | 2008 | Kielce | 2 | ND | − | 1 | A | II |
| 171/08 | 2008 | Warszawa | ND | Bacteremia | + | 2 | A1 | II |
| 172/08 | 2008 | Warszawa | 10 | Diarrhea | − | 1 | A | II |
| 175/08 | 2008 | Zabrze | 5 | ND | − | 1 | A | II |
| 180/08 | 2008 | Bydgoszcz | 3 | Diarrhea | + | 3 | A | III |
| 181/08 | 2008 | Bydgoszcz | 6 | Pseudoappendicitis | − | 1 | A | II |
Presence (+) or absence (−) of the Yersinia virulence plasmid.
Groups of isolates indistinguishable by combined NotI and XbaI PFGE genotyping.
NA, not applicable.
ND, not determined.
Grodzisk Maz., Grodzisk Mazowiecki.
The clinical Y. enterocolitica 1B/O8 isolates described in this study were obtained from sanitary-epidemiological units and hospitals in Poland. (Geographic distribution of the isolates is shown in Fig. S1 in the supplemental material). Biotypes and serotypes were determined as described previously (5, 23). Since Y. enterocolitica biotype 1A serotype O7,8 strains, which are widespread in the environment and have the O8 antigenic component, may be confused with the highly pathogenic strains of bioserotype 1B/O8, molecular investigations are recommended to confirm bioserotyping results. (17). The major chromosomal Y. enterocolitica virulence marker genes ystA, ail, and myfA, encoding heat-stable enterotoxin, attachment invasion locus protein, and Myf fimbriae, respectively (for a review, see reference 2), were examined by PCR to confirm the virulence of the tested isolates. Hence, the genes irp1 and irp2 of the yersiniabactin biosynthesis cluster and the siderophore receptor fyuA, which are located on the Y. enterocolitica high-pathogenicity island, were investigated to demonstrate that the Polish 1B/O8 isolates belong to the highly pathogenic Y. enterocolitica lineage (2, 3). PCRs were carried out as described previously (7) with primers listed in Table S1 in the supplemental material. All of the tested isolates yielded PCR amplicons for the Y. enterocolitica 16S rRNA gene and the aforementioned virulence marker genes. There is strong evidence suggesting that the Polish 1B/O8 isolates belong to highly pathogenic Y. enterocolitica.
Congo red magnesium oxalate agar medium (14) and the PCR assay with primers for the yadA gene, which encodes Yersinia adhesin A (2, 6), were used to detect the Yersinia virulence plasmid (pYV). The virulence plasmid is considered the gold standard for Yersinia virulence determination. However, in contrast to the chromosomal virulence markers, pYV can easily be lost when bacteria are cultured at 37°C (1, 2, 17). In this study, pYV was detected in nine isolates only (Table 1). Given the high sensitivity of PCR, this result may suggest that the majority of tested isolates lost pYV during manipulations in a routine diagnostic laboratory.
In order to gain better insight into the pathogenic potential of Polish Y. enterocolitica 1B/O8 isolates, the PCR assay was carried out for the genes yts1M and ysrS, which encode components of the Yersinia chromosomal type II and III secretion systems, termed Yts1 and Ysa, respectively (12, 21). In addition, the putative virulence genes YE2407 and YE2447 of Y. enterocolitica 1B/O8 strain 8081 were investigated (10, 20). All of these genes have been reported to occur exclusively in highly pathogenic Y. enterocolitica. In this study, PCR amplicons of yts1M, ysrS, YE2407, and YE2447 were detected for all the tested isolates. Our data demonstrate that Y. enterocolitica 1B/O8 isolates from sporadic cases in Poland carry the Yts1 and Ysa gene clusters and share the high-pathogenicity-Y. enterocolitica-specific virulence traits reported elsewhere (2, 3, 17, 20).
