Abstract
Four multiplex PCR assays for detection of 19 enterotoxigenic Escherichia coli (ETEC) colonization factors and an improved ETEC toxin multiplex PCR were developed and tested on Bangladeshi and Bolivian ETEC strain collections. The assays will be useful for surveillance of ETEC infections in diagnostic laboratories that have access to PCR.
Infection with enterotoxigenic Escherichia coli (ETEC) is one of the main causes of childhood diarrhea in developing countries and in travelers to these areas (7). ETEC strains express one or both of two different enterotoxins, the heat-stable toxins (STh and STp) and a heat-labile toxin (LT), and more than 23 different colonization factors (CFs) that are named CFA/I and CS1 to CS22 (2). The LT enterotoxin and the CF antigens are immunogenic, and one or both of these components are included in most vaccines that are being developed against ETEC diarrhea (14, 15). To evaluate the putative impact of different vaccine compositions on ETEC diarrhea, it is important to characterize clinical ETEC isolates from different parts of the world with regard to toxin and CF profiles, since they have been observed to vary from one geographic region to another (4, 7, 8, 10, 12).
In order to simplify and accelerate the detection of the different CFs and toxins by PCR, we have established a set of multiplex PCR assays for detection of the most-prevalent CFs and the toxins using previously established primers (11), as well as new primers when appropriate. The CF primers were assembled into four panels designed to amplify 19 of the most common CFs, and a new set of STh primers was included in the previously established toxin multiplex PCR (Table 1).
TABLE 1.
Reference strains, primer sequences, and multiplex PCR panel assignment for ETEC toxins and colonization factors
| Straina | PCR panel | Virulence factor | Product size (bp) | Primer sequence
|
Reference or source | |
|---|---|---|---|---|---|---|
| Forward (5′-3′) | Reverse (5′-3′) | |||||
| 286 C2 | Toxin | LT | 273 | ACGGCGTTACTATCCTCTC | TGGTCTCGGTCAGATATGTG | 11 |
| VX 67356 | Toxin | STp | 166 | TCTTTCCCCTCTTTTAGTCAG | ACAGGCAGGATTACAACAAAG | 1 |
| ST 64111 | Toxin | STh | 120 | TTCACCTTTCCCTCAGGATG | CTATTCATGCTTTCAGGACCA | This study |
| 60R936 | I | CS1 | 243 | TCCGTTCGGCTAAGTCAGTT | CCGCACATTTCCTGTGTTCT | 11 |
| E11881/9 | I | CS4 | 198 | ACCTGCGGCAAGTCGTTT | TCTGCAGGTTCAAAAGTCACA | 11 |
| E 29101A | I | CS7 | 154 | CGCCGGTTACACGTAGTGAT | CCATTTAAAGTGATTGCGACTT | 11 |
| 350C1A | I | CS12 | 137 | CCAGTCTATGCCAGGTTGCT | TGTGGGGTCACAGTTTACCA | 11 |
| E19446 | I | CS3 | 100 | CTAGCTTTGCCACCACCATT | GGCAACTGACTCCCATTTGT | 11 |
| E 9034A | II | CS21 | 617 | TCATGAGCCTGCTGGAAGTTATCA | TCCGGCTACCTAAAGTAATTGAGT | 4 |
| 58R957 | II | CS2 | 368 | AGTGGTGGCAGCGAAACTAT | TTCCTCTGTGGGTTCTCAGG | This study |
| E17018/A | II | CS5 | 226 | TCCGCTCCCGTTACTCAG | GAAAAGCGTTCACACTGTTTATATT | 11 |
| 258909-3 | II | CFA/I | 170 | GCTTATTCTCCCGCATCAAA | ACTTGTCCTCCCCATGACAC | 11 |
| E20738A | II | CS17 | 130 | GGAGACGCTGAATACAACTGA | CTCAGGCGCAGTTCCTTGT | 11 |
| E 20738A, CS19 | III | CS17/19 | 195 | CGGTGCGTTTAACACAGCTA | TCGATACACTCGCATTCGTT | 11 |
| E34420A | III | CS8 | 166 | ATCCGGATTATCAAGCTCCA | GAAGATGTTATTGCACCACCAA | 11 |
| E11881/14 | III | CS6 | 152 | CTGTGAATCCAGTTTCGGGT | CAGGAACTTCCGGAGTGGTA | 11 |
| 8786 | III | CS15 | 130 | CGAAATTGGACAAGCGATG | TCCAGCAGGGATATTATTCG | 11 |
| CS20 Ws 7179A | III | CS20 | 114 | AGGTATCCAAATCCGCACTG | CATCAGCCAGCACATAGGAA | 11 |
| CS18 | IV | CS18 | 362 | ATCCGTCAGGTGTTTGTGGT | CACCTGAATTCCTCGACAGG | This study |
| PCFO9 | IV | CS13 | 178 | GGGACTGCCACAATGAATTT | CAGCACCACCTGCTGATTTA | 11 |
| E7476A | IV | CS14 | 162 | TTTGCAACCGACATCTACCA | CCGGATGTAGTTGCTCCAAT | 11 |
| ARG-3 | IV | CS22 | 127 | ATTGGACAAGCGTCCAACAC | TTCCAGCAGGGATATTATCATTTT | 11 |
All strains belong to the reference strain collection at Department of Microbiology and Immunology, Institute of Biomedicine, University of Gothenburg, Göteborg, Sweden.
To establish the multiplex assays for the CFs and toxins, 10- to 100-ng amounts of DNA from ETEC reference strains boiled in water (Table 1) were mixed with master mix as described previously (11), except that 400 nM deoxynucleoside triphosphates and 8 to 10 forward and reverse primers at a final concentration of 200 nM each were added to the mix. The PCRs were amplified by an initial denaturation at 94°C (1 min), followed by 35 cycles of amplification (94°C for 30 s, 52°C for 30 s, and 72°C for 1 min), and finally, 5 min at 72°C. The products were run on 3% agarose gels or 3% MetaPhor gels (MetaPhor agarose; Cambrex Bio Science Rockland, Inc.) for better resolution and visualized under UV light (Fig. 1). Only the correct CF and toxin PCR products were amplified, and by mixing several strains and performing multiplex PCR on the mixed DNA, we verified that the primers in each panel could detect the presence of several CFs in one reaction.
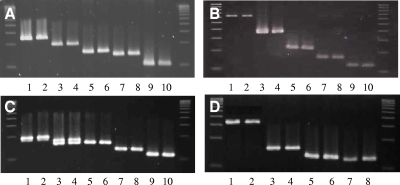
FIG. 1.
ETEC CF reference strains amplified in duplicate with the indicated multiplex PCR panel (panels I to IV; see Table 1 for strains). In panel C, note that strain E34420A contains both CS6 and CS8 and generates two bands, while only the CS6 band is amplified in the next strain, E11881/14. (A) Panel I. Lanes: 1 and 2, CS1 (243 bp); 3 and 4, CS4 (198 bp); 5 and 6, CS7 (154 bp); 7 and 8, CS12 (137 bp); 9 and 10, CS3 (100 bp). (B) Panel II. Lanes: 1 and 2, CS21 (630 bp); 3 and 4, CS2 (368 bp); 5 and 6, CS5 (226 bp); 7 and 8, CFA/I (170 bp); 9 and 10, CS17 (130 bp). (C) Panel III. Lanes: 1 and 2, CS19 (195 bp); 3 and 4, CS8 (166 bp); 5 and 6, CS6 (152 bp); 7 and 8, CS15 (130 bp); 9 and 10, CS20 (114 bp). (D) Panel IV. Lanes: 1 and 2, CS18 (362 bp); 3 and 4, CS13 (178 bp); 5 and 6, CS14 (162 bp); 7 and 8, CS22 (127 bp). Molecular size ladders are on each side of each panel.
To evaluate the new multiplex PCR methods for ETEC toxins and CFs, 106 clinical ETEC isolates were analyzed by multiplex PCR and the results were compared with results obtained by using the previously established ETEC toxin multiplex PCR and with the dot blot method using available monoclonal antibodies (11). We used two different sets of ETEC strains that were isolated in two geographically different developing countries: 65 Bolivian strains isolated between 2002 and 2008 and 41 ETEC strains randomly selected from a large birth cohort study in Bangladesh (6). All strains were collected from samples from children less than 3 years old with diarrhea or asymptomatic ETEC infections.
The results obtained when testing the Bolivian and Bangladeshi strains with both the new and the previously reported ETEC toxin multiplex PCR assay were identical, although the new toxin multiplex PCR results were easier to interpret (Fig. 2).
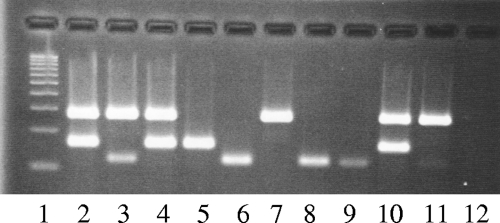
FIG. 2.
Improved multiplex PCR for the toxins using the new STh primers that amplify a longer, 120-bp fragment. Lanes: 1, 100-bp ladder; 2, E57 (LT/STp); 3, E88 (LT/STh); 4, E133 (LT/STp); 5, E382 (STp); 6, E865 (STh); 7, 286C2 (LT); 8, ST64111 (STh); 9, Re117a (STh); 10, XC110a (LT/STp); 11, E640 (LT/STh); 12, master mix.