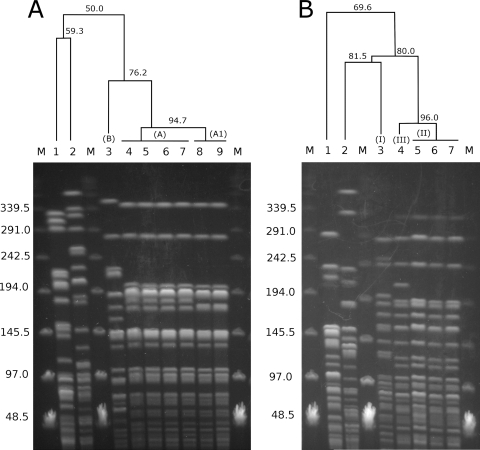
To determine the genetic relatedness of the Polish Y. enterocolitica 1B/O8 isolates described in this study, we carried out pulsed-field gel electrophoresis (PFGE), which is the genotyping standard for Y. enterocolitica (4, 16). PFGE was conducted as described previously (22) using the Chef-DR II system (Bio-Rad) and the endonuclease NotI (Fermentas, Lithuania) with a switching time of 5 to 24 s for 26 h at 14°C and a voltage gradient of 6 V/cm. Since all of the tested isolates were collected within a 4-year period from distinct patients with no epidemiological link, we initially expected diverse genotypes. Surprisingly, the resulting PFGE patterns were homogeneous (Fig. 1A). Only two highly similar NotI patterns (genotypes A and A1) could be distinguished in the Polish isolates. Twenty-eight and five isolates were classified as genotypes A and A1, respectively (Table 1). In contrast, the reference 1B/O8 strain WA-314 revealed a distinct NotI pattern (genotype B). Isolates of the A1 genotype could be reproducibly distinguished from those of the predominating genotype A, even under various PFGE conditions (data not shown).
FIG. 1.
NotI (A) or XbaI (B) PFGE profiles of Y. enterocolitica 1B/O8 clinical isolates from Poland (lanes 4 to 9) compared with profiles of bioserotype 1A/O7,8 (lanes 1 and 2) and 1B/O8 (lane 3) strains. The corresponding dendrograms illustrate genetic similarity of the NotI and XbaI profiles. The similarity values are shown in the dendrograms. PFGE genotypes are indicated in parentheses. Lanes in panels A and B: M, bacteriophage λ DNA ladder; 1, Y. enterocolitica 1A/O7,8 strain 323; 2, Y. enterocolitica 1A/O7,8 strain UG55; 3, reference 1B/O8 strain WA-314; 7, isolate 51/07; lanes in panel A: 4, 27/04; 5, 152/05; 6, 82/06; 8, 84/07; 9, 93-1/08; lanes in panel B: 4, 180/08; 5, 27/04; 6, 152/05. Sizes of DNA fragments are given in kilobases.
Interestingly, in the study by Saken and coworkers (16), we found NotI PFGE patterns that strongly resemble patterns of the here-described genotypes A and A1. The patterns showing resemblance were observed for the Y. enterocolitica 1B/O8 isolates 893/87 and 900/90, collected in Italy and Japan, respectively. Notably, Saken and coworkers used the reference strain WA-314 and comparable PFGE conditions (pulse ramp, 8 to 23; time period, 26 h). These findings might suggest that the Polish Y. enterocolitica 1B/O8 clinical isolates are related to the isolates from Italy and Japan. Further studies, however, are required to confirm this thesis.
The remarkable homogeneity of the genetic backbone shown in this study by PFGE may suggest that Polish Y. enterocolitica 1B/O8 isolates are clonal. Struelens and coworkers emphasize that identification of clones must be based on monitoring of several markers of sufficient discriminatory power (18). For this reason, we performed additional PFGE analyses using the enzyme XbaI (Fermentas, Lithuania). Even though application of an additional endonuclease has been reported to enhance the genotyping resolution of PFGE (4), all but one of the Polish 1B/O8 clinical isolates were indistinguishable (genotype II). The only exception was the isolate 180/08 (genotype III), which exhibited a single additional band in the XbaI pattern (Fig. 1B). The reference strain, WA-314, revealed a distinct XbaI pattern (genotype I).
To determine the similarity of the genotypes, PFGE patterns were analyzed using the GelCompar II software, version 5.10 (Applied Maths, Saint-Matins-Latem, Belgium). Similarity clustering analyses were performed using the single-linkage algorithm and the Dice correlation coefficient with a tolerance of 1.0%. The similarity of the PFGE genotypes distinguished in this study was high and ranged from 94.7% to 96.0% (Fig. 1) for NotI and XbaI, respectively. When NotI and XbaI PFGE profiles were analyzed simultaneously, the Polish 1B/O8 isolates were diversified into three highly similar (97.7%) types (clones), 1, 2, and 3, which comprised 27 and 5 isolates and a single isolate, respectively (Table 1). In conclusion, our results demonstrate that all of the tested Y. enterocolitica 1B/O8 isolates from Poland are closely related.
In accordance with the criteria proposed by Tenover and coworkers (19) and recommendations for genotyping data interpretation described by Struelens and colleagues (18), our results may indicate that the tested Y. enterocolitica 1B/O8 isolates collected in Poland constitute the same strain comprising three highly related PFGE clones. The genetic homogeneity of the tested isolates may result from the recent emergence of Y. enterocolitica 1B/O8 in Poland. Notably, the majority of the tested isolates were collected within the last 2 years, including all of the isolates classified as clones 2 and 3 (Table 1). This might reflect recent speciation events in the predominant clone 1. It is also noteworthy that we have recently observed in Poland a dramatic increase in the number of patients seropositive for both the Y. enterocolitica O8 lipopolysaccharide and the Yersinia outer protein antigens (13). Taken together, these findings show that the emergence and rapid dissemination of Y. enterocolitica 1B/O8 may be a challenge for the public health authorities in Poland.
To date, only a few sporadic isolates of the highly pathogenic Y. enterocolitica bioserotype 1B/O8 have been found in Europe (16, 17). The genetic similarity of these isolates has not yet been defined. In this study, we characterized a number of clinical isolates of Y. enterocolitica 1B/O8 by PFGE. To the best of our knowledge, this is the first report of the dissemination of the closely related clones of highly pathogenic Y. enterocolitica 1B/O8 in Europe. 1B/O8 isolates indistinguishable by PFGE were reported in Japan (11, 15), where an epidemiological link between patients and contaminated food or direct contact with infected animals has been documented. In light of these reports, the strong clonality of Polish Y. enterocolitica 1B/O8 isolates may suggest a common origin. Further epidemiological investigations supported by local veterinary authorities are required to elucidate the reservoir of the pathogen in Poland and its route of transmission. Our findings also indicate the need to develop novel, high-resolution genotyping approaches devoted to Yersinia enterocolitica 1B/O8 subtyping.
Supplementary Material
Acknowledgments
We thank all of the colleagues from sanitary-epidemiological units and hospitals in Poland for the submission of Y. eneterocolitica isolates.
This study was supported by the MNiSW of Poland (grant no. NN401-235834).
Footnotes
Published ahead of print on 9 February 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bercovier, H., and H. H. Mollaret. 1984. Genus XIV. Yersinia Van Loghem 1944, 15AL, p. 498-506. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 2.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3561-569. [DOI] [PubMed] [Google Scholar]
- 4.Fredriksson-Ahoma, M., A. Stolle, and H. Korkeala. 2006. Molecular epidemiology of Yersinia enterocolitica infections. FEMS Immunol. Med. Microbiol. 47315-329. [DOI] [PubMed] [Google Scholar]
- 5.Gierczyński, R., M. Jagielski, and W. Rastawicki. 2002. Molecular virulence attributes and occurrence of pYV-bearing strains among human clinical isolates of Yersinia enterocolitica in Poland. Eur. J. Clin. Microbiol. Infect. Dis. 21158-159. [DOI] [PubMed] [Google Scholar]
- 6.Gierczyński, R., M. Jagielski, and W. Rastawicki. 2002. Evaluation of usefulness of selected virulence markers for identifying pathogenic Yersinia enterocolitica strains. IV. Genes myfA and ureC. Med. Dośw. Mikrobiol. 54347-355. (In Polish.) [PubMed] [Google Scholar]