The strain collections were further tested for CFs with dot blotting and the new multiplex PCR panels in parallel (Table 2). The most-common CF detected was CS21 (longus) that was detected by multiplex PCR in a total of 17 of the 65 Bolivian strains and 19 of the 41 Bangladeshi strains; CS21 was coexpressed with other CFs in most isolates. Of the total of 37 strains that were positive for CS21 by PCR, only 18 were positive both in the dot blot assay and by PCR (100% sensitivity, 78% specificity, 50% positive predictive value, and 100% negative predictive value). There was complete agreement between dot blotting and CF multiplex PCR results for CFA/I, CS2 plus CS3, CS4 plus CS6, CS5 plus CS6, CS7, CS12, and CS14, while CS1 plus CS3 and CS17 were detected by PCR but not by dot blotting in 2 and 1 of the strains, respectively. A total of 40 of the 106 strains tested did not express any CFs in the dot blot assay; of these strains, 7 were found to harbor the genes for CS21 (5 strains), CS17 plus CS21 (1 strain), and CS1 plus CS3 (1 strain), but 33 strains remained CF negative, and the less-common CFs CS8, CS13, CS15, CS18, CS19, CS20, and CS22 were not detected in any of the strains analyzed.
TABLE 2.
Comparison between the multiplex PCR for the CFs and dot blot assay in analysis of ETEC strains from Bolivia and Bangladesh
| Colonization factor(s) | No. of strains (no. which coexpress CS21) from indicated set identified by indicated assay
|
|||
|---|---|---|---|---|
| Bolivian (2002-2008; n = 65)
|
Bangladeshi (2003-2005; n = 41)
|
|||
| Multiplex PCR | Dot blot | Multiplex PCR | Dot blot | |
| CFA/I | 9 (8) | 9 (7) | 5 (5) | 5 (2) |
| CS1+CS3 | 6 (5) | 5 (3) | 6 (5) | 5 (5) |
| CS2+CS3 | 1 (0) | 1 (0) | 3 (3) | 3 (1) |
| CS4+CS6 | 0 | 0 | 1 (1) | 1 |
| CS5+CS6 | 0 | 0 | 3 | 3 |
| CS6 | 1 (0) | 1 (0) | 3 (2) | 3 (0) |
| CS7 | 3 | 3 | 1 | 1 |
| CS12 | 3 (0) | 3 (0) | 3 (1) | 3 (0) |
| CS14 | 5 | 5 | 5 | 5 |
| CS17 | 11 (1) | 10 (0) | 0 | 0 |
| CS21 | 3 | 0 | 3 | 1 |
| None | 23 | 28 | 8 | 11 |
The multiplex PCR for the detection of the CFs was established to provide a time-efficient method that is easy to perform and interpret. The multiplex PCR presented provides data for 19 CFs within 3 to 4 h, including PCR amplification, gel run, and staining, provided that all samples can be loaded onto gels simultaneously. The interpretation is facilitated by using high-resolution gels or 3% agarose gels and by always loading a reference ladder at both sides in each gel. Amplification of reference strains to verify that the correct amplified band is present and verification by single PCR is recommended. In addition, we tested an improved version of our previously published ETEC toxin multiplex PCR that uses a set of STh primers that amplify a larger part of the estA gene than the previously described primers (1, 11). We favor the use of the new toxin multiplex PCR due to the longer and more-distinct STh PCR product that will reduce the possibility for false-negative STh scoring in surveillance analyses and recommend that this modified multiplex PCR assay be performed on E. coli cultures to verify the presence of ETEC toxins.
The results obtained with the CF multiplex PCR indicate that in general, the same CFs were found in ETEC strains from children in Bolivia and Bangladesh. The most-common CF was found to be the CS21 pilus, also known as longus (3).
We found that CS21 was commonly coexpressed with CFA/I, CS1 plus CS3, and CS2 plus CS3 and that these CF profiles were the most-common ones both in Bolivian and Bangladeshi strains. CFA/I and CS1 to CS3 are, together with CS4 to CS6, the most-commonly identified ETEC CFs worldwide, e.g., in Bangladesh, Guinea-Bissau, India, and Egypt (5, 8, 12, 13). The present results for the Bolivian strains suggest that CFA/I and CS1 to CS3 are also the most-prevalent ones in Bolivia, while CS4, CS5, and CS6 were found less frequently than expected, and instead, CS14 and CS17 were prevalent in Bolivian strains.
However, the relatively large number of ETEC isolates without identifiable CFs found in this and other studies suggests that additional CFs may still be undetected, although new and interesting putative CFs have been proposed recently (9).
In summary, the CF multiplex PCR panels reported in this study will allow rapid and sensitive identification of the most-prevalent ETEC CFs identified and will allow inclusion of primers targeting new important ETEC CFs when their biological significance has been confirmed. The methodology is as sensitive as dot blotting but will in some cases, i.e., with CS21, detect more positive samples than the phenotypic dot blot test. Hence the new multiplex PCR assays will aid in epidemiological surveys of ETEC and its CFs and will provide a basis for the development of ETEC vaccines based on CF immunity.
Acknowledgments
This work was supported by the Swedish Agency for Research and Economic Cooperation (Sida-SAREC) and the Swedish Research Council, grant no. 6X-09084. Å.S. acknowledges the Swedish Society for Medical Research (SSMF) for their support.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Bölin, I., G. Wiklund, F. Qadri, O. Torres, A. L. Bourgeois, S. Savarino, and A.-M. Svennerholm. 2006. Enterotoxigenic Escherichia coli with STh and STp genotypes is associated with diarrhea both in children in areas of endemicity and in travelers. J. Clin. Microbiol. 443872-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaastra, W., and A. M. Svennerholm. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4444-452. [DOI] [PubMed] [Google Scholar]
- 3.Gutiérrez-Cázarez, Z., F. Qadri, M. J. Albert, and J. A. Girón. 2000. Identification of enterotoxigenic Escherichia coli harboring longus type IV pilus gene by DNA amplification. J. Clin. Microbiol. 381767-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pichel, M. G., N. Binsztein, F. Qadri, and J. A. Giron. 2002. Type IV longus pilus of enterotoxigenic Escherichia coli: occurrence and association with toxin types and colonization factors among strains isolated in Argentina. J. Clin. Microbiol. 40694-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qadri, F., S. K. Das, A. S. Faruque, G. J. Fuchs, M. J. Albert, R. B. Sack, and A. M. Svennerholm. 2000. Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. J. Clin. Microbiol. 3827-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qadri, F., A. Saha, T. Ahmed, A. Al Tarique, Y. A. Begum, and A. M. Svennerholm. 2007. Disease burden due to enterotoxigenic Escherichia coli in the first 2 years of life in an urban community in Bangladesh. Infect. Immun. 753961-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qadri, F., A. M. Svennerholm, A. S. Faruque, and R. B. Sack. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18465-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao, M. R., R. Abu-Elyazeed, S. J. Savarino, A. B. Naficy, T. F. Wierzba, I. Abdel-Messih, H. Shaheen, R. W. Frenck, Jr., A. M. Svennerholm, and J. D. Clemens. 2003. High disease burden of diarrhea due to enterotoxigenic Escherichia coli among rural Egyptian infants and young children. J. Clin. Microbiol. 414862-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy, K., D. Hamilton, K. P. Allen, M. P. Randolph, and J. M. Fleckenstein. 2008. The EtpA exoprotein of enterotoxigenic Escherichia coli promotes intestinal colonization and is a protective antigen in an experimental model of murine infection. Infect. Immun. 762106-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaheen, H. I., I. A. Abdel Messih, J. D. Klena, A. Mansour, Z. El-Wakkeel, T. F. Wierzba, J. W. Sanders, S. B. Khalil, D. M. Rockabrand, M. R. Monteville, P. J. Rozmajzl, A. M. Svennerholm, and R. W. Frenck. 2009. Phenotypic and genotypic analysis of enterotoxigenic Escherichia coli (ETEC) in samples obtained from Egyptian children presenting to referral hospitals. J. Clin. Microbiol. 47189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sjöling, Å., G. Wiklund, S. J. Savarino, D. I. Cohen, and A. M. Svennerholm. 2007. Comparative analyses of phenotypic and genotypic methods for detection of enterotoxigenic Escherichia coli toxins and colonization factors. J. Clin. Microbiol. 453295-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sommerfelt, H., H. Steinsland, H. M. Grewal, G. I. Viboud, N. Bhandari, W. Gaastra, A. M. Svennerholm, and M. K. Bhan. 1996. Colonization factors of enterotoxigenic Escherichia coli isolated from children in north India. J. Infect. Dis. 174768-776. [DOI] [PubMed] [Google Scholar]
- 13.Steinsland, H., P. Valentiner-Branth, H. M. Grewal, W. Gaastra, K. K. Molbak, and H. Sommerfelt. 2003. Development and evaluation of genotypic assays for the detection and characterization of enterotoxigenic Escherichia coli. Diagn. Microbiol. Infect. Dis. 4597-105. [DOI] [PubMed] [Google Scholar]
- 14.Svennerholm, A. M., and J. Tobias. 2008. Vaccines against enterotoxigenic Escherichia coli. Expert Rev. Vaccines 7795-804. [DOI] [PubMed] [Google Scholar]
- 15.Walker, R. I., D. Steele, and T. Aguado. 2007. Analysis of strategies to successfully vaccinate infants in developing countries against enterotoxigenic E. coli (ETEC) disease. Vaccine 252545-2566. [DOI] [PubMed] [Google Scholar]