- 7.Gierczyński, R., A. Golubov, H. Neubaurer, J. N. Pham, and A. Rakin. 2007. Development of multiple-locus variable-number tandem-repeat analysis for Yersinia enterocolitica subspecies palearctica and its application to bioserogroup 4/O3 subtyping. J. Clin. Microbiol. 452508-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashidani, H., Y. Ohtomo, Y. Toyokawa, M. Saito, K.-I. Kaneko, J. Kosuge, M. Kato, M. Ogawa, and G. Kapperud. 1995. Potential sources of sporadic human infection with Yersinia enterocolitica serovar O:8 in Amori prefecture, Japan. J. Clin. Microbiol. 331253-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosaka, S., M. Uchiyama, M. Ishikawa, T. Akahoshi, H. Kondo, C. Shimauchi, T. Sasahara, and M. Inoue. 1997. Yersinia enterocolitica serotype O:8 septicemia in an otherwise healthy adult: analysis of chromosome DNA pattern by pulsed-field gel electrophoresis. J. Clin. Microbiol. 353346-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard, S. L., M. W. Gaunt, J. Hinds, A. A. Witney, R. Stabler, and B. W. Wren. 2006. Application of comparative phylogenomics to study the evolution of Yersinia enterocolitica and to identify genetic differences relating to pathogenicity. J. Bacteriol. 1883645-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwata, T., Y. Une, A. T. Okatani, S. Kaneko, S. Namai, S. Yoshida, T. Horisaka, T. Horikita, A. Nakadai, and H. Hayashidani. 2005. Yersinia enterocolitica serovar O:8 infection in breeding monkeys in Japan. Microbiol. Immunol. 491-7. [DOI] [PubMed] [Google Scholar]
- 12.Iwobi, A., J. Heesemann, E. Garcia, E. Igwe, C. Noelting, and A. Rakin. 2003. Novel virulence-associated type II secretion system unique to high-pathogenicity Yersinia enterocolitica. Infect. Immun. 711872-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rastawicki, W., J. Szych, R. Gierczyński, and N. Rokosz. A dramatic increase of Yersinia enterocolitica serogroup O:8 infections in Poland. Eur. J. Clin. Microbiol. Infect. Dis., in press. [DOI] [PubMed]
- 14.Riley, G., and S. Toma. 1989. Detection of pathogenic Yersinia enterocolitica by using Congo red-magnesium oxalate agar medium. J. Clin. Microbiol. 27213-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakai, T., A. Nakayama, M. Hashida, Y. Yamamoto, H. Takebe, and S. Imai. 2005. Outbreak of food poisoning by Yersinia enterocolitica serotype O8 in Nara Prefecture: the first case report in Japan. Jpn. Infect. Dis. 58257-258. [PubMed] [Google Scholar]
- 16.Saken, E., A. Roggenkamp, S. Aleksic, and J. Heesemann. 1994. Characterisation of pathogenic Yersinia enterocolitica serogroups by pulsed-field gel electrophoresis of genomic NotI restriction fragments. J. Med. Microbiol. 41329-338. [DOI] [PubMed] [Google Scholar]
- 17.Schubert, S., J. Bockemühl, U. Brendler, and J. Heesemann. 2003. First isolation of virulent Yersinia enterocolitica O8, biotype 1B in Germany. Eur. J. Clin. Microbiol. Infect. Dis. 2266-68. [DOI] [PubMed] [Google Scholar]
- 18.Struelens, M. J., A. Bauernfeind, A. van Belkum, D. Blanc, B. D. Cookson, L. Dijkshoorn, N. El Solh, J. Etienne, J. Garaizar, P. Gerner-Smidt, N. Legakis, H. De Lencastre, M. H. Nicolas, T. L. Pitt, U. Romling, V. Rosdahl, and W. Witte. 1996. Consensus guidelines for appropriate use and evaluation of microbial typing systems. Clin. Microbiol. Infect. 22-11. [DOI] [PubMed] [Google Scholar]
- 19.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomson, N. R., S. Howard, B. W. Wren, M. T. Holden, L. Crossman, G. L. Challis, C. Churcher, K. Mungall, K. Brooks, T. Chillingworth, T. Feltwell, Z. Abdellah, H. Hauser, K. Jagels, M. Maddison, S. Moule, M. Sanders, S. Whitehead, M. A. Quail, G. Dougan, J. Parkhill, and M. B. Prentice. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLOS Genet. 2e206. http://www.plosgenetics.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venecia, K., and G. M. Young. 2005. Environmental regulation and virulence attributes of the Ysa type III secretion system of Yersinia enterocolitica biovar 1B. Infect. Immun. 735961-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wardak, S., J. Szych, A. A. Zasada, and R. Gierczyński. 2007. Antibiotic resistance of Campylobacter jejuni and Campylobacter coli clinical isolates from Poland. Antimicrob. Agents Chemother. 511123-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wauters, G., K. Kandolo, and M. Janssens. 1987. Revised biogrouping scheme of Yersinia enterocolitica. Contrib. Microbiol. Immunol. 914-21. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